Abstract
PAR proteins are key regulators of cellular polarity and have links to the endocytic machinery and the actin cytoskeleton. Our data suggest a unique role for PAR proteins in cytokinesis. We have found that at the onset of cytokinesis, anterior PAR-6 and posterior PAR-2 proteins are redistributed to the furrow membrane in a temporal and spatial manner. PAR-6 and PAR-2 localize to the furrow membrane during ingression but PAR-2-GFP is distinct in that it is excluded from the extreme tip of the furrow. Once the midbody has formed, PAR-2-GFP becomes restricted to the midbody region (the midbody plus the membrane flanking it). Depletion of both anterior PAR proteins, PAR-3 and PAR-6, led to an increase in multinucleate embryos, suggesting that the anterior PAR proteins are necessary during cytokinesis and that PAR-3 and PAR-6 function in cytokinesis may be partially redundant. Lastly, anterior PAR proteins play a role in the maintenance of DYN-1 in the cleavage furrow. Our data indicate that the PAR proteins are involved in the events that occur during cytokinesis and may play a role in promoting the membrane trafficking and remodeling events that occur during this time.
Keywords: cytokinesis, Ray Rappaport, PAR proteins, polarity, DYN-1, PAR-6, PAR-2
Introduction
Cytokinesis is a dynamic process involving interactions between astral and spindle midzone microtubules, cortical microfilaments, and cellular membranes to permit cleavage furrow ingression and daughter cell separation [Bringmann and Hyman, 2005; Glotzer, 2005, 2010]. The astral microtubules and spindle midzone, the region of microtubules between the separating chromatids, appear to play a key role in the initiation and completion of cytokinesis [White and Borisy, 1983; Cao and Wang, 1996; Wheatley and Wang, 1996], although the exact nature of their roles have been debated. Plasma membrane and actin dynamics also play necessary roles in the formation of the contractile ring [Balasubramanian et al., 1994, 2004; Takeda et al., 2004; Ng et al., 2005; Logan and Mandato, 2006], yet there is very little evidence about what factors signal and regulate this event. The insertion and removal of membrane at the cleavage furrow is also an important component of cytokinesis [Shuster and Burgess, 2002; Montagnac et al., 2008b; Horgan and McCaffrey, 2012], as it appears that membrane donated from the cell surface alone is not sufficient to support cleavage furrow ingression and the separation of daughter cells [Danilchik and Brown, 2008]. There is also critical interplay between the cell division machinery and polarity cues to properly segregate not only chromosomes and organelles but also cytoplasmic determinants to newly formed daughter cells [Cheeks et al., 2004; Munro et al., 2004]. The role of polarity proteins in cell division events is unclear.
The PAR proteins are a widely conserved group of proteins necessary for various cell polarization events [Nance and Zallen, 2011]. The PAR proteins were originally identified in a genetic screen in Caenorhabditis elegans embryos [Kemphues et al., 1988]. PAR-3 and PAR-6 localize to the anterior cortex while PAR-1 and PAR-2 localize to the posterior domain [Cuenca et al., 2003; Nance and Zallen 2011]. Acto-myosin based transport of PAR-3/PAR-6/PKC-3 has been shown to play a crucial role in establishing polarity. The anterior localization of the PAR proteins is established by cortical flows regulated by the sperm centrosome [Cheeks et al., 2004; Munro et al. 2004]. PAR-3, PAR-6, and PKC-3 protein distribution is also controlled by the acto-myosin regulators CDC-42 and RHO-1 [Schonegg et al., 2007]. Our lab has shown that C. elegans dynamin, DYN-1, participates in the maintenance of anterior–posterior cues by mediating the localization of PAR-6 to the cortex via endocytosis. [Nakayama et al., 2009]. Here, the spatial and temporal regulation of endocytosis during the cell cycle contributes to the maintenance of PAR proteins within a dynamic plasma membrane. Subsequently, we have found that the endocytic recycling of PAR-6 is disrupted in the absence of Arp2/3 complex [Shivas and Skop, 2012]. Endocytic recycling mechanisms appear to be an important step in maintaining PAR proteins in the plasma membrane throughout development. Overall, the importance of PAR proteins in the generation of cell asymmetry is obvious in the C. elegans embryo and have equally important and conserved functions in Drosophila and vertebrate embryos [Betschinger and Knoblich, 2004; Cowan and Hyman, 2004; Macara, 2004; Shivas et al., 2010], but their function during cell division remains elusive.
Cell polarity is naturally coupled with many events that occur during cell division. As a tissue proliferates and develops, each cell must maintain its polarization state upon each division event [Devenport et al., 2011]. Failure to coordinate these events can lead to over proliferation and loss of tissue polarity, as seen in a variety of epithelial cancers [Aranda et al., 2008; Nolan et al., 2008; Sagona and Stenmark, 2010; Schenk et al., 2010]. Polarity factors are differentially targeted and maintained at distinct sites in the membrane cortex before cleavage furrow formation. In C. elegans, cleavage furrow placement occurs at the exact boundary between anterior and posterior PAR protein domains [Schenk et al., 2010]. Prior to cellularization in Drosophila, the plasma membrane is polarized and remains so during cleavage furrow invagination [Mavrakis et al., 2009]. Polarity is not only important during animal cell division. In S. pombe polarity factors are essential for proper cellular axis determination and placement of cytokinetic factors prior to division [Chang, 2001; Tavares et al., 2012]. Polarity and cytokinesis also require polarized vesicle trafficking [Skop et al., 2001; Lu and Bilder, 2005; Wilson et al., 2005; Montagnac et al., 2008; Prekeris and Gould, 2008; Qi et al., 2011; Hehnly and Doxsey, 2012]. As the cleavage furrow ingresses, polarized vesicle trafficking is important for the insertion of new membrane along the furrow and in the midbody [Prekeris and Gould, 2008]. Properly polarizing cellular factors prior to and during cytokinesis is a key step that must be precisely regulated throughout development
In this study, we examined the role of the PAR proteins in cytokinesis. We found that double depletion of the anterior PAR proteins, PAR-3 and PAR-6, led to a variety of early embryonic phenotypes, including multinucleate embryos. PAR proteins display a dynamic localization pattern at the furrow membrane that is disrupted when other PAR proteins are perturbed. PAR proteins play a role in the maintenance of DYN-1 at the cleavage furrow. We postulate that PAR proteins play a necessary role during cytokinesis by targeting membrane trafficking and remodeling factors necessary for the events that occur during this time.
Results
Anterior and Posterior PAR Proteins Temporally and Spatially Localize Along the Cleavage Furrow
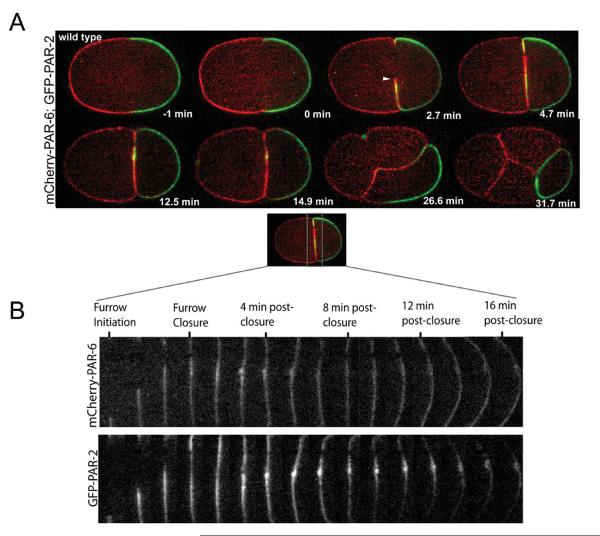
Anterior and posterior PAR proteins localize to respective cortical domains by the end of polarity establishment [Boyd et al., 1996; Guo and Kemphues, 1996; Cowan and Hyman, 2004]. Although PAR proteins were first shown to localize to the cleavage furrow in fixed C. elegans embryos [Etemad-Moghadam et al., 1995; Guo and Kemphues, 1995], it is unclear if there is a temporal and spatial pattern during furrow formation. To characterize the dynamic localization patterns of anterior and posterior PAR proteins during cleavage furrow ingression, we examined PAR-6-mCherry and PAR-2-GFP localization in early C. elegans embryos. In wild-type embryos, PAR-6-mCherry and PAR-2-GFP localize to the anterior and posterior poles prior to cleavage furrow initiation (Fig. 1A, 0 min). As the furrow starts to ingress (Fig. 1A 2.7 min), both anterior and posterior PAR proteins appear along the furrow membrane, but only PAR-6-mCherry is found at the leading edge of the furrow membrane. As the furrow continues to ingress (Fig. 1A, 4.7 min), this localization pattern is maintained. At 12.5 min after the initiation of furrow ingression, the flanking membrane along the furrow is predominately labeled with PAR-6-mCherry. The midbody region is the only place where PAR-2-GFP co-localizes with PAR-6-mCherry. As the embryo continues to divide to become a four-celled embryo, the localization of the anterior and posterior PAR proteins segregate to the membrane boundaries on opposite sides of the embryo (Fig. 1A). Here, PAR-6 is found predominantly along the membrane boundaries between the ABa, Abp, and EMS blastomeres (Fig. 1A, 31.7 min). PAR-2-GFP localizes exclusively to the P2 blastomere in the 4-cell embryo. To take a closer look at the localization patterns of the anterior and posterior PAR proteins during cytokinesis, we followed PAR-6-mCherry and PAR-2-GFP dynamics every 80 s and split the channels to look at the differences between them (Fig. 1B). Anterior and posterior PAR protein localization at the furrow is observed until 8-min postcleavage furrow closure (Fig. 1B). Between 8-and 16-min postcleavage furrow closure, PAR-2-GFP localization becomes restricted to the membrane region in vicinity of the midbody (Fig. 1B, 8–16-min postclosure). PAR-6-mCherry localizes along the entire furrow membrane during cytokinesis, whereas, PAR-2 is temporally and spatially restricted along the furrow. We suspect that during cytokinesis the PAR proteins establish a polarity along the furrow, such that the temporal and spatial targeting of distinct membrane trafficking and remodeling machinery would occur.
Fig. 1. Dynamics of PAR-6-mCherry and PAR-2-GFP proteins during cell division.
(A) Embryos expressing both PAR-6-mCherry and PAR-2-GFP were imaged using spinning disk confocal microscopy from one minute prior to furrow formation through the four-cell stage. Localization of PAR-6-mCherry and PAR-2-GFP are temporally and spatially localized along the cleavage furrow membrane. PAR-6 and PAR-2 localize to the furrow membrane during ingression but are distinct in that PAR-2-GFP is excluded from the extreme tip of the furrow (white arrowhead, Fig. 1A, 2.7 min), where PAR-6-mCherry is only found. Once the midbody has formed, PAR-2-GFP becomes restricted to the midbody region (the midbody membrane accumulation plus the membrane flanking it) postfurrow closure (Fig 1A, 12.5–14.9 min and Fig 1B). During the next division the localization of PAR-2-GFP disappears from the midbody region and localizes to the entire cortex of the P2 blastomere (Fig 1A, 26.6–31.7 min). At this time, PAR-6-mCherry is now found at the cortex and adjoining membranes of the anterior blastomeres (ABa, ABp, and EMS) at the four-cell stage (Fig 1A, 31.7 min). The adjoining membrane adjacent to the P2 blastomere in EMS and ABp is labeled with PAR-2-GFP. (B) View of PAR-6-mCherry and PAR-2-GFP dynamics along the cleavage furrow at the mid-focal plane. Corresponding red and green channels were separated and a montage of furrow ingression was built from frames 80 s apart from furrow initiation to 16-min post-closure. At 4-min postclosure, PAR-6 and PAR-2 are evenly distributed along the furrow membrane and accumulate at the midbody. After the 4-min postclosure, PAR-6-mCherry remains evenly distributed along the furrow membrane whereas, PAR-2-GFP becomes restricted to the midbody (a bolus of membrane) and regions flanking the midbody by the end of the time course.
Depletion of PAR-3 and PAR-6 Leads to Multinucleate Embryos
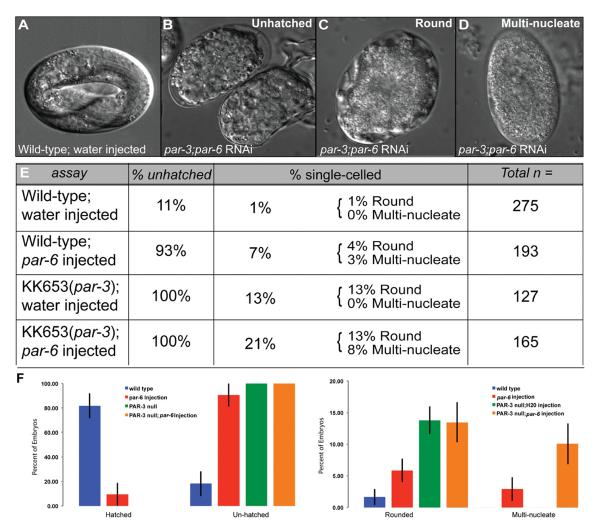
The role of polarity proteins in cytokinesis is unclear. Work by the Glotzer lab has shown that when depleted singly, PAR proteins (PAR-3 and PAR-2) do not have cytokinesis defects. In addition, depletion of opposing PAR proteins (PAR-3 and PAR-2) together does not lead to defects in cell division [Dechant and Glotzer, 2003]. However, depletion of nonopposing PAR proteins was not explored. To determine if nonopposing PAR proteins play a role in cytokinesis, we explored the contribution of both PAR-3 and PAR-6 in the early C. elegans embryo. To do this, we injected par-3 mutant worms with par-6 double-stranded RNA and assessed the terminal phenotypes 24–30-h postinjection (Fig. 2A). We characterized the embryos into two groups: unhatched (embryos that arrested later in development that did not hatch) and single-celled embryos (Fig. 2). Then, the single-cell embryos were categorized into two groups: rounded and multinucleate embryos (Figs. 2C and 2D). The rounded, single-cell embryos were small and possessed puckered membranes (Fig. 2B), often indicative of membrane trafficking defects that have been observed previously [Thompson et al., 2002a]. The multinucleate embryos were not morphologically different than wild-type embryos, but multiple nuclei were present in the cytoplasm with no distinct cell boundaries separating them (Fig. 2D). The rounded and multinucleate embryos were separated to distinguish the difference between them in their membrane formation. It is possible that the round, single-celled embryos could also be multinucleate. We quantified the phenotypes and compared them to control worms injected with water. In wild-type worms injected with par-6 double stranded RNA, 93% of embryos failed to hatch and 4% were rounded, single-celled embryos and 3% were multinucleate (Fig. 2E, n = 193). In par-3 mutant worms injected with water, 100% of the embryos failed to hatch and 13% of the unhatched embryos were rounded, single-celled embryos (n = 127). When the par-3 mutant worms were injected with par-6 double stranded RNA, 100% of the embryos failed to hatch, similar to the par-3 mutants injected with water alone. However, we saw an increase in the percentage of embryos that were single-celled (21%). Of these embryos, 13% were rounded and 8% were multinucleate (n = 165). So, par-3 mutants injected with par-6 RNA showed an increase in multinucleated embryos. Given the number of nuclei observed, these embryos attempted multiple rounds of cytokinesis but failed (Fig. 2D). To account for variability between the experimental replicates, we also analyzed the terminal phenotypes as a percentage of each individual experiment, and then averaged them together (Fig. 2F). Percentages of phenotypes were similar to that observed in Fig. 2E, but include error bars to indicate reproducibility and variance. Our attempt to catch failures in cytokinesis in young embryos was quite difficult due to the sickness of the hermaphrodites; so terminal phenotypes were determined and quantified. We observed a large increase in the numbers of multinucleated embryos in cases where both anterior PAR proteins, PAR-3, and PAR-6, are diminished. This data suggest that the anterior PAR proteins play a possible role in the progression through cell division.
Fig. 2. Phenotypes observed in double par-3 mutant and PAR-6 RNAi-depleted embryos.
Depletion of both anterior PAR proteins (PAR-3 and PAR-6) yields a smaller brood size and a variety of embryonic lethal phenotypes. The total number of embryos from each particular assay was quantified (Total n). These embryos were first divided into groups based on gross embryonic phenotypes observed: wild type (hatched) and unhatched embryos. Unhatched embryos are embryos that did not arrest at the one-cell stage but arrested later in development. Of the wild type embryos injected with water, 11% of them did not hatch and 89% of them developed normally (A). For the par-6 injected, par-3 mutant and par-3 mutant; par-6 injected embryos their percent lethality was between 93 and 100%. A majority of these embryos resembled (B), where the embryos died at later stages of development and did not hatch (i.e., unhatched). All of the unhatched embryos that did not die at later stages in development, we identified these embryos as having a single-celled phenotype (% single-celled). Of the wild-type; par-6 injected animals, 7% of the unhatched embryos were single-celled. Of the KK653 (par-3; water injected animals, 13% of the unhatched embryos were single-celled. Of the par-3; par-6 RNAi-treated embryos, 21% of these unhatched embryos arrested at the one-cell stage and were either (C) round or (D) multinucleate. It is not clear what percentage of the sickly embryos in (C) were also multinucleate. (E) Table showing the percentages of embryos with particular embryonic phenotypes. The percentage of unhatched column designates the percent of embryonic lethality for the total number of embryos from all four experiments. Of the lethal embryos observed, we then calculated the percentage of those embryos that were multinucleate (percentage of single-celled column). Total n is the total number of embryos from multiple experiments (four each). (F) These graphs are a representation of the average percentage of each phenotype observed (hatched, unhatched, rounded, multinucleate) in the experimental replicates (four times). Each phenotype was quantified as a percentage of each individual experiment and then averaged together. Error bars indicate standard error. The percentages are similar to that of the table in (E), but this graph conveys the reproducibility of the phenotypes.
Localization of Anterior and Posterior PAR Proteins to the Cleavage Furrow is Dependent on Each Other
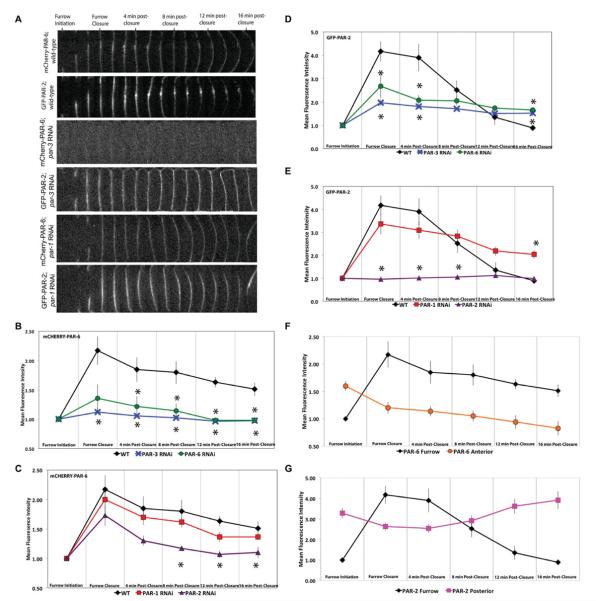
To determine if PAR proteins are required for the stable localization of PAR-6 or PAR-2 to the cleavage furrow during cytokinesis, we examined PAR-6-mCherry and PAR-2-GFP expression at the furrow in either anterior (PAR-3) or posterior (PAR-1) RNAi-treated embryos using spinning disk confocal microscopy (Fig. 3A). Next, we quantified the fluorescence intensity of PAR-2-GFP and PAR-6-mCherry during cytokinesis by taking the fold change value of the mean fluorescence intensity and plotted it over time (Figs. 3B–3E, wild type).
Fig. 3. PAR-6 and PAR-2 localization and maintenance at the cleavage furrow membrane is dependent on other PAR proteins.
(A) Single, mid-focal plane confocal images of the cell equator from time-lapse sequences of embryos expressing both PAR-6-mCherry and PAR-2-GFP in the absence of PAR-3 or PAR-1. (B) and (C) show the mean fluorescence intensity of PAR-6-mCherry expression along the cleavage furrow in wild type embryos, par-3, par-6, par-1 and par-2 RNAi-treated animals. (D) and (E) show the mean fluorescence intensity of PAR-2-GFP along the cleavage furrow in wild-type, anterior and posterior PAR RNAi-treated embryos. (F) and (G) show the differences between PAR-6-mCherry and PAR-2-GFP localization at the cortex versus cleavage furrow during cytokinesis. All time points were normalized to the mean fluorescence intensity of cytoplasmic levels measured during ingression. The number of embryos for each experiment is listed as: wild type, n = 8; par-1 RNAi, n = 6; par-2 RNAi n = 7; par-3 RNAi n = 9; par-6 RNAi n = 5. Asterisks indicate time points that displayed a significant difference (A P-value of <0.05) between corresponding PAR depletion and wild type fluorescence intensity values. Statistical significance was determined by ANOVA followed by post hoc testing.
In wild-type embryos, during initiation and furrow closure, PAR-6-mCherry and PAR-2-GFP are present along the entire furrow membrane (Fig. 3A, wild type) and the fluorescence intensity peaks at furrow closure (Figs. 3B and 3D, wild type). PAR-6-mCherry fluorescence changes from 2.5-fold above cytoplasmic levels at furrow closure to 1.3-fold above cytoplasmic levels at 16-min postfurrow closure (Fig. 3B, wild type). Throughout cytokinesis, the fluorescence intensity of PAR-6-mCherry remains above the cytoplasmic levels at furrow initiation. This observation is disrupted when either anterior or posterior proteins are depleted (Fig. 3). Depletion of anterior PAR proteins (PAR-3 and PAR-6) leads to a 1.5-to 2.0-fold decrease in PAR-6-mCherry fluorescence at the cleavage furrow at all time points, with fluorescence intensity barely rising above the initial cytoplasmic level of one (Fig. 3B). Depletion of posterior PAR-1 had little influence on PAR-6-mCherry at the cleavage furrow with no statistically significant difference at any time point (Fig. 3C). Knockdown of posterior PAR-2 had a more significant affect on PAR-6-mCherry fluorescence at the furrow with a 1.5-fold decrease in fluorescence intensity observed between 8- and 16-min postfurrow closure (Fig. 3C).
PAR-2-GFP fluorescence at the cleavage furrow also decreases after membrane closure, like PAR-6-mCherry, but to a greater extent. The fluorescence intensity of PAR-2-GFP is highest during furrow closure at a fold change of 4.2 above cytoplasmic levels, but by 16-min postclosure, PAR-2-GFP fluorescence intensity is at 0.9, below initial cytoplasmic levels; which is approximately a fourfold decrease (Fig. 3D). This clearance of PAR-2-GFP is less severe when either anterior or posterior PAR proteins are knocked-down. Depletion of either PAR-3 or PAR-6 leads to a decrease in initial PAR-2-GFP accumulation at the cleavage furrow, from a fourfold increase above cytoplasmic levels in wild type embryos to a 2.0- and 2.7-fold increase, respectively, between furrow initiation and furrow closure (Fig. 3D). Although PAR-2-GFP fluorescence is less intense at early time points in PAR-3 and PAR-6 depleted embryos, at 16-min postfurrow closure PAR-2-GFP fluorescence is approximately twofold higher than that of wild-type embryos (Fig. 3D). Anterior PAR proteins may be necessary for the removal of PAR-2 from the cleavage furrow over time. PAR-2-GFP fluorescence was minimally affected by depletion of PAR-1, except at 16-min postfurrow closure where fluorescence intensity was twofold higher than in wild type (Figs. 3A and 3E). RNAi depletion of PAR-2 had a greater affect on PAR-2-GFP fluorescence at the cleavage furrow, reducing fluorescence to cytoplasmic levels from furrow initiation through 16-min postclosure (Fig. 3E), which is not surprising given that we are knocking down PAR-2 expression.
Overall, our data suggest that anterior and posterior PAR protein localization is dynamic during and well after the furrow has fully ingressed. PAR-6 is maintained at a constant level at the cleavage furrow, while PAR-2-GFP is being removed from the membrane flanking the midbody. The anterior PAR proteins are necessary for the removal of PAR-2 from the cleavage furrow membrane. The role of PAR proteins at the mid-body is unclear, but they may mediate the membrane trafficking events that are occurring during this time.
The Dynamics of PAR Proteins During Cytokinesis
We have shown that PAR protein localization to the cleavage furrow is dynamic. To determine if the distribution of PAR-6 and PAR-2 at the anterior and posterior cortex of the embryo are redistributed at the onset of cytokinesis, we measured the fluorescence intensity of both anterior and posterior cortex and compared it to the furrow localization (Figs. 3F and 3G). As the furrow has formed, cortical anterior PAR-6-mCherry fluorescence intensity decreases (1.6 at furrow ingression to 1.2 at furrow closure) (Fig. 3F) while it increases at the cleavage furrow (mean intensity value change of 1–2.2). The anterior levels of PAR-6-mCherry then steadily decrease at beginning of furrow closure (Fig. 3F). PAR-6 is most likely being directed from the anterior of the embryo to the furrow membrane during cytokinesis. PAR-6 could also be degraded over time, since fluorescence intensity decreases in both the anterior and at the furrow at this time, yet bleaching during microscopy could also be the cause.
In the posterior cortex of the single-cell embryo, PAR-2-GFP fluorescence intensity decreases at furrow initiation to furrow closure (mean fold change of 3.3–2.6 at furrow closure), yet increases in intensity at the cleavage furrow (Fig. 3G). The posterior cortex fluorescence of PAR-2-GFP continues to decrease after furrow closure. After 4-min postclosure, posterior PAR-2-GFP fluorescence starts to increase until 16-min postclosure resulting in an 18% increase in cortical fluorescence intensity. The increase in the cortical posterior fluorescence correlates with the clearance of PAR-2-GFP at the cleavage furrow (Figs. 3D and 3E). Our data suggests that anterior and posterior PAR proteins are spatially and temporally regulated throughout cytokinesis. This dynamic localization may reflect the requirement of PAR proteins during furrow formation and completion, possibly by mediating the targeting or removal of factors along the furrow membrane during cytokinesis.
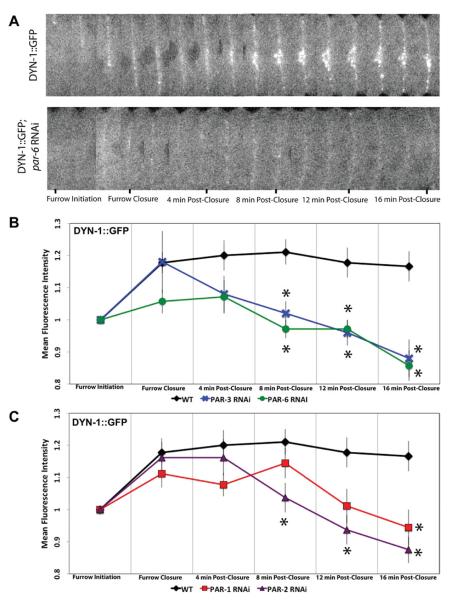
Depletion of Anterior and Posterior PAR Proteins Leads to Loss of DYN-1 at the Cleavage Furrow
Our lab has shown that dynamin/DYN-1 localizes to the cleavage furrow and mediates the localization and maintenance of PAR-6 in the anterior of the embryo [Thompson et al., 2002; Nakayama et al., 2009]; however, it is not known if PAR proteins mediate DYN-1 localization to the furrow. First, we determined the dynamics of DYN-1-GFP during cytokinesis. DYN-1-GFP localizes to the furrow membrane right at furrow initiation and remains along the furrow membrane even 16-min postclosure (Figs. 4A and 4B). The localization pattern is similar to the PAR proteins (Fig. 3) except that the intensity of DYN-1 remains high well after furrow closure. To determine if the PAR proteins play a role in mediating DYN-1-GFP localization at the cleavage furrow, we used RNAi to knockdown anterior and posterior PAR proteins in DYN-1-GFP expressing embryos. In embryos depleted of PAR-6, a decrease in fluorescence intensity (~12%) is observed at furrow closure and continues to decrease until 16-min post-closure. After furrow closure the localization of DYN-1-GFP at furrow dramatically disappears in PAR-6 RNAi-treated embryos. Here, we observe a 26% decrease in the mean fluorescence intensity compared to wild-type embryos (Fig. 4B). A similar phenotype is seen in embryos depleted of PAR-3, but the decrease in DYN-1-GFP intensity does not occur until 4-min postfurrow closure. PAR-3 does not appear to have an affect on the initial DYN-1-GFP localization to the furrow. In PAR-3 RNAi-treated embryos there is a 10% decrease in fluorescence intensity that occurs at 4-min postfurrow closure that eventually drops to a 25% decrease by 16-min post-closure (Fig. 4B).
Fig. 4. Anterior and posterior PAR proteins influence DYN-1 localization to the furrow.
Single, mid-focal plane confocal images of the cell equator from time-lapse sequences of embryos expressing DYN-1-GFP. (A) Montages show a control embryo and an embryo in which PAR-6 was depleted. DYN-1-GFP is maintained along the furrow throughout cytokinesis and 16-min postfurrow closure concentrating at the midbody. In PAR-6 RNAi-treated embryos, a reduction in recruitment of DYN-1-GFP to the cleavage furrow is observed. (B) Analysis of DYN-1-GFP intensity at the furrow in wild-type and anterior PAR (PAR-3 and PAR-6) depleted embryos. (C) Analysis of DYN-1-GFP fluorescence intensity at the cleavage furrow in posterior PAR depleted embryos. Depletion of both anterior and posterior PAR proteins leads to a decrease in fluorescence intensity of DYN-1-GFP at the cleavage furrow. The number of embryos for each experiment is listed as: wild type, n = 9, par-1 RNAi n = 9, par-2 RNAi n = 8, par-3 RNAi n = 5, par-6 RNAi n = 7. Asterisks indicate time points that displayed a significant difference (A P-value of <0.05) between corresponding PAR depletion and wild type fluorescence intensity values. Statistical significance was determined by ANOVA followed by post hoc testing.
We also determined if the posterior PAR proteins, PAR-1 and PAR-2, have an affect on DYN-1-GFP localization at the furrow. In embryos depleted of PAR-1, a 10% decrease in fluorescence intensity is seen at 4-min postclosure. By 16-min postclosure, DYN-1-GFP fluorescence decreases to 19%. PAR-2 depleted embryos do not show a significant decrease in fluorescence intensity until 8-min postclosure where a 14% decrease in fluorescence intensity is observed. Fluorescence intensity of DYN-1-GFP continues to decrease in PAR-2 depleted embryos until 16-min postclosure where it is 25% lower than that of the control DYN-1-GFP expressing embryos. Our data suggest that both anterior and posterior PAR proteins are necessary for the maintenance of DYN-1 at the cleavage furrow. Absence of PAR-6 leads to a 12% decrease in fluorescence intensity of DYN-1-GFP localization at the furrow, suggesting that it may play some role in the localization of DYN-1 to the furrow or PAR-6 could solely function in the maintenance of DYN-1 at the furrow. PAR-3, PAR-1 and PAR-2 appear to play a role in the maintenance of DYN-1 at the furrow. Future research will uncover these possibilities.
Discussion
PAR-6 and PAR-2 Localize to the Cleavage Furrow During Cytokinesis
PAR proteins have been documented to localize to the cleavage furrow [Etemad-Moghadam et al., 1995; Watts et al., 2000; Li et al., 2010; Schenk et al., 2010], but the importance and dynamics of their localization patterns to the cleavage furrow was unclear. To determine the significance of PAR protein localization during cytokinesis, we have taken advantage of live-cell imaging in the C. elegans embryo. First, we discovered that both anterior and posterior PAR proteins are found in the cleavage furrow and their localization is spatially and temporally regulated. PAR-6-mCherry and PAR-2-GFP are found in the early cleavage furrow, with only PAR-6-mCherry at the very tip of the ingressing furrow early. Eventually, PAR-2-GFP eventually becomes restricted to the midbody region, whereas PAR-6-mCherry is found along the entire furrow membrane and midbody during abscission. Upon the next division, the germline precursor (P4) is the only blastomere where the PAR-2-GFP was found, whereas, PAR-6-mCherry was found along the cortex and junctions between ABa and ABp and also ABa and EMS blastomeres. What is currently unknown is whether PAR proteins play a specific function during cytokinesis.
As in cell polarity, membrane-cytoskeletal dynamics plays a very important role in the cell division process. Numerous signaling molecules found at the equatorial membrane are important for mediating the furrowing process and assembly of the actomyosin ring [Skop et al., 2004; Piekny et al., 2005]. The polarity factors Cdc42/CDC-42 and Rho1/RHO-1 also play major roles in cytokinesis and spindle alignment [Gotta et al., 2001; Ohno, 2001; Cowan and Hyman, 2007), and CDC-42 directly associates with PAR-6 [Aceto et al., 2006]. PAR proteins have long been known to associate with the actin cytoskeleton [Munro et al., 2004; Patalano et al., 2006; Georgiou and Baum, 2010; Dawes and Munro, 2011; Nance and Zallen, 2011; Xiong et al., 2011], and actin ring assembly or dynamics may be mediated in part by their association with the PAR proteins. In support of this idea, PAR-4 has been shown to affect actomyosin ring dynamics [Chartier et al., 2011], yet 10 years earlier was shown to localize to the cleavage furrow membrane [Watts et al., 2000]. The function of the PARs in cell polarity and cytokinesis may also be a separable one as truncating the second PDZ domain in PAR-3 caused a loss of PAR-3-GFP at the anterior cortex but not at the cleavage furrow [Li et al., 2010]. Our understanding of the nature of the PARs during mitosis and early embryonic development are just beginning to be understood.
PAR-3 and PAR-6 Function Redundantly in Cytokinesis
We examined the contribution of nonopposing anterior PAR proteins (PAR-3 and PAR-6) during cytokinesis. Previous work from the Glotzer lab had shown that single and double depletions of opposing PAR proteins (PAR-3 and PAR-2) do not give rise to cytokinesis defects [Dechant and Glotzer, 2003]. We found that single PAR-3 mutant embryos did not give rise to multinucleate embryos, but a small proportion of these singly depleted embryos arrested as round, single-celled embryos (Fig. 2B). The round PAR-3 mutant embryos were very sick and never attempted to divide. Loss of PAR-6 function enhances the multinucleate phenotype of par-3 mutants. These results suggest that PAR-3 and PAR-6 may function redundantly in cytokinesis. Genetic redundancy of the anterior PDZ-containing PAR proteins, PAR-3, and PAR-6, is not surprising given that they are in a complex, localize similarly and that they function together to regulate a number of conserved cellular processes [Pellettieri and Seydoux, 2002; Macara, 2004; Goldstein and Macara, 2007]. We did observe an increase in single-celled embryos (7%) after par-6 RNAi injection, 3% of these embryos were multinucleate, suggesting that par-6 depletions alone can give rise to cytokinesis phenotypes. But this number is enhanced when PAR-6 is depleted in the par-3 mutant embryos (Fig. 2E). It is possible that PAR-6 may act independently as it has been shown to be dispensable for cell polarization in epithelial cells [Totong et al., 2007]. In Drosophila, Par-6 and aPKC/PKC-3 localize to distinct apical areas in epithelial cells than that of Bazooka/PAR-3 [Harris and Peifer, 2005], suggesting that they may function in distinct ways. It is becoming clear that polarity cues may function together and independently at different points in the cell cycle and throughout development.
Do PAR-3 and PAR-6 make distinct contributions to cytokinesis? Given published work [Balklava et al., 2007; Hyenne et al., 2012; Shivas and Skop, 2012], and our data presented here, it is likely that PAR-3, PAR-6 and the rest of anterior PAR complex (PKC-3 and CDC-42) contribute to cytokinesis as a complex and distinctly. The entire anterior PAR complex was identified in a screen for proteins that regulate endocytosis [Balklava et al., 2007], suggesting that their role in regulating endocytosis as a complex is likely. PAR-6 and the early endosomal protein, RAB-5, have been shown to play a distinct role in regulating endosomal trafficking in the anterior of the early embryo [Nakayama et al., 2009; Hyenne et al., 2012]. In addition, PAR-3 also appears to affect the maintenance of DYN-1-GFP at the furrow (Fig. 4). We cannot rule out the possibilities that the posterior PAR proteins similarly play a role in cytokinesis given that posterior PAR proteins, PAR-1 and PAR-2 also affect DYN-1-GFP postfurrow closure, albeit much less than the anterior PAR proteins do (Fig. 4). Recent work has shown that par-4 mutants have a reduced efficiency of actomyosin contractility and affect the localization of ANI-2/anilin during both polarity and cytokinesis [Chartier et al., 2011], suggesting that PAR proteins can play necessary yet distinct roles in both events. In Drosophila, mutations in several tumor suppressors, lethal giant larvae (lgl), scribble (scrib), discs large (dlg), and rab5, which also play key roles in cell polarity, lead to massive polyploid tumors [Bilder et al., 2000; Bilder, 2004; Vaccari and Bilder, 2009]. These tumors could arise from numerous failures in cytokinesis. Thus, many of the genes necessary for cell polarity could also function during cell division. The specific roles of all of the PAR proteins in cytokinesis are unclear but future research will uncover these possibilities.
The Importance of a Polarized Furrow Membrane During Cytokinesis
How is the trafficking machinery spatially organized along the furrow during division? Work from several labs has shown that membrane trafficking and remodeling factors, such as the ESCRT machinery, FIP3-Rab11, dynamin/DYN-1, RACK-1, ARF6/Rab35, and PITP/GIOTTO, localize along the cleavage furrow and midbody during abscission [Thompson et al., 2002; Horgan et al., 2004; Wilson et al., 2005; Dukes et al., 2008; Montagnac et al., 2008; Ai et al., 2009; Dambournet et al., 2011; Chesneau et al., 2012]. The loss of any of these proteins leads to multinucleate embryos and abscission failures, not unlike the multinucleate embryos we observed after double depletion of PAR-3 and PAR-6 (Fig. 2). The molecular mechanisms that mediate the precise targeting of the trafficking machinery to the furrow membrane or intercellular canal are unclear. Membrane machinery can be trafficked to several places during cytokinesis: back to the plasma membrane, to cytoplasmic membrane stores, along the furrow membrane and to the midbody. In the C. elegans embryo, we have shown that the PAR proteins localize in a very specific manner along the cleavage furrow (Fig. 1), likely establishing a polarity along the furrow membrane. Not unlike what we have observed in C. elegans with the localization of the PARs (Fig. 1), the Drosophila membrane during cellularization is polarized in an apical and basal manner by the end of cellularization. Here, the furrow invaginates between adjacent nuclei in the syncytium forming a membrane that lies perpendicular to the cell surface. Basal adherens junctions initially form and follow the furrow canal during the initial phase of cellularization. Apical adherens junctions form along the apical lateral membrane during late cellularization [Sisson et al., 1999; Tepass et al., 2001]. It is unclear if the orthologs of C. elegans PAR-6/PAR-3/PKC-3 (Bazooka, PAR-6, aPKC) and the posterior markers PAR-1 and Lgl temporally and spatially define the incomplete furrows and junctions in Drosophilia, but we do know that proteins necessary for cytokinesis localize along the membrane formed during cytokinesis in a polarized fashion [Riggs et al., 2003; Papoulas et al., 2005; Giansanti et al., 2006; Dyer et al., 2007; Giansanti et al., 2007]. Actin-associated proteins, such as Spaghetti squash and Pavarotti are found at the tip of the fully ingressed membrane [Dyer et al., 2007]. Trans-membrane proteins such as Neurotactin (Nrt) and Toll become apically inserted [Lecuit et al., 2002). Interestingly, in slam mutants the accumulation of Nrt and Toll to the apical membrane is inhibited [Lecuit et al., 2002], suggesting that membrane targeting to the furrow is mediated in a polarized fashion. This result is very similar to what we observe in PAR-6 depleted embryos, where DYN-1 fails to properly localize the furrow membrane (Fig. 4). PAR-6 may direct membrane traffic to and from the membrane flanking the midbody similar to the proposed function of Slam in Drosophila. At the midbody, PAR-6 and PAR-2 may be necessary for the specific targeting and removal of membrane necessary for furrow ingression and abscission. Thus, we propose that during cytokinesis the PARs establish a polarity to the furrow membrane so that factors necessary for membrane remodeling and trafficking can be shuttled to and from their proper place. The relationship between the PAR proteins and DYN-1 during cytokinesis is unclear, but our findings may prove useful in our understanding of how membrane trafficking is regulated during cytokinesis.
PAR Proteins, Dynamin, and Cytokinesis
We have shown that PAR proteins are found in the cleavage furrow and appear to mediate the maintenance of DYN-1 at the furrow. Could these proteins influence the assembly or dynamics of the actin ring in any way? PAR proteins and dynamin/DYN-1 have long been known to influence actomyosin dynamics [Schafer, 2002; Macara, 2004; Munro et al., 2004; Cestra et al., 2005]. Could their role at the furrow play a role in ring assembly? PAR-4 has been recently found to mediate actin dynamics via the proper positioning of ANI-2 (anillin). Defects in PAR-4 lead to defects in ring ingression [Chartier et al., 2011]. In animal cells, dynamin plays a conserved role during early and late cytokinesis events [Wienke et al., 1999; Thompson et al., 2002; Kang et al., 2003]. Plant dynamins also play a role in cytokinesis in addition to cell and tissue polarization [Kang et al., 2003] and microfilaments in conjunction with the dynamins play a necessary function in both of these events [Bednarek and Falbel, 2002; Konopka et al., 2006]. Given this we suspect that in animal cells the PAR proteins and DYN-1 may facilitate ring assembly or actin dynamics by influencing the specific targeting of actin regulators or other factors to and from the furrow membrane. Further research into this hypothesis may provide more clues into the significance the PAR proteins and DYN-1 at the furrow during cytokinesis.
Conclusion
In summary, we have found that both anterior and posterior PAR proteins localize to the cleavage furrow and midbody in a spatial and temporal manner during cytokinesis. Double depletion of PAR-3 and PAR-6 leads to multinucleate embryos, suggesting that anterior PAR proteins play a role in cytokinesis. PAR protein localization to the cleavage furrow is dependent on the other PAR proteins. PAR proteins are redistributed to the furrow at the onset of cytokinesis. PAR-6 plays a role in the maintenance of DYN-1 at the cleavage furrow. Together, these data indicate that the PAR proteins are involved in the events that occur during cytokinesis. We suggest that the anterior PAR protein PAR-6 mediates dynamin/DYN-1 localization to and from the furrow thus promoting the membrane trafficking events that occur during this time. The PAR proteins and DYN-1 may also play a necessary role in mediating actin assembly and dynamics at the equatorial membrane during cytokinesis. Our data underscores the importance of a polarized furrow membrane during cytokinesis.
Material and Methods
Nematode Strains
The following strains were used: TH120 (PAR-2-GFP; mCherry-PAR-6) [Schonegg et al., 2007] MAD3 (DYN-1-GFP) [Nakayama et al., 2009], KK653 (unc-32(e189)par-3(it71)/qC1 III) [Li et al., 2010] and N2 (Wild type) [Brenner, 1974]. Worms were maintained and cultured at 25°C as described by Brenner [1974].
RNAi
RNAi was performed by the feeding method [Fraser et al., 2000] and by injection of double-stranded RNA into the gonads of multiple N2 hermaphrodites per experiment [Fire et al., 1998]. Single depletions of PAR-3 and PAR-6 were conducted by feeding bacteria expressing double-stranded RNA to L4-stage hermaphrodites for 24–30 h at 25°C. par-3 and par-6 RNAi feeding bacteria were obtained from the Ahringer RNAi library [Kamath and Ahringer, 2003], and sequence verified. Depletion of PAR-3 and PAR-6 simultaneously was achieved through injection of par-6 double-stranded RNA into the germline of par-3 mutant (strain) KK653 worms. Worms were maintained at 25°C for 24–30-h postinjection. Double-stranded RNA for injection was synthesized using MEGA-script® T7 Kit from Ambion.
Imaging
For Fig. 2: worms were dissected in a watch glass in 30 μl of Shelton's Growth Media [Shelton and Bowerman, 1996] and the embryos were transferred to a 22 × 22 mm2 coverslip coated with poly-lysine. The coverslip was mounted to a slide, using vacuum grease feet and sealed with silicon oil. For Figs. 1, 3, and 4 worms were dissected in egg salts (118 mM NaCl, 40 mM KCl, 3.4 mM CaCl2, 3.4 mM MgCl2, 5 mM HEPES (pH 7.2) [Munro et al., 2004] and the embryos were transferred to a 22 × 22 mm2 coverslip. A 2% agarose pad, in egg salts, was placed on top of the coverslip and sealed with Vaseline. Time-lapse videos were recorded using a Zeiss 200M inverted Axioskop microscope equipped with a spinning disk confocal scan head (QLC100, Visitech International). The motorized filter turret and focus, external shutters, and a 12-bit camera (Orca ER; Hamamatsu) were controlled using OpenLab software (Improvision, Inc.). Images were taken every 8 s using a 63×, 1.4 NA Plan-Apochromat objective. For the DYN-1-GFP strain, the exposure time was 700 ms. For imaging the PAR-2-GFP; PAR-6-mCherry strain, the exposure time for PAR-2-GFP was 500 ms, and 1500 ms for mCherry-PAR-6. Images for Fig. 2 were collected using Differential Interference Contrast microscopy. All images were acquired at mid-focal plane and processed in ImageJ (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, http://imagej.nih.gov/ij/, 1997–2011). Image montages were created using Adobe PhotoShop and Illustrator (San Jose, CA).
Quantification
Kymographs were collected by cropping uniform areas (19,038 × 64,128 pixels) at specific time points (furrow initiation, furrow closure, 4-min postclosure, 8-min postclosure, 12-min postclosure, and 16-min postclosure). Fluorescence intensity of PAR-2-GFP and PAR-6-mCherry was measured using ImageJ. Taking individual frames of the kymographs and tracing the cleavage furrow, using the line tool in ImageJ, and measuring the fluorescence intensity of the selected area, accomplished this. Fluorescence intensity of the cytoplasmic area was also measured prior to cleavage furrow ingression. All other measurements were standardized to this fluorescence intensity, and treated as a fold-change value. Measurements were taken from each embryo and an average was obtained, seen in the graphs of Figs. 3 and 4. In Fig. 2, phenotypes were quantified by counting embryos under a 10x, 1.4 NA Plan-Apochromat objective while suspended in Shelton's Growth Media. Each experiment in Fig. 2 was conducted four separate times. The results are displayed in two ways (E and F). In both E and F, embryos were quantified based on terminal phenotype. In E, embryos from all four experiments were pooled together for analysis where n is the total number of embryos. Percentages of each phenotype were quantified together from all four experiments in E. In F, percentages of the terminal phenotypes were calculated separately from each of the four experiments and averaged together. The bars shown indicate standard error. This accounts for reproducibility and variability between experiments.
Statistical Analysis
To examine the effects of PAR protein depletion, through fRNAi, on PAR-6-mCherry, PAR-2-GFP, and DYN-1-GFP fluorescence intensity at the cleavage furrow was determined. We treated all fluorescence intensity values as a fold change value, as describe above in Quantification. An average fold change value was calculated for wild type and PAR depleted embryos. The average fold change values of wild type PAR-1, PAR-2, PAR-3, and PAR-6 depletions were compared at every time point by analysis of variation (ANOVA). The time points that yielded a significant difference were then subjected to post hoc testing to determine which PAR depletions lead to a significant change in PAR-6-mCherry, PAR-2-GFP, or DYN-1-GFP fluorescence. A P-value of <0.05 was considered statistically significant. Time points that were considered significantly different from wild type mean fluorescence values were marked with asterisks. Error bars on graphs indicate standard error.
Acknowledgments
The authors want to especially thank Daniel Poole and Jessica Shivas for his help and advice with the manuscript. They thank Tony Hyman, Ken Kemphues and Bruce Bowerman for strains and advice. ARS is supported by a NIH K01 Career Award (K01 HL092583) and an NSF (MCB1158003). KJP is supported by an NSF Doctoral Fellowship (DGE-0718123). The funders had no role in study design, data collection and analysis or preparation of the manuscript.
References
- Aceto D, Beers M, Kemphues KJ. Interaction of PAR-6 with CDC-42 is required for maintenance but not establishment of PAR asymmetry in C. elegans. Dev Biol. 2006;299:386–397. doi: 10.1016/j.ydbio.2006.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai E, Poole DS, Skop AR. RACK-1 directs dynactin-dependent RAB-11 endosomal recycling during mitosis in Caenorhabditis elegans. Mol Biol Cell. 2009;20:1629–1638. doi: 10.1091/mbc.E08-09-0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda V, Nolan ME, Muthuswamy SK. Par complex in cancer: a regulator of normal cell polarity joins the dark side. Oncogene. 2008;27:6878–87. doi: 10.1038/onc.2008.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian MK, Bi E, Glotzer M. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr Biol. 2004;14:R806–R818. doi: 10.1016/j.cub.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Balasubramanian MK, Hirani BR, Burke JD, Gould KL. The Schizosaccharomyces pombe cdc3+ gene encodes a profilin essential for cytokinesis. J Cell Biol. 1994;125:1289–1301. doi: 10.1083/jcb.125.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balklava Z, Pant S, Fares H, Grant BD. Genome-wide analysis identifies a general requirement for polarity proteins in endocytic traffic. Nat Cell Biol. 2007;9:1066–1073. doi: 10.1038/ncb1627. [DOI] [PubMed] [Google Scholar]
- Bednarek SY, Falbel TG. Membrane trafficking during plant cytokinesis. Traffic. 2002;3:621–629. doi: 10.1034/j.1600-0854.2002.30904.x. [DOI] [PubMed] [Google Scholar]
- Betschinger J, Knoblich JA. Dare to be different: asymmetric cell division in Drosophila, C. elegans and vertebrates. Curr Biol. 2004;14:R674–R685. doi: 10.1016/j.cub.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Bilder D, Li M, Perrimon N. Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science. 2000;289:113–116. doi: 10.1126/science.289.5476.113. [DOI] [PubMed] [Google Scholar]
- Bilder D. Epithelial polarity and proliferation control: links from the Drosophila neoplastic tumor suppressors. Genes Dev. 2004;18:1909–1925. doi: 10.1101/gad.1211604. [DOI] [PubMed] [Google Scholar]
- Boyd L, Guo S, Levitan D, Stinchcomb DT, Kemphues KJ. PAR-2 is asymmetrically distributed and promotes association of P granules and PAR-1 with the cortex in C. elegans embryos. Development. 1996;122:3075–3084. doi: 10.1242/dev.122.10.3075. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann H, Hyman AA. A cytokinesis furrow is positioned by two consecutive signals. Nature. 2005;436:731–734. doi: 10.1038/nature03823. [DOI] [PubMed] [Google Scholar]
- Cao LG, Wang YL. Signals from the spindle midzone are required for the stimulation of cytokinesis in cultured epithelial cells. Mol Biol Cell. 1996;7:225–232. doi: 10.1091/mbc.7.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cestra G, Kwiatkowski A, Salazar M, Gertler F, De Camilli P. Tuba, a GEF for CDC42, links dynamin to actin regulatory proteins. Methods Enzymol. 2005;404:537–545. doi: 10.1016/S0076-6879(05)04047-4. [DOI] [PubMed] [Google Scholar]
- Chang F. Establishment of a cellular axis in fission yeast. Trends Genet. 2001;17:273–278. doi: 10.1016/s0168-9525(01)02279-x. [DOI] [PubMed] [Google Scholar]
- Chartier NT, Salazar Ospina DP, Benkemoun L, Mayer M, Grill SW, Maddox AS, Labbe JC. PAR-4/LKB1 mobilizes non-muscle myosin through anillin to regulate C. elegans embryonic polarization and cytokinesis. Curr Biol. 2011;21:259–269. doi: 10.1016/j.cub.2011.01.010. [DOI] [PubMed] [Google Scholar]
- Cheeks RJ, Canman JC, Gabriel WN, Meyer N, Strome S, Goldstein B. C. elegans PAR proteins function by mobilizing and stabilizing asymmetrically localized protein complexes. Curr Biol. 2004;14:851–862. doi: 10.1016/j.cub.2004.05.022. [DOI] [PubMed] [Google Scholar]
- Chesneau L, Dambournet D, Machicoane M, Kouranti I, Fukuda M, Goud B, Echard A. An ARF6/Rab35 GTPase cascade for endocytic recycling and successful cytokinesis. Curr Biol. 2012;22:147–153. doi: 10.1016/j.cub.2011.11.058. [DOI] [PubMed] [Google Scholar]
- Cowan CR, Hyman AA. Asymmetric cell division in C. elegans: cortical polarity and spindle positioning. Annu Rev Cell Dev Biol. 2004;20:427–453. doi: 10.1146/annurev.cellbio.19.111301.113823. [DOI] [PubMed] [Google Scholar]
- Cowan CR, Hyman AA. Acto-myosin reorganization and PAR polarity in C. elegans. Development. 2007;134:1035–1043. doi: 10.1242/dev.000513. [DOI] [PubMed] [Google Scholar]
- Cuenca AA, Schetter A, Aceto D, Kemphues K, Seydoux G. Polarization of the C. elegans zygote proceeds via distinct establishment and maintenance phases. Development. 2003;130:1255–1265. doi: 10.1242/dev.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambournet D, Machicoane M, Chesneau L, Sachse M, Rocan-court M, El Marjou A, Formstecher E, Salomon R, Goud B, Echard A. Rab35 GTPase and OCRL phosphatase remodel lipids and F-actin for successful cytokinesis. Nat Cell Biol. 2011;13:981–988. doi: 10.1038/ncb2279. [DOI] [PubMed] [Google Scholar]
- Danilchik MV, Brown EE. Membrane dynamics of cleavage furrow closure in Xenopus laevis. Dev Dyn. 2008;237:565–579. doi: 10.1002/dvdy.21442. [DOI] [PubMed] [Google Scholar]
- Dawes AT, Munro EM. PAR-3 oligomerization may provide an actin-independent mechanism to maintain distinct par protein domains in the early Caenorhabditis elegans embryo. Biophys J. 2011;101:1412–22. doi: 10.1016/j.bpj.2011.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechant R, Glotzer M. Centrosome separation and central spindle assembly act in redundant pathways that regulate microtubule density and trigger cleavage furrow formation. Dev Cell. 2003;4:333–344. doi: 10.1016/s1534-5807(03)00057-1. [DOI] [PubMed] [Google Scholar]
- Devenport D, Oristian D, Heller E, Fuchs E. Mitotic internalization of planar cell polarity proteins preserves tissue polarity. Nat Cell Biol. 2011;13:893–902. doi: 10.1038/ncb2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukes JD, Richardson JD, Simmons R, Whitley P. A dominant-negative ESCRT-III protein perturbs cytokinesis and trafficking to lysosomes. Biochem J. 2008;411:233–239. doi: 10.1042/BJ20071296. [DOI] [PubMed] [Google Scholar]
- Dyer N, Rebollo E, Dominguez P, Elkhatib N, Chavrier P, Daviet L, Gonzalez C, Gonzalez-Gaitan M. Spermatocyte cytokinesis requires rapid membrane addition mediated by ARF6 on central spindle recycling endosomes. Development. 2007;134:4437–4447. doi: 10.1242/dev.010983. [DOI] [PubMed] [Google Scholar]
- Etemad-Moghadam B, Guo S, Kemphues KJ. Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. elegans embryos. Cell. 1995;83:743–752. doi: 10.1016/0092-8674(95)90187-6. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Georgiou M, Baum B. Polarity proteins and Rho GTPases cooperate to spatially organise epithelial actin-based protrusions. J Cell Sci. 2010;123(Pt 7):1089–1098. doi: 10.1242/jcs.060772. [DOI] [PubMed] [Google Scholar]
- Giansanti MG, Belloni G, Gatti M. Rab11 is required for membrane trafficking and actomyosin ring constriction in meiotic cytokinesis of Drosophila males. Mol Biol Cell. 2007;18:5034–5047. doi: 10.1091/mbc.E07-05-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti MG, Bonaccorsi S, Kurek R, Farkas RM, Dimitri P, Fuller MT, Gatti M. The class I PITP giotto is required for Drosophila cytokinesis. Curr Biol. 2006;16:195–201. doi: 10.1016/j.cub.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–1739. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- Glotzer M. Cytokinesis: integrating signaling, the cytoskeleton, and membranes to create new daughter cells. Semin Cell Dev Biol. 2010;21:865. doi: 10.1016/j.semcdb.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Goldstein B, Macara IG. The PAR proteins: fundamental players in animal cell polarization. Dev Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M, Abraham MC, Ahringer J. CDC-42 controls early cell polarity and spindle orientation in C. elegans. Curr Biol. 2001;11:482–488. doi: 10.1016/s0960-9822(01)00142-7. [DOI] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. Molecular genetics of asymmetric cleavage in the early Caenorhabditis elegans embryo. Curr Opin Genet Dev. 1996;6:408–415. doi: 10.1016/s0959-437x(96)80061-x. [DOI] [PubMed] [Google Scholar]
- Harris TJ, Peifer M. The positioning and segregation of apical cues during epithelial polarity establishment in Drosophila. J Cell Biol. 2005;170:813–823. doi: 10.1083/jcb.200505127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehnly H, Doxsey S. Polarity sets the stage for cytokinesis. Mol Biol Cell. 2012;23:7–11. doi: 10.1091/mbc.E11-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horgan CP, McCaffrey MW. Endosomal trafficking in animal cytokinesis. Front Biosci (Schol Ed) 2012;4:547–555. doi: 10.2741/284. [DOI] [PubMed] [Google Scholar]
- Horgan CP, Walsh M, Zurawski TH, McCaffrey MW. Rab11-FIP3 localises to a Rab11-positive pericentrosomal compartment during interphase and to the cleavage furrow during cytokinesis. Biochem Biophys Res Commun. 2004;319:83–94. doi: 10.1016/j.bbrc.2004.04.157. [DOI] [PubMed] [Google Scholar]
- Hyenne V, Tremblay-Boudreault T, Velmurugan R, Grant BD, Loerke D, Labbe JC. RAB-5 controls the cortical organization and dynamics of PAR proteins to maintain C. elegans early embryonic polarity. PLoS One. 2012;7:e35286. doi: 10.1371/journal.pone.0035286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kang BH, Busse JS, Bednarek SY. Members of the Arabidopsis dynamin-like gene family, ADL1, are essential for plant cytokinesis and polarized cell growth. Plant Cell. 2003;15:899–913. doi: 10.1105/tpc.009670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues KJ, Priess JR, Morton DG, Cheng NS. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 1988;52:311–320. doi: 10.1016/s0092-8674(88)80024-2. [DOI] [PubMed] [Google Scholar]
- Konopka CA, Schleede JB, Skop AR, Bednarek SY. Dynamin and cytokinesis. Traffic. 2006;7:239–247. doi: 10.1111/j.1600-0854.2006.00385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit T, Samanta R, Wieschaus E. slam encodes a developmental regulator of polarized membrane growth during cleavage of the Drosophila embryo. Dev Cell. 2002;2:425–436. doi: 10.1016/s1534-5807(02)00141-7. [DOI] [PubMed] [Google Scholar]
- Li B, Kim H, Beers M, Kemphues K. Different domains of C. elegans PAR-3 are required at different times in development. Dev Biol. 2010;344:745–757. doi: 10.1016/j.ydbio.2010.05.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan MR, Mandato CA. Regulation of the actin cytoskeleton by PIP2 in cytokinesis. Biol Cell. 2006;98:377–388. doi: 10.1042/BC20050081. [DOI] [PubMed] [Google Scholar]
- Lu H, Bilder D. Endocytic control of epithelial polarity and proliferation in Drosophila. Nat Cell Biol. 2005;7(12):1232–1239. doi: 10.1038/ncb1324. [DOI] [PubMed] [Google Scholar]
- Macara IG. Par proteins: partners in polarization. Curr Biol. 2004;14:R160–R162. [PubMed] [Google Scholar]
- Mavrakis M, Rikhy R, Lippincott-Schwartz J. Cells within a cell: insights into cellular architecture and polarization from the organization of the early fly embryo. Commun Integr Biol. 2009;2:313–314. doi: 10.4161/cib.2.4.8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnac G, Echard A, Chavrier P. Endocytic traffic in animal cell cytokinesis. Curr Opin Cell Biol. 2008;20:454–461. doi: 10.1016/j.ceb.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Munro E, Nance J, Priess JR. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev Cell. 2004;7:413–424. doi: 10.1016/j.devcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Nakayama Y, Shivas JM, Poole DS, Squirrell JM, Kulkoski JM, Schleede JB, Skop AR. Dynamin participates in the maintenance of anterior polarity in the Caenorhabditis elegans embryo. Dev Cell. 2009;16:889–900. doi: 10.1016/j.devcel.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nance J, Zallen JA. Elaborating polarity: PAR proteins and the cytoskeleton. Development. 2011;138:799–809. doi: 10.1242/dev.053538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng MM, Chang F, Burgess DR. Movement of membrane domains and requirement of membrane signaling molecules for cytokinesis. Dev Cell. 2005;9:781–790. doi: 10.1016/j.devcel.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Nolan ME, Aranda V, Lee S, Lakshmi B, Basu S, Allred DC, Muthuswamy SK. The polarity protein Par6 induces cell proliferation and is overexpressed in breast cancer. Cancer Res. 2008;68:8201–8209. doi: 10.1158/0008-5472.CAN-07-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Intercellular junctions and cellular polarity: the PAR-aPKC complex, a conserved core cassette playing fundamental roles in cell polarity. Curr Opin Cell Biol. 2001;13:641–648. doi: 10.1016/s0955-0674(00)00264-7. [DOI] [PubMed] [Google Scholar]
- Papoulas O, Hays TS, Sisson JC. The golgin Lava lamp mediates dynein-based Golgi movements during Drosophila cellularization. Nat Cell Biol. 2005;7:612–618. doi: 10.1038/ncb1264. [DOI] [PubMed] [Google Scholar]
- Patalano S, Pruliere G, Prodon F, Paix A, Dru P, Sardet C, Chenevert J. The aPKC-PAR-6-PAR-3 cell polarity complex localizes to the centrosome attracting body, a macroscopic cortical structure responsible for asymmetric divisions in the early ascidian embryo. J Cell Sci. 2006;119(Pt 8):1592–1603. doi: 10.1242/jcs.02873. [DOI] [PubMed] [Google Scholar]
- Pellettieri J, Seydoux G. Anterior-posterior polarity in C. elegans and Drosophila--PARallels and differences. Science. 2002;298:1946–1950. doi: 10.1126/science.1072162. [DOI] [PubMed] [Google Scholar]
- Piekny A, Werner M, Glotzer M. Cytokinesis: welcome to the Rho zone. Trends Cell Biol. 2005;15:651–658. doi: 10.1016/j.tcb.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Prekeris R, Gould GW. Breaking up is hard to do—membrane traffic in cytokinesis. J Cell Sci. 2008;121(Pt 10):1569–1576. doi: 10.1242/jcs.018770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Kaneda M, Chen J, Geitmann A, Zheng H. A specific role for Arabidopsis TRAPPII in post-Golgi trafficking that is crucial for cytokinesis and cell polarity. Plant J. 2011;68:234–248. doi: 10.1111/j.1365-313X.2011.04681.x. [DOI] [PubMed] [Google Scholar]
- Riggs B, Rothwell W, Mische S, Hickson GR, Matheson J, Hays TS, Gould GW, Sullivan W. Actin cytoskeleton remodeling during early Drosophila furrow formation requires recycling endosomal components Nuclear-fallout and Rab11. J Cell Biol. 2003;163:143–154. doi: 10.1083/jcb.200305115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagona AP, Stenmark H. Cytokinesis and cancer. FEBS Lett. 2010;584:2652–2661. doi: 10.1016/j.febslet.2010.03.044. [DOI] [PubMed] [Google Scholar]
- Schafer DA. Coupling actin dynamics and membrane dynamics during endocytosis. Curr Opin Cell Biol. 2002;14:76–81. doi: 10.1016/s0955-0674(01)00297-6. [DOI] [PubMed] [Google Scholar]
- Schenk C, Bringmann H, Hyman AA, Cowan CR. Cortical domain correction repositions the polarity boundary to match the cytokinesis furrow in C. elegans embryos. Development. 2010;137:1743–1753. doi: 10.1242/dev.040436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonegg S, Constantinescu AT, Hoege C, Hyman AA. The Rho GTPase-activating proteins RGA-3 and RGA-4 are required to set the initial size of PAR domains in Caenorhabditis elegans one-cell embryos. Proc Natl Acad Sci USA. 2007;104:14976–14981. doi: 10.1073/pnas.0706941104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton CA, Bowerman B. Time-dependent responses to glp-1-mediated inductions in early C. elegans embryos. Development. 1996;122:2043–2050. doi: 10.1242/dev.122.7.2043. [DOI] [PubMed] [Google Scholar]
- Shivas JM, Morrison HA, Bilder D, Skop AR. Polarity and endocytosis: reciprocal regulation. Trends Cell Biol. 2010;20:445–452. doi: 10.1016/j.tcb.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivas JM, Skop AR. Arp2/3 mediates early endosome dynamics necessary for the maintenance of PAR asymmetry in Caenorhabditis elegans. Mol Biol Cell. 2012;23:1917–1927. doi: 10.1091/mbc.E12-01-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster CB, Burgess DR. Targeted new membrane addition in the cleavage furrow is a late, separate event in cytokinesis. Proc Natl Acad Sci USA. 2002;99:3633–3638. doi: 10.1073/pnas.052342699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisson JC, Rothwell WF, Sullivan W. Cytokinesis: lessons from rappaport and the Drosophila blastoderm embryo. Cell Biol Int. 1999;23:871–876. doi: 10.1006/cbir.1999.0484. [DOI] [PubMed] [Google Scholar]
- Skop AR, Bergmann D, Mohler WA, White JG. Completion of cytokinesis in C. elegans requires a brefeldin A-sensitive membrane accumulation at the cleavage furrow apex. Curr Biol. 2001;11:735–746. doi: 10.1016/s0960-9822(01)00231-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skop AR, Liu H, Yates J, 3rd, Meyer BJ, Heald R. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science. 2004;305:61–66. doi: 10.1126/science.1097931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T, Kawate T, Chang F. Organization of a sterol-rich membrane domain by cdc15p during cytokinesis in fission yeast. Nat Cell Biol. 2004;6:1142–1144. doi: 10.1038/ncb1189. [DOI] [PubMed] [Google Scholar]
- Tavares A, Goncalves J, Florindo C, Tavares AA, Soares H. Mob1: defining cell polarity for proper cell division. J Cell Sci. 2012;125(Pt 2):516–527. doi: 10.1242/jcs.096610. [DOI] [PubMed] [Google Scholar]
- Tepass U, Tanentzapf G, Ward R, Fehon R. Epithelial cell polarity and cell junctions in Drosophila. Annu Rev Genet. 2001;35:747–784. doi: 10.1146/annurev.genet.35.102401.091415. [DOI] [PubMed] [Google Scholar]
- Thompson HM, Skop AR, Euteneuer U, Meyer BJ, McNiven MA. The large GTPase dynamin associates with the spindle midzone and is required for cytokinesis. Curr Biol. 2002;12:2111–2117. doi: 10.1016/s0960-9822(02)01390-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totong R, Achilleos A, Nance J. PAR-6 is required for junction formation but not apicobasal polarization in C. elegans embryonic epithelial cells. Development. 2007;134:1259–1268. doi: 10.1242/dev.02833. [DOI] [PubMed] [Google Scholar]
- Vaccari T, Bilder D. At the crossroads of polarity, proliferation and apoptosis: the use of Drosophila to unravel the multifaceted role of endocytosis in tumor suppression. Mol Oncol. 2009;3:354–365. doi: 10.1016/j.molonc.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JL, Morton DG, Bestman J, Kemphues KJ. The C. elegans par-4 gene encodes a putative serine-threonine kinase required for establishing embryonic asymmetry. Development. 2000;127:1467–1475. doi: 10.1242/dev.127.7.1467. [DOI] [PubMed] [Google Scholar]
- Wheatley SP, Wang Y. Midzone microtubule bundles are continuously required for cytokinesis in cultured epithelial cells. J Cell Biol. 1996;135:981–989. doi: 10.1083/jcb.135.4.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Borisy GG. On the mechanisms of cytokinesis in animal cells. J Theor Biol. 1983;101:289–316. doi: 10.1016/0022-5193(83)90342-9. [DOI] [PubMed] [Google Scholar]
- Wienke DC, Knetsch ML, Neuhaus EM, Reedy MC, Manstein DJ. Disruption of a dynamin homologue affects endocytosis, organelle morphology, and cytokinesis in Dictyostelium discoideum. Mol Biol Cell. 1999;10:225–243. doi: 10.1091/mbc.10.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GM, Fielding AB, Simon GC, Yu X, Andrews PD, Hames RS, Frey AM, Peden AA, Gould GW, Prekeris R. The FIP3-Rab11 protein complex regulates recycling endosome targeting to the cleavage furrow during late cytokinesis. Mol Biol Cell. 2005;16:849–860. doi: 10.1091/mbc.E04-10-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H, Mohler WA, Soto MC. The branched actin nucleator Arp2/3 promotes nuclear migrations and cell polarity in the C. elegans zygote. Dev Biol. 2011;357:356–369. doi: 10.1016/j.ydbio.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]