Abstract
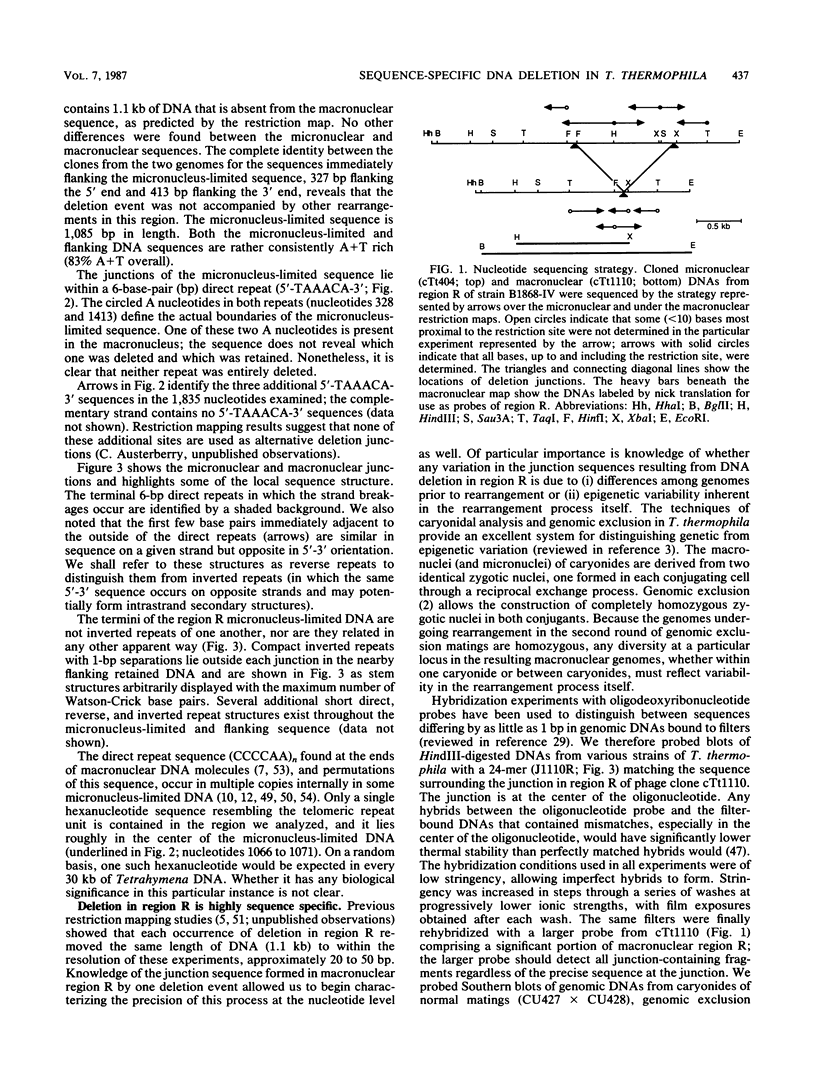
DNA deletion by site-specific chromosome breakage and rejoining occurs extensively during macronuclear development in the ciliate Tetrahymena thermophila. We have sequenced both the micronuclear (germ line) and rearranged macronuclear (somatic) forms of one region from which 1.1 kilobases of micronuclear DNA are reproducibly deleted during macronuclear development. The deletion junctions lie within a pair of 6-base-pair direct repeats. The termini of the deleted sequence are not inverted repeats. The precision of deletion at the nucleotide level was also characterized by hybridization with a synthetic oligonucleotide matching the determined macronuclear (rejoined) junction sequence. This deletion occurs in a remarkably sequence-specific manner. However, a very minor degree of variability in the macronuclear junction sequences was detected and was shown to be inherent in the mechanism of deletion itself. These results suggest that DNA deletion during macronuclear development in T. thermophila may constitute a novel type of DNA recombination and that it can create sequence heterogeneity on the order of a few base pairs at rejoining junctions.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Allen S. L. Cytogenetics of genomic exclusion in Tetrahymena. Genetics. 1967 Apr;55(4):797–822. doi: 10.1093/genetics/55.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammermann D., Steinbrück G., von Berger L., Hennig W. The development of the macronucleus in the ciliated protozoan Stylonychia mytilus. Chromosoma. 1974 May 10;45(4):401–429. doi: 10.1007/BF00283386. [DOI] [PubMed] [Google Scholar]
- Austerberry C. F., Allis C. D., Yao M. C. Specific DNA rearrangements in synchronously developing nuclei of Tetrahymena. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7383–7387. doi: 10.1073/pnas.81.23.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn E. H., Gall J. G. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J Mol Biol. 1978 Mar 25;120(1):33–53. doi: 10.1016/0022-2836(78)90294-2. [DOI] [PubMed] [Google Scholar]
- Brunk C. F., Conover R. K. Elimination of micronuclear specific DNA sequences early in anlagen development. Mol Cell Biol. 1985 Jan;5(1):93–98. doi: 10.1128/mcb.5.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunk C. F., Tsao S. G., Diamond C. H., Ohashi P. S., Tsao N. N., Pearlman R. E. Reorganization of unique and repetitive sequences during nuclear development in Tetrahymena thermophila. Can J Biochem. 1982 Sep;60(9):847–853. doi: 10.1139/o82-107. [DOI] [PubMed] [Google Scholar]
- Callahan R. C., Shalke G., Gorovsky M. A. Developmental rearrangements associated with a single type of expressed alpha-tubulin gene in Tetrahymena. Cell. 1984 Feb;36(2):441–445. doi: 10.1016/0092-8674(84)90237-x. [DOI] [PubMed] [Google Scholar]
- Cherry J. M., Blackburn E. H. The internally located telomeric sequences in the germ-line chromosomes of Tetrahymena are at the ends of transposon-like elements. Cell. 1985 Dec;43(3 Pt 2):747–758. doi: 10.1016/0092-8674(85)90248-x. [DOI] [PubMed] [Google Scholar]
- Cupples C. G., Pearlman R. E. Isolation and characterization of the actin gene from Tetrahymena thermophila. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5160–5164. doi: 10.1073/pnas.83.14.5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels S. B., McCarron M., Love C., Chovnick A. Dysgenesis-induced instability of rosy locus transformation in Drosophila melanogaster: analysis of excision events and the selective recovery of control element deletions. Genetics. 1985 Jan;109(1):95–117. doi: 10.1093/genetics/109.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Döring H. P., Starlinger P. Barbara McClintock's controlling elements: now at the DNA level. Cell. 1984 Dec;39(2 Pt 1):253–259. doi: 10.1016/0092-8674(84)90002-3. [DOI] [PubMed] [Google Scholar]
- Egner C., Berg D. E. Excision of transposon Tn5 is dependent on the inverted repeats but not on the transposase function of Tn5. Proc Natl Acad Sci U S A. 1981 Jan;78(1):459–463. doi: 10.1073/pnas.78.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P. J., Schmeissner U., Hofer M., Miller J. H. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J Mol Biol. 1978 Dec 25;126(4):847–857. doi: 10.1016/0022-2836(78)90023-2. [DOI] [PubMed] [Google Scholar]
- Foster T. J., Lundblad V., Hanley-Way S., Halling S. M., Kleckner N. Three Tn10-associated excision events: relationship to transposition and role of direct and inverted repeats. Cell. 1981 Jan;23(1):215–227. doi: 10.1016/0092-8674(81)90286-5. [DOI] [PubMed] [Google Scholar]
- Georgiev G. P. Mobile genetic elements in animal cells and their biological significance. Eur J Biochem. 1984 Dec 3;145(2):203–220. doi: 10.1111/j.1432-1033.1984.tb08541.x. [DOI] [PubMed] [Google Scholar]
- Golden J. W., Robinson S. J., Haselkorn R. Rearrangement of nitrogen fixation genes during heterocyst differentiation in the cyanobacterium Anabaena. Nature. 1985 Apr 4;314(6010):419–423. doi: 10.1038/314419a0. [DOI] [PubMed] [Google Scholar]
- Gorovsky M. A. Macro- and micronuclei of Tetrahymena pyriformis: a model system for studying the structure and function of eukaryotic nuclei. J Protozool. 1973 Feb;20(1):19–25. doi: 10.1111/j.1550-7408.1973.tb05995.x. [DOI] [PubMed] [Google Scholar]
- Gorovsky M. A., Yao M. C., Keevert J. B., Pleger G. L. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 1975;9(0):311–327. doi: 10.1016/s0091-679x(08)60080-1. [DOI] [PubMed] [Google Scholar]
- Hanyu N., Kuchino Y., Nishimura S., Beier H. Dramatic events in ciliate evolution: alteration of UAA and UAG termination codons to glutamine codons due to anticodon mutations in two Tetrahymena tRNAs. EMBO J. 1986 Jun;5(6):1307–1311. doi: 10.1002/j.1460-2075.1986.tb04360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Hood L., Kronenberg M., Hunkapiller T. T cell antigen receptors and the immunoglobulin supergene family. Cell. 1985 Feb;40(2):225–229. doi: 10.1016/0092-8674(85)90133-3. [DOI] [PubMed] [Google Scholar]
- Horowitz S., Gorovsky M. A. An unusual genetic code in nuclear genes of Tetrahymena. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2452–2455. doi: 10.1073/pnas.82.8.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard E. A., Blackburn E. H. Reproducible and variable genomic rearrangements occur in the developing somatic nucleus of the ciliate Tetrahymena thermophila. Mol Cell Biol. 1985 Aug;5(8):2039–2050. doi: 10.1128/mcb.5.8.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura K., Rossi J. J., Wallace R. B. Synthesis and use of synthetic oligonucleotides. Annu Rev Biochem. 1984;53:323–356. doi: 10.1146/annurev.bi.53.070184.001543. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Klobutcher L. A., Jahn C. L., Prescott D. M. Internal sequences are eliminated from genes during macronuclear development in the ciliated protozoan Oxytricha nova. Cell. 1984 Apr;36(4):1045–1055. doi: 10.1016/0092-8674(84)90054-0. [DOI] [PubMed] [Google Scholar]
- Kraut H., Lipps H. J., Prescott D. M. The genome of hypotrichous ciliates. Int Rev Cytol. 1986;99:1–28. doi: 10.1016/s0074-7696(08)61422-9. [DOI] [PubMed] [Google Scholar]
- Landy A., Ross W. Viral integration and excision: structure of the lambda att sites. Science. 1977 Sep 16;197(4309):1147–1160. doi: 10.1126/science.331474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauth M. R., Spear B. B., Heumann J., Prescott D. M. DNA of ciliated protozoa: DNA sequence diminution during macronuclear development of Oxytricha. Cell. 1976 Jan;7(1):67–74. doi: 10.1016/0092-8674(76)90256-7. [DOI] [PubMed] [Google Scholar]
- Martindale D. W., Allis C. D., Bruns P. J. Conjugation in Tetrahymena thermophila. A temporal analysis of cytological stages. Exp Cell Res. 1982 Jul;140(1):227–236. doi: 10.1016/0014-4827(82)90172-0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- O'Hare K., Rubin G. M. Structures of P transposable elements and their sites of insertion and excision in the Drosophila melanogaster genome. Cell. 1983 Aug;34(1):25–35. doi: 10.1016/0092-8674(83)90133-2. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Saedler H., Nevers P. Transposition in plants: a molecular model. EMBO J. 1985 Mar;4(3):585–590. doi: 10.1002/j.1460-2075.1985.tb03670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Tondravi M. M., Yao M. C. Transformation of Tetrahymena thermophila by microinjection of ribosomal RNA genes. Proc Natl Acad Sci U S A. 1986 Jun;83(12):4369–4373. doi: 10.1073/pnas.83.12.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983 Apr 14;302(5909):575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Voelker R. A., Greenleaf A. L., Gyurkovics H., Wisely G. B., Huang S. M., Searles L. L. Frequent Imprecise Excision among Reversions of a P Element-Caused Lethal Mutation in Drosophila. Genetics. 1984 Jun;107(2):279–294. doi: 10.1093/genetics/107.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace R. B., Shaffer J., Murphy R. F., Bonner J., Hirose T., Itakura K. Hybridization of synthetic oligodeoxyribonucleotides to phi chi 174 DNA: the effect of single base pair mismatch. Nucleic Acids Res. 1979 Aug 10;6(11):3543–3557. doi: 10.1093/nar/6.11.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T. C., Allen S. L. Alternative processing of sequences during macronuclear development in Tetrahymena thermophila. J Protozool. 1986 Feb;33(1):30–38. doi: 10.1111/j.1550-7408.1986.tb05551.x. [DOI] [PubMed] [Google Scholar]
- Yao M. C., Blackburn E., Gall J. Tandemly repeated C-C-C-C-A-A hexanucleotide of Tetrahymena rDNA is present elsewhere in the genome and may be related to the alteration of the somatic genome. J Cell Biol. 1981 Aug;90(2):515–520. doi: 10.1083/jcb.90.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M. C., Choi J., Yokoyama S., Austerberry C. F., Yao C. H. DNA elimination in Tetrahymena: a developmental process involving extensive breakage and rejoining of DNA at defined sites. Cell. 1984 Feb;36(2):433–440. doi: 10.1016/0092-8674(84)90236-8. [DOI] [PubMed] [Google Scholar]
- Yao M. C. Elimination of specific DNA sequences from the somatic nucleus of the ciliate Tetrahymena. J Cell Biol. 1982 Mar;92(3):783–789. doi: 10.1083/jcb.92.3.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M. C., Gorovsky M. A. Comparison of the sequences of macro- and micronuclear DNA of Tetrahymena pyriformis. Chromosoma. 1974;48(1):1–18. doi: 10.1007/BF00284863. [DOI] [PubMed] [Google Scholar]
- Yao M. C., Yao C. H. Repeated hexanucleotide C-C-C-C-A-A is present near free ends of macronuclear DNA of Tetrahymena. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7436–7439. doi: 10.1073/pnas.78.12.7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R., Yao M. C. Internal micronuclear DNA regions which include sequences homologous to macronuclear telomeres are deleted during development in Tetrahymena. Nucleic Acids Res. 1984 Aug 10;12(15):6103–6116. doi: 10.1093/nar/12.15.6103. [DOI] [PMC free article] [PubMed] [Google Scholar]