Abstract
Human papillomaviruses (HPVs) amplify in differentiated strata of a squamous epithelium. The HPV E7 protein destabilizes the p130/retinoblastoma susceptibility protein family of tumor suppressors and reactivates S-phase reentry, thereby facilitating viral DNA amplification. The high-risk HPV E6 protein destabilizes the p53 tumor suppressor and many other host proteins. However, the critical E6 targets relevant to viral DNA amplification have not been identified, because functionally significant E6 mutants are not stably maintained in transfected cells. Using Cre-loxP recombination, which efficiently generates HPV genomic plasmids in transfected primary human keratinocytes, we have recapitulated a highly productive infection of HPV-18 in organotypic epithelial cultures. By using this system, we now report the characterization of four HPV-18 E6 mutations. An E6 null mutant accumulated high levels of p53 and amplified very poorly. p53 siRNA or ectopic WT E6 partially restored amplification, whereas three missense E6 mutations that did not effectively destabilize p53 complemented the null mutant poorly. Unexpectedly, in cis, two of the missense mutants amplified, albeit to a lower extent than the WT and only in cells with undetectable p53. These observations and others implicate p53 and additional host proteins in regulating viral DNA amplification and also suggest an inhibitory effect of E6 overexpression. We show that high levels of viral DNA amplification are critical for late protein expression and report several previously undescribed viral RNAs, including bicistronic transcripts predicted to encode E5 and L2 or an alternative form of E1^E4 and L1.
Keywords: human papillomavirus DNA amplification, trans complementation, HPV transcripts
Papillomaviruses are small DNA viruses with a protein capsid harboring a double-stranded circular genome of ∼7,900 bp. Dozens of human papillomavirus (HPV) types are tropic for the anogenital tract (1). HPV-16 and -18 and closely related genotypes are high-risk (HR) types and at a low frequency, can cause cancers, in which the viral E6 and E7 oncogenes are invariably overexpressed. HPV-6 and -11 are low-risk (LR) types and induce benign anogenital warts and laryngeal papillomas (review in ref. 2). Typically, viral DNA is maintained as low copy nuclear plasmids in basal and parabasal keratinocytes, and vegetative amplification depends on squamous differentiation (review in ref. 3). Because viral DNA replication requires the host DNA replication machinery, the role of the HPV E7 protein is to promote S-phase reentry in differentiated cells that have withdrawn from the cell cycle. It does so by destabilizing the p130 protein (4, 5), a pocket protein related to the retinoblastoma susceptibility protein, a major tumor suppressor. The HR but not the LR HPV E7 protein also destabilizes retinoblastoma susceptibility protein (6). Both HR and LR HPV E6 proteins inactivate the transcription regulatory activities of another major tumor suppressor, p53 (7–10), abrogating its control over cell cycle arrest, DNA repair, apoptosis, or senescence (11). However, only the HR HPV E6 protein also destabilizes p53 (12). The HR HPV E6 protein and a truncated E6*I peptide destabilize several additional host proteins that regulate cell growth and differentiation (13). These unique properties of HR HPV oncoproteins form the basis of viral oncogenesis.
The purpose of E6 during the productive lifecycle is not completely understood. Ectopically expressed p53 can inhibit transient viral DNA replication in transfected cells (14–16). In as much as E7 expression causes p53 stabilization in the absence of E6 (17–19), E6 may well counteract this defensive response of the host cells. However, this hypothesis has not been tested in the sequence context of the entire HPV genome in differentiating squamous epithelium, because functionally significant E6 mutants cannot be maintained stably as plasmids in transfected cells, and they do not immortalize primary human keratinocytes (PHKs) (20, 21).
We have reported a system enabling robust production of infectious HPV-18 in organotypic raft cultures of PHKs (22). HPV-18 genomic plasmids with a 34-bp loxP site in the upstream regulatory region (URR; termed WT for simplicity) are efficiently generated from a recombinant plasmid by excisional recombination mediated by Cre-loxP in transfected PHKs. In contrast, an HPV-18 mutant, which expresses the truncated E6*I peptide but not the full-length E6 protein, does not amplify or produce progeny virus. Rather, p53 protein stabilized by E7 accumulates in a high fraction of basal and suprabasal cells. A retrovirus that expresses WT HPV-18 E6 (under the control of the HPV-18 URR) qualitatively rescues this mutant, restoring the WT phenotypes. Thus, the E6 protein has a critical role in viral DNA amplification. For a better understanding of the E6 functions, it became important to examine E6 missense mutations.
In this study, we established a trans complementation system to examine missense E6 mutations in PHK raft cultures. First, we constructed an HPV-18 E6 null mutant, in which the translation initiation codon was mutated. It had the same phenotype as the E6*I mutant and was similarly complemented by ectopic WT E6. We then examined the abilities of retroviruses expressing three E6 missense mutations or a p53 siRNA to complement the E6 null mutant genome. These mutations were selected based on their homology to mutated HPV-16 E6 proteins that are deficient in destabilizing p53 protein. As an alternative approach, full-length mutant genomes harboring the same missense mutations were examined. Our results suggest that p53 and additional host proteins are E6 targets in productive infection and that a balance between the E6 and E7 oncoproteins is important to maximize virus DNA amplification. Moreover, we show that late viral RNA levels correlate with the extent of viral DNA amplification and describe spliced viral RNAs predicted to encode known or unique viral proteins.
Results
Phenotypes of the E6 Null Mutant and Its Complementation in Trans by WT E6.
As we reported (22), histology of tissue sections showed that day 13 raft cultures containing WT HPV-18 were hyperplastic, with many more cell layers than the normal PHK raft cultures (Fig. S1). Abundant major capsid protein L1 signals were readily detected on day 16 in the cornified envelopes across the surface of the cultures, indicative of a highly productive program (Fig. 1, Top). Incorporation of BrdU in numerous suprabasal differentiated keratinocytes marked efficient induction of S-phase reentry, whereas very few p53-positive cells were observed (Fig. S2). Fluorescent in situ hybridization (FISH) revealed that viral DNA amplification increased with time, and on day 13, a large fraction of the upper spinous cells was positive, the majority of which were negative for p53 (Fig. 2A, Top). In contrast, cultures containing the HPV-18 E6 null mutant exhibited a less hyperplastic histology relative to the WT (Fig. S1), lacked L1 signals (Fig. 1, Top), and had a reduction in BrdU-positive suprabasal cells (Fig. S2 and Table S1). p53 protein was detected throughout the epithelium (Fig. 2A, Top and Fig. S2), whereas few spinous cells had amplified viral DNA (Fig. 2A, Top). These phenotypes are identical to the E6*I mutant (22). In normal PHK raft cultures, BrdU incorporation and p53 signals were detected only in basal and occasional parabasal cells (Fig. S2).
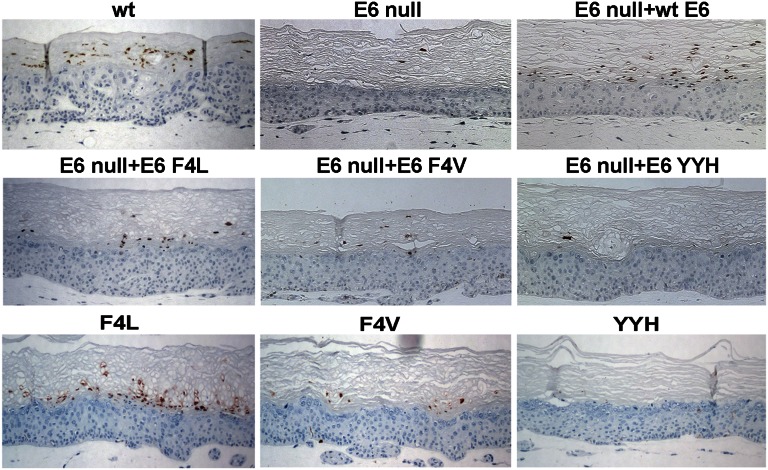
Fig. 1.
HPV-18 major capsid protein (L1) detection. HPV-18 L1 was probed by immunohistochemistry with an L1 antibody in raft cultures containing HPV-18 WT or E6 mutant genomes (WT, F4L, F4V, YYH, or E6 null) or the E6 null mutant trans complemented by pBabe Puro HPV-18 URR-E6 or E6 mutations (E6 null + WT E6, E6 F4L, E6 F4V, or E6 YYH).
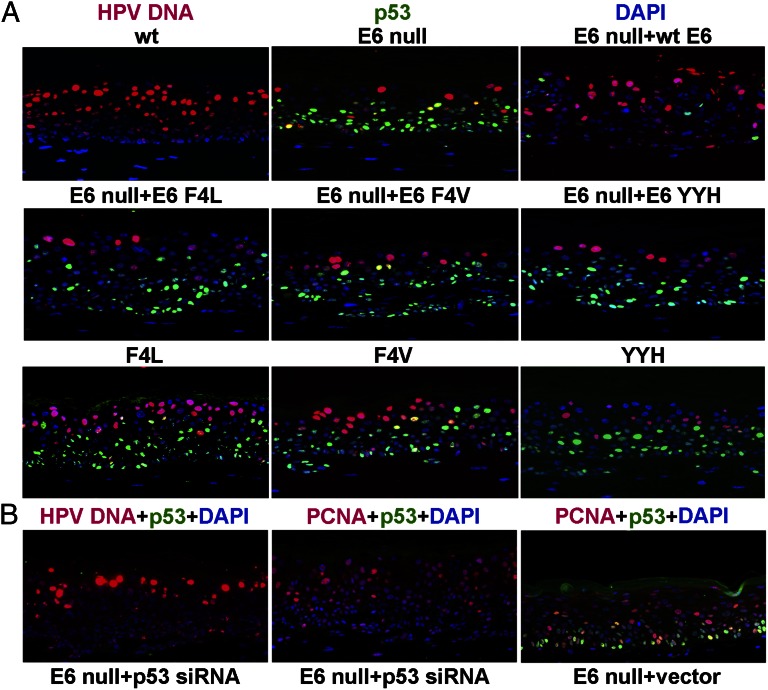
Fig. 2.
In situ analysis of HPV-18 viral DNA amplification and p53 stabilization or PCNA induction. (A) Raft cultures containing HPV-18 WT, E6 mutant genomes, or E6 null mutant in the presence of ectopic E6 WT or missense mutations were simultaneously probed for viral DNA amplification (red, Alexa Fluor 555) by DNA FISH and for p53 (green, Alexa Fluor 488) by indirect immunofluorescence. These experiments were conducted on multiple raft cultures, and they gave the same qualitative results. (B) Raft cultures of PHKs harboring HPV-18 E6 null genome were transduced with retroviruses expressing a p53 siRNA (Left and Center) or an empty vector (Right). Sections were probed for HPV-18 viral DNA amplification (red, Tyramide Cy3; Left), p53 stabilization (green, Tyramide, FITC; Left, Center, and Right), or PCNA induction (red, Alexa Fluor 555; Center and Right). Nuclei were stained with DAPI (blue).
We revised our previous protocol for trans complementation studies (22) to ensure that all PHKs harbored both the URR-E6 provirus and the E6 null mutant (Materials and Methods). As with the E6*I mutant (22), the E6 null mutant was partially rescued by WT E6 in trans, with respect to tissue histology (Fig. S1), an increase in suprabasal BrdU-positive cells, loss of p53, and increased viral DNA amplification (Fig. 2A, Top and Fig. S2) and L1 expression (Fig. 1, Top). Thus, the incomplete complementation of E6*I by E6 (22) cannot be attributed to the dominant negative effect of E6*I over E6 in p53 destabilization (23).
Trans Complementation of the E6 Null Mutant by E6 Missense Mutations.
We selected three HPV-18 E6 missense mutations to study based on sequence homology to mutant forms of HPV-16 E6 that have a deficiency in destabilizing p53 because of impaired binding to p53 or E6-associated protein (9, 24–26). The second residue phenylalanine (F2) in HPV-16 E6 protein is conserved among HR HPV types. HPV-16 E6 F2V is deficient in destabilizing p53 in vitro and in human mammary epithelial cells. HPV-16 E6 F2L partially degraded p53 in vitro, but it was not examined in mammary epithelial cells. The HPV-16 E6 F45,F47,D49 residues are also conserved among HR HPV E6 proteins. When substituted with Y,Y,H, respectively (the corresponding residues in LR HPV E6 proteins), the mutant form of E6 does not destabilize p53. HPV-16 E6 F2V and E6 YYH retain the ability to inactivate the transactivation function of p53 (8–10), whereas there was no report for HPV-16 E6 F2L. The comparable residues in HPV-18 are numerically offset by 2 aa. We, therefore, constructed pBabe Puro retroviral vectors expressing HPV-18 E6 F4L, E6 F4V, and E6 F47,49Y,D51H (E6 YYH), each under the transcriptional control of the HPV-18 URR.
Ectoptic WT E6 or E6 mutations restored the histology of the E6 null mutant-containing raft culture to cultures harboring the WT virus (Fig. S1). As before, WT E6 partially restored L1 expression. However, very little L1 signal was detected in cultures with ectopically expressed E6 F4V or E6 YYH. Only the E6 F4L elicited signals in a small region of the raft culture in only one of several experiments (Fig. 1, Middle). p53 accumulated in numerous cells throughout the epithelium in the presence of ectopic missense E6, and none were able to increase the fraction of nuclei positive for viral DNA amplification significantly (Fig. 2A, Middle).
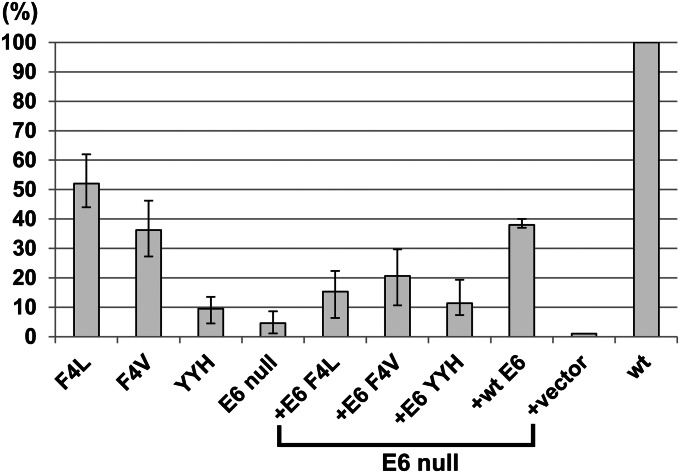
For a more objective comparison, we performed real-time quantitative PCR (qPCR) to quantify viral DNA copy numbers per cell from raft cultures not subjected to formalin fixation. In five independent experiments, the copy numbers on day 13 of the WT or the E6 null mutant were examined. The WT DNA ranged from 3,600 to 5,900 plasmids per cell, with an average of about 4,300 plasmids per cell, whereas the E6 null mutant averaged 4.5% of the WT, ranging from 0.3% to 9% (Fig. 3). In one of these experiments, the copy numbers of the WT and E6 null mutant genomes from a fraction of the PHKs were determined just before the remaining cells were used to develop raft cultures. They contained 82 and 35 copies per cell, respectively. Thus, the inability of the E6 null mutant to amplify to high copy numbers could not be entirely attributed to a significantly reduced basal copy number. In three of five experiments, the copy numbers of the E6 null mutant in the presence of ectopic WT or missense E6 were determined in parallel. With WT E6, E6 F4L, E6 F4V, or E6 YYH in trans, the E6 null mutant achieved 38%, 15%, 20%, or 10%, respectively, of the WT virus copy number on day 13 (Fig. 3). Thus, the mutant forms of E6 that are unable to destabilize p53 effectively are deficient in supporting viral DNA amplification. The effect of the empty vector will be elaborated later.
Fig. 3.
Real-time qPCR to compare HPV-18 DNA copy numbers per cell. The average HPV-18 genomic plasmid copy numbers per diploid cell were determined by real-time qPCR in raft cultures containing HPV-18 WT or E6 mutants in five sets of independent experiments, each with a different batch of PHKs. In three of five experiments, the copy numbers of the E6 null mutant complemented by ectopic WT E6, E6 missense mutations, or empty vector in trans were conducted in parallel. The WT genome copy number averaged about 4,300 and was set as 100%. Normal PHK raft cultures were negative (0.021%) for HPV DNA.
Amplification of HPV-18 E6 Missense Mutants in Spinous Cells with Undetectable p53 Protein.
We then constructed and tested mutant genomes, each harboring the same E6 missense mutations. Raft cultures containing mutant genomes were hyperplastic, similar to the WT virus (Fig. S1). The fractions of stochastically distributed suprabasal BrdU-positive cells increased but never reached the number induced by the WT virus (Fig. S2 and Table S1). Many basal and suprabasal cells remained positive for p53. Colocalized p53 and BrdU were observed only in a small fraction of the cells (Fig. S2).
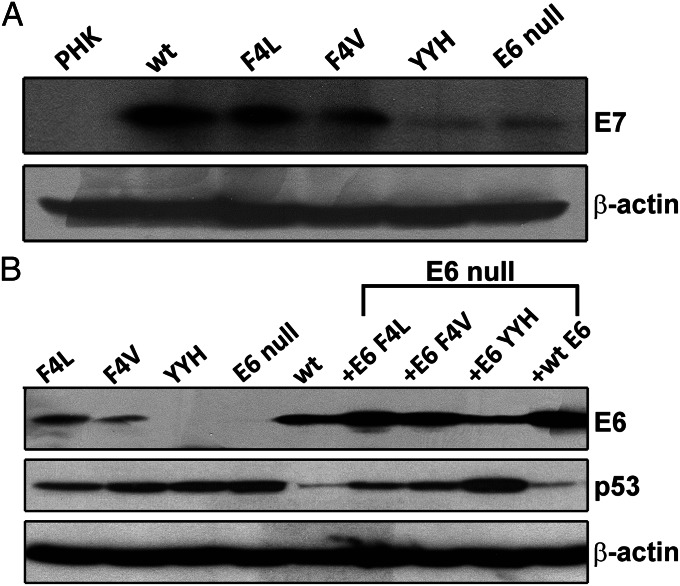
To understand the causes for the reduction in S-phase cells, we probed for but did not detect cleaved caspase 3 by immunofluorescence (22), consistent with the absence of apoptotic cells based on histology (Fig. S1). We then examined whether the stabilized p53 up-regulated p21cip1 expression. Raft culture lysates were analyzed using real-time RT-qPCR and Western blots. p53 mediated up-regulation of p21cip1, an inhibitor of cdk2/cyclin E and cdk2/cyclin A, prevents S-phase entry upon DNA damage. Previously, we have shown that p21cip1 RNA is constitutively transcribed in differentiated keratinocytes independent of HPV infection or E7 gene expression and that p21cip1 protein is stabilized by posttranslational mechanisms (17, 27). The results showed that the variations in p21cip1 RNA did not correlate with p21cip1 protein (Fig. S3) or p53 protein (Fig. 4B). These findings strongly suggest that the E7-stabilized p53 protein is transcriptionally inactive and unable to activate p21cip1 transcription to a significant extent. Importantly, the levels of p21cip1 protein did not correlate with the numbers of suprabasal BrdU-positive cells (Table S1). Thus, the reduction in suprabasal S-phase cells cannot be attributed to elevated p21cip protein. Interestingly, the number of suprabasal S-phase cells did correlate with the relative amounts of the E7 protein in these cultures in three independent experiments (Fig. 4A).
Fig. 4.
Western blot detections of E7, E6, and p53 in raft cultures. (A) Comparison of HPV-18E7 protein levels in the lysates of raft cultures containing HPV-18 WT and E6 mutant genomes. (B) Lysates from raft cultures containing HPV-18 WT, E6 mutant genomes, or E6 null mutant complemented by WT E6 or E6 missense mutations in trans were probed for viral E6 and p53 proteins; β-actin was used as internal loading reference for all of the experiments.
We then examined viral DNA amplification relative to p53 accumulation (Fig. 2A, Bottom). Cells with amplified E6 YYH mutant remained scarce. To our surprise, differentiated cells with amplified F4L and F4V mutants significantly increased relative to cells with amplified E6 null mutant when the same mutations were expressed in trans. However, the frequencies of viral DNA-positive cells remained reduced relative to cultures harboring the WT virus. Very few nuclei were positive for both amplified viral DNA and stabilized p53. Real-time qPCR from five independent experiments showed that F4L, F4V, and YYH had a copy number, on average, about 50%, 35%, and 10% of the WT genome, respectively (Fig. 3). These experiments were conducted in parallel with the trans complementation assays described above. This side-by-side comparison allowed us to evaluate the two assay systems directly without experimental variability introduced by using different batches of PHKs or the WT plasmid DNA. We conclude that E6 F4L and F4V mutations in cis support viral DNA amplification significantly better than when expressed in trans. Immunohistochemistry revealed that only the F4L mutant exhibited L1 capsid protein signal, which was only slightly reduced relative to the WT genome (Fig. 1).
Partial Rescue of the E6 Null Mutant by p53 siRNA.
The above observations strongly suggest a correlation between elevated p53 and inhibition of viral DNA amplification. We then performed p53 knockdown experiments. A retrovirus expressing p53 siRNA (28) was transduced into PHKs that harbored the E6 null genome. p53 was below detection in situ. In three independent experiments, differentiated cells containing amplified viral DNA increased (Fig. 2B, Left) but remained fewer than the number of cells observed with the WT virus (Fig. 2A). To investigate the possibility that the E6 null mutant genome was lost in some PHKs during p53 siRNA retrovirus transduction, we examined the E7-induced proliferating cell nuclear antigen (PCNA). PCNA was diffusely induced throughout the epithelium (Fig. 2B, Center). Raft cultures harboring the E6 null genome and transduced with empty retroviral vector were also positive for PCNA, p53, or both in numerous cells (Fig. 2B, Right). Thus, E6 null mutant DNA amplification took place only in a reduced fraction of spinous cells relative to the raft cultures containing WT virus, despite an effective p53 knockdown by p53 siRNA. These results confirm a role of p53 and also implicate additional factors in controlling viral DNA amplification.
Modulation of Viral DNA Amplification by E6 and p53.
To confirm that p53 accumulation is attributable to a deficiency of the mutant forms of E6 to destabilize p53, Western blots were conducted to detect viral and host proteins in the same lysates. The F4L and F4V mutant genomes had slightly reduced levels of E6 protein relative to E6 protein from the WT E6, whereas the YYH showed very weak or no E6 signal (Fig. 4B). The E6 null mutant was negative for E6 as expected. p53 protein levels were higher in cultures harboring E6 mutant genomes than p53 in cultures with the WT genome. E6 null mutant had the highest p53 followed by the YYH mutant. Thus, the levels of p53 protein are inversely correlated with the levels of E6 protein. Unexpectedly, ectopic WT and all three mutant forms of the E6 protein in E6 null-containing raft cultures were significantly higher than the corresponding E6 proteins expressed in cis. Interestingly, elevated WT E6 expressed in trans did not abolish the residual p53. Despite the very high levels of ectopic mutant forms of E6, the p53 protein levels remained highly elevated relative to p53 present in cultures expressing the WT E6 (Figs. 2A and 4B). These results show that the mutant forms of E6 are deficient in destabilizing p53. In addition, the level of E6 YYH protein remained significantly reduced relative to WT E6, E6 F4L, or E6 F4V.
To examine why elevated levels of the WT or mutant forms of E6 protein in trans led to reduced E6 null amplification relative to E6 WT or mutations expressed in cis, we examined whether retrovirus infection had a negative effect on viral DNA amplification. Retroviral transduction before viral DNA transfection necessitated one additional passage of PHKs relative to transfection of mutant genomes. In two experiments, the use of passage 2 PHKs (PHK2) for transfection of the WT viral plasmid slightly reduced the virus copy number relative to passage 1 PHKs (PHK1), an effect similar to introduction of an empty retrovirus into PHK1. When WT E6 was ectopically expressed, the copy number of the WT virus was significantly reduced relative to the copy number in PHK1 (Table S2). These observations support the notion that overexpression of E6 is detrimental to viral DNA amplification. Retrovirus transduction per se does not have a negative effect other than increasing the PHK passage number.
Late Viral Transcripts.
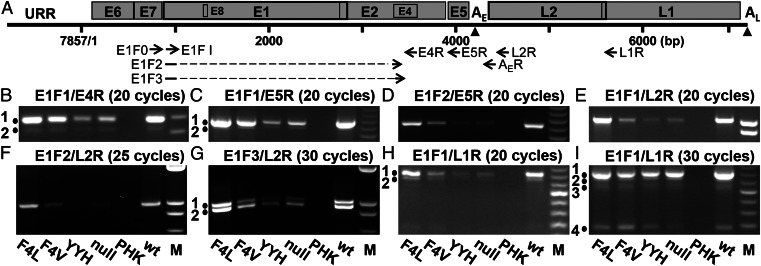
HPV mRNA splicing patterns are conserved among different HPVs (review in ref. 29). To investigate the reasons for the scarce late protein expression by E6 mutants, we compared viral RNA levels by using semi quantitative RT-PCR followed by agarose gel electrophoresis (Fig. 5). The locations of PCR primers are indicated in Fig. 5A, and their sequences are presented in Table S3. Major and minor bands obtained from the WT or F4L mutant viruses were directly sequenced to establish their identities. The results are summarized in Table 1 and elaborated below. All together, we identified five previously undescribed spliced transcripts in addition to known transcripts. With only one exception (Fig. 5I, band 4 and Table 1, species j), the relative abundance for each of the RNA species is WT and F4L >> F4V > YYH and E6 null, correlating with the levels of viral DNA amplification. β-actin cDNA was detected as an internal control (Fig. S4A).
Fig. 5.
RT-PCR detection of HPV-18 viral transcripts in HPV-18 WT or E6 mutants. (A) The circular HPV-18 genome of 7,857 bp was linearized in the URR. Triangles denote early (AE) and late (AL) polyadenylation sites. ORFs are represented by open boxes, with primers for RT-PCR indicated below. E1F2 and E1F3 primers spanned the splice junctions nucleotides 929^3465 and 929^3506, respectively. Primer sequences are shown in Table S3. (B–I) RT-PCR reactions were conducted on total RNAs extracted from the raft cultures containing HPV-18 WT or E6 mutant genomes as described in Materials and Methods. Primer pairs and PCR cycles are indicated above each panel. Primer annealing was conducted at 58 °C, except for E1F3/L2R (G) at 65 °C. The samples in each panel are indicted at the bottom. Length markers in B–D, H, and I are comprised of 1 µg 50-bp DNA ladders (Invitrogen); 1 µg 1-kb plus ladders (Invitrogen) was used in E–G. Gel images were captured using different exposures to visualize minor bands. The lengths of the PCR products are (B) band 1, 190 bp; band 2, 159 bp; (C) band 1, 584 bp; band 2, 553 bp; (D) 551 bp; (E) 1,005 bp; (F) 972 bp; (G) band 1, 1,003 bp; band 2, 931 bp. The intensity of band 1 in (G) increased relative to band 2 if the primer annealing temperature was decreased from 65 °C to 58 °C. (H) Band 1, 450 bp; band 2, 419 bp; (I) bands 1–4, 450, 419, 378, and 187 bp.
Table 1.
Splice species, cDNA lengths, and inferred ORFs
| Species | Figure panel and band (cDNA length; bp) | Splice(s) | Inferred ORFs |
| a | Fig. 5B, band 1 and Fig. S4C, band 3 (190) | 929^3434 | E1^E4, E5 |
| a | Fig. 5C, band 1 (584) | 929^3434 | E1^E4, E5 |
| b | Fig. 5B, band 2 and Fig. S4C, band 4 (159) | 929^3465 | E5 |
| b | Fig. 5C, band 2 (553) | 929^3465 | E5 |
| b | Fig. 5D (551) | 929^3465 | E5 |
| C | Fig. S4C, band 5 (118) | 929^3506 | E1^E4s, E5 |
| d | Fig. 5E (1,005) | 929^3434 | E1^E4, E5, L2, L1 |
| E | Fig. 5F (972) | 929^3465 | E5, L2, L1 |
| F | Fig. 5G, band 2 (931) | 929^3506 | E1^E4s, E5, L2, L1 |
| g | Fig. 5 H, band 1 and I, band 1 (450) | 929^3434 to 3696^5613 | E1^E4, L1 |
| H | Fig. 5 H, band 2 and I, band 2 (419) | 929^3465 to 3696^5613 | L1 |
| I | Fig. 5I, band 3 (378) | 929^3506 to 3696^5613 | E1^E4s, L1 |
| j | Fig. 5I, band 4 (187) | 929^5613 | L1? |
RT-PCR amplification was performed on total RNA isolated from day 13 raft cultures containing HPV-18 WT or E6 mutant genomes. cDNA species and length are presented in Fig. 5 and Fig. S4C. Species not previously described are labeled with bold capital letters. Species a–C use the early poly(A) site (AE), which was revealed by experiments presented in Fig. S4C. Species d–j are presumed to use the late poly(A) site (AL) by virtue of the L-region sequences.
L1 transcripts.
Using primer set E1F1/L1R (Fig. 5A), four bands were detected. The major L1 ORF-containing band 1 (Fig. 5 H and I and Table 1, species g) had two splices from nucleotide 929 to nucleotide 3434 and then from nucleotide 3696 to nucleotide 5613 at the L1 initiation codon. It is identical to a previous report (30) and predicted to encode both E1^E4 and L1. Band 2 (Fig. 5 H and I and Table 1, species H) had two splices (nucleotides 929^3465 to 3696^5613), connecting the short E1 ORF to a closed frame within the E2/E4 overlap region; it could encode L1 only (30). An early E5 RNA with the same or comparable first splice only has been reported for HPV-18 and -11 (30, 31). A twice-spliced band 3 had a first splice (nucleotides 929^3506) not previously reported (Fig. 5I and Table 1, species I); it could encode an internally deleted, shortened E1^E4 protein (designated as E1^E4s) and the L1 protein. Band 4 (Fig. 5I) had one splice (nucleotides 929^5613) (Table 1, species j) as reported (30). This splice brings the short E1 exon into a closed frame. Unless the ribosome skips the strong E1 initiation codon, this RNA species cannot encode a full-length L1 protein (hence, it is designated as L1?).
L2 transcripts.
With primer set E1F1/L2R for PCR, a single product of 1,005 bp was obtained (Fig. 5E); it had a single splice of nucleotides 929^3434 (Table 1, species d). Because of the long cDNA fragment, it was not possible to discern minor species of similar lengths. We, thus, used the L2R primer in conjunction with forward primers E1F2 or E1F3 (Table S3), which spans the splice junctions just described. E1F2/L2R produced one band of 972 bp (Fig. 5F) with a splice at nucleotides 929^3465; it has the potential to encode E5, L2, and L1 (Table 1, species E). Control experiments showed that this product is not an artifact caused by the splice junction primer (Fig. S4B). E1F3/L2R generated two bands, each with one splice (Fig. 5G). The more prominent cDNA fragment of 931 bp (band 2) had the splice (nucleotides 929^3506) and would encode E1^E4s, E5, L2, and L1 (Table 1, species F). The less prominent band 1 of 1,003 bp was identical to species d and attributable to mispriming on the abundant species d cDNA, which has sequence homology with species F at the splice acceptor.
Early Viral Transcripts.
We reexamined the early region transcripts to determine whether the splice, nucleotides 929^3506, in the E1^E4s RNA could also be found. Using E1F1/E4R or E1F1/E5R primer set, a major band and a minor band (Fig. 5 B and C, bands 1 and 2) were detected; they correspond to the E1^E4, E5 transcript with a splice at nucleotides 929^3434 (Table 1, species a) and the E5 transcript with the splice at nucleotides 929^3465 (Table 1, species b), respectively (29, 30). A product with the splice nucleotides 929^3506 was not observed. For a better assessment of the relative abundance of species b among WT and mutants, we performed PCR using the splice junction primer E1F2 and E5R primer set. The WT and F4L mutant had the highest signal, whereas the other mutants had much weaker signals (Fig. 5D and Table 1, species b). We then verified that species a and b were primarily derived from early RNA rather than the late RNAs just described. RT-PCR was performed using E1F0 coupled with AER, an oligo dT ending with several adjacent viral-encoded bases (30), followed by reamplification with E1F1/E4R. We recovered primarily species a and b. The E1^E4s, E5 cDNA (species C) was also recovered but only on additional rounds of PCR, indicative of its rarity (Fig. S4C).
Discussion
The use of Cre-loxP recombination to generate HPV-18 genomic plasmids in vivo from a vector in acutely transfected PHKs allows the recapitulation of a robust HPV-18 productive program in organotypic raft cultures. Because selection of transfected cells does not require the immortalization functions of the HR HPV oncogenes, we were able to perform genetic analyses of the E6 mutants not previously possible. We showed that p53 accumulated to high levels in raft cultures harboring an E6 null mutant. The mutant was unable to amplify efficiently but was partially complemented in trans by ectopic WT E6 (Figs. 1, 2, and 3 and Figs. S1 and S2). This ability forms the basis for rapid screening of E6 mutations in trans to assess their impact on virus DNA amplification. We successfully tested three HPV-18 E6 missense mutations, F4L, F4V, and F47,49Y,D51H (YYH), selected according to the deficiencies of the corresponding HPV-16 E6 in destabilizing p53 in vitro or in vivo. Indeed, p53 accumulated in numerous cells when the E6 mutations were ectopically expressed (Fig. 2). E6 F4L or F4V increased E6 null mutant copy number significantly less efficiently than the WT E6, whereas the E6 YYH was largely inactive (Fig. 3). Unexpectedly, when expressed in cis from mutant genomes, F4L and F4V were able to amplify in a fraction of spinous cells, in which p53 was below detection (Fig. 2A). Real-time qPCR revealed that F4L was least defective among the mutants and achieved a DNA copy number about 50% of the WT genome. YYH was most defective, achieving no better than 10% of the WT (Fig. 3). Nevertheless, relative to the E6 null mutant, the YYH mutant was able to induce a more hyperplastic histology (Fig. S1) and slightly higher viral copy number (Fig. 3), consistent with the increase in suprabasal S-phase cells (Table S1). The HPV-31 E6 YYH mutant was previously reported to replicate weakly in transient replication, but it was not assessed in raft cultures (20).
The expression of E7 stabilized p53 (17–19), which is attributable to E7 activation of DNA damage response kinases that not only phosphorylate p53 but also cause a prolonged G2 phase (32), during which the viral DNA amplifies (22). HPV missense E6 mutants destabilized the accumulated p53 much less effectively than the WT E6 (Fig. 4B) and induced fewer suprabasal S-phase cells (Fig. S2 and Table S1); consequently, they supported lower viral DNA amplification (Fig. 3). Our data showed that the accumulated p53 was not transcriptionally active (Fig. S3), and the levels of p21cip1 protein did not correlate with the reduction in S-phase reentry. Rather, the levels of E7 protein seemed to correlate with the number of suprabasal BrdU-positive cells (Fig. 4A, Fig. S2, and Table S1) and viral DNA amplification (Figs. 2 and 3). However, the differences in E7 protein levels could, in part, have been influenced by the viral DNA copy numbers. It is also possible that E6 normally conditions the cells to increase E7 protein expression or stability but the mutant forms of E6 are deficient in these activities. However, this hypothetical E6 activity alone cannot explain why the high levels of ectopic WT E6 were only able to rescue the E6 null mutant partially (Fig. 3). In this regard, it is interesting to note our unexpected observation: overexpression of WT E6 protein (and presumably, mutant forms of E6) was inhibitory to high levels of viral DNA amplification (Figs. 3 and 4B). The negative effect might be attributable to an imbalance between E6 and E7 proteins (33). Moreover, a threshold level of the E6-targeted host proteins could be necessary for optimal viral DNA amplification. This hypothesis may explain why a significant fraction of the E6 transcripts are spliced. Intra- and intergenic E6 mRNA splicing not only facilitates the expression of downstream genes (34), but it also reduces the amount of full-length E6 protein, thereby maintaining optimal levels of host targets. The coordination between E6 and E7 proteins during the viral lifecycle remains to be investigated.
The extent of mutant DNA amplification was inversely correlated with the p53 levels, and amplification was observed in cells in which p53 was below detection (Figs. 2, 3, and 4). These observations implicate p53 in negatively controlling viral DNA amplification. Interestingly, all small DNA tumor viruses encode proteins to target p53, perhaps to alleviate its inhibitory effect to viral DNA amplification. For HPV, p53 has been proposed to inhibit viral DNA amplification by interaction with the E2 origin binding protein (15, 16). However, this latter interpretation alone cannot account for our observation in that, despite an effective knockdown of p53 protein with siRNA (Fig. 2), viral DNA amplification was not restored to the levels of the WT virus. Thus, there could be additional E6-targeted host proteins that modulate viral DNA amplification. This conclusion is consistent with the observation that the YYH mutant, which expressed little or no E6 protein, is more defective than E6 F4L and E6 F4V mutants that expressed reduced levels of E6 protein, although all three were deficient in destabilizing p53.
In the context of the whole HPV genome, E6 coordinates with E7 in supporting HPV genome maintenance in transfected cells (33). Under our experimental conditions, the E6/E7 coordination seems to increase S-phase reentry and viral DNA amplification but is not critical for genome persistence. A possible explanation for the role of E6 is its ability to activate mammalian target of rapamycin (mTOR). In submerged cultures of human keratinocytes, HPV-16 E6 activates protein kinase B (PKB)/AKT, leading to elevated mTOR activity (35). Another study reported that HPV-16 E6 destabilizes tuberous sclerosis complex 2 (36), a negative regulator of the mTOR pathway. mTOR up-regulates cap-dependent protein translation, linking cell growth to cell cycle entry (37). Elevated mTOR activity has previously been implicated in replication of adenoviruses (38). Interestingly, E7 alone also up-regulates activating phosphorylation of PKB/AKT (39, 40). It will be interesting to examine the role of mTOR in the viral productive lifecycle.
By using RT coupled to unsaturated cycles of PCR, our studies provide approximate relative abundances among different RNA species for the WT and E6 mutant viruses. For instance, compared with the E1^E4 message (Table 1, species a), the E5 transcript (Table 1, species b) is relatively minor. This finding is in accordance with our understanding that E5 is a signal transduction regulatory protein (41), whereas E1^E4 is an abundant cytoplasmic protein that interacts with cytokeratins (42). Interestingly, the E6 F4L mutant and the WT virus were virtually indistinguishable in abundance of each of the RNA species, whereas the other mutants had significantly reduced RNA levels (Fig. 5). Our results show that late RNAs and proteins are expressed to high levels only when viral DNA has amplified to certain copy numbers achieved by WT and the E6 F4L mutant. However, E6 may also play additional roles in late RNA transcription or processing. For instance, the E6 F4V mutant genome amplified to levels similar to the levels achieved by E6 null trans complemented by WT E6 (Fig. 3), but it had sparse L1 signals in repeated experiments (Fig. 1). We also note that E6 F4V had the highest relative abundance of RNA species j (Table 1), which may not be able to encode the L1 protein.
Although not meant to be exhaustive in our characterization of the viral transcripts, we nevertheless identified five minor transcripts, resulting from alternative splicing to acceptor sites not previously reported (Table 1). The most significant species E (Table 1), with splice nucleotides 929^3465, is predicted to encode E5, L2, and L1. This late E5 coding species is less abundant than the early E5 RNA (Table 1, species b). Interestingly, in productive lesions, bovine papillomavirus type 1 E5 protein is detected in both the lower strata and the superficial strata, where late protein is expressed (43). Until now, L2 mRNA has never been satisfactorily identified. The relatively low abundance of this RNA is in accordance with L2 being a minor capsid protein. In this regard, we hypothesize that the more abundant singly spliced transcript (nucleotides 929^3434), which spans E1^E4, E5, and the entire L region (29, 30), is not a message for L2, because L2 would be the third ORF, an unfavorable position for protein translation. We suggest that this long transcript is the splicing intermediate for the twice-spliced (nucleotides 929^3434 to 3696^5613) mature bicistronic mRNA encoding E1^E4 and L1 (Table 1, species g). Another interesting species I is twice spliced (nucleotides 929^3506 to 3696^5613) to encode E1^E4s and L1. E1^E4s has an internal deletion of 24 aa entirely within the E4 ORF. An amino–terminus truncated form of the E4 protein has been attributed to calpain cleavage (42). The splice site in the E1^E4s RNA is located just downstream of the calpain cleavage site. Thus, E1^E4s may have properties similar to the N terminus-truncated E4 peptide in inducing E4 amyloid filaments, which are thought to facilitate virus release (42). The E1^E4s transcript is primarily a late RNA, consistent with the proposed functions of E4. This RNA provides a different mechanism to produce this shortened E1^E4s protein. The singly spliced species F (nucleotides 929^3506) that spans ORFs E1^E4s, E5, L2, and L1 would then represent the splicing intermediate of species I just described.
In summary, the ability to express HPV E6 mutations in trans and in cis allows us to investigate papillomavirus–host cell interactions in the context of differentiating squamous epithelia developed from PHKs. Trans complementation of a null mutant more readily revealed the deficiencies of the E6 mutations than direct assays of the mutant genomes, because overexpression of E6 protein in trans is inhibitory, and hence, it accentuated the defects of the mutations. Our studies reveal that the p53 protein stabilized by E7 is transcriptionally inactive, but it and additional host proteins are important E6 targets to enable high levels of viral DNA amplification.
Materials and Methods
Plasmid Construction.
In the E6 null mutation, the initiation and only AUG codon in the E6 ORF were mutated (nucleotide substitution T106A), and missense mutations E6 F4L (T116G), F4V (T114G and T116A) and F47Y, F49Y, and D51H (abbreviated as YYH; T244A, T250A, and G255C) were generated by PCR mutagenesis. Each of the fragments spanning HPV-18 URR E6 (nucleotides 6929 to 7857/1 to 581) containing the WT sequence or mutation was cloned into the retroviral vector pBabe Puro (44). pNeo-loxP vectors harboring the corresponding HPV-18 E6 mutant genome were similarly constructed as described (22).
Retrovirus Transduction and DNA Transfection into Primary Human Keratinocytes and Development of Organotypic Raft Cultures.
PHKs isolated from neonatal foreskins after elective circumcision were obtained at the University of Alabama at Birmingham Hospital and grown in keratinocyte serum-free medium (Invitrogen). DNA transfection with pNeo-loxP HPV-18 WT or mutant together with pCAGGS-nlsCre expression plasmid (45) and establishment of raft cultures were conducted as described (22). pBabe Puro retroviruses were prepared as described (46). For trans complementation, PHKs were transduced with various retroviruses and selected with puromycin (Sigma) for 2 d before cotransfection with DNA plasmids carrying the neomycin (G418) -resistance gene. PHKs were then selected with G418 for 4 d to obtain transfected cells for raft cultures. For p53 knockdown, the HPV-18 E6 null mutant-containing plasmid was first transfected into PHKs, and the G418-selected PHKs were then transduced with the pBabe Puro retroviruses, in which the U6 promoter drives the expression of a p53 siRNA (28). A pBabe Puro vector containing the U6 promoter without siRNA was used to produce empty control retroviruses. For the final 12 h before harvest, all raft cultures for in situ assays were exposed to media containing 100 μg/mL BrdU (Sigma) for incorporation into replicating cellular DNA. The raft cultures were harvested by fixation in 10% (wt/vol) buffered formalin (Fisher Scientific) and then embedded in paraffin. Additional sets of raft cultures were processed for biochemical analyses without fixation.
In Situ Analyses.
Four-micrometer tissue sections were cut and deparaffinized. Day 16 raft cultures were used for evaluating L1 expression and p53 siRNA experiments, whereas all other experiments were performed with day 13 raft cultures. In situ analyses were performed as described (22, 47). Primary antibodies were L1 (K1H8, 1:100 dilution; Dako Cytomation), PCNA (PC-10, 1:1,000 dilution; Dako Cytomation), BrdU (1:100 dilution; Abcam or ZBU 30, 1:100 dilution; Invitrogen), and p53 (DO7, 1:50 dilution; Novocastra).
Western Blotting.
The epithelial portions of day 13 raft cultures were harvested and sonicated in mammalian cell lysis buffer supplemented with protease inhibitor mixture (Roche) as described (32); 200 µg lysates were Western-blotted. p53 antibody (DO7, 1:200 dilution; Novocastra), E6 antibody (2 µg/mL; Arbor Vita Corp), E7 antibody (N-19, 1:500 dilution; Santa Cruz), p21 antibody (1:1,000 dilution; Abcam), and β-actin antibody (mAbcam 8226, 1:7,000 dilution; Abcam) were used to probe the membranes.
Real-Time qPCR to Determine Viral DNA Copy Numbers.
Except for one set of raft cultures, which was from day 16 (Table S2), all others were from day 13 cultures. The epithelia were minced, lysed in buffer (50 mM Tris⋅HCl, pH 8.0, 100 mM EDTA, 100 mM NaCl, 0.5% SDS), homogenized, and then digested with Proteinase K (Sigma); 50 ng extracted DNA were analyzed by real-time qPCR as described (22). To determine the DNA copy numbers of G418-selected PHKs before raft culture development, 1 µg total DNA was digested by DpnI (New England Biolabs) at 37 °C overnight; 50 ng of the digested DNA were used for real-time qPCR using the qPCR primers that bracket DpnI sites in HPV-18 DNA (Table S3).
Viral Transcript Analyses by RT-PCR.
The epithelia of day 13 raft cultures were minced, mixed with TRIzol (Invitrogen), and homogenized on ice. Extracted RNA was treated with DNA-Free (Ambion); 5 µg total RNA were reverse-transcribed with oligo dT primers using SuperScript III (Invitrogen) in 40 µL reaction volume, and 1 µL RT product was then PCR-amplified in 35 µL reaction volume using primer sets and primer annealing conditions as described in Fig. 5 and Table S3. Fifteen microliters PCR product were loaded into agarose gels for electrophoretic separation and then stained with ethidium bromide. Bands from WT virus and the E6F4L mutant were extracted for direct sequence determination.
Real-Time RT-qPCR to Determine p21cip mRNA.
cDNA from day 13 raft cultures was prepared using random hexamers or oligo dT primers (5 µg total RNA in 50 µL reaction); 1 µL 1:20 diluted cDNA was used for real-time qPCR reaction using primers for p21cip1 (48) or human β-actin (forward: 5′-AATCTGGCACCACACCTTCTAC-3′; reverse: 5′-ATAGCACAGCCTGGATAGCAAC-3′). The results were normalized against β-actin, and the various E6 mutants were compared with WT HPV-18 (set as 100%).
Supplementary Material
Acknowledgments
We thank Dr. Xinbin Chen (University of California, Davis, CA) for the retroviral vector of p53 siRNA. We also thank Dr. Carolyn Ashworth and the nurses in the University of Alabama at Birmingham Well-Baby Nursery for collection of neonatal foreskins from elective circumcision with informed consent of the parents. We thank Arbor Vita for supplying the antibody to HPV-18 E6. We also thank Dr. Clinton Grubbs of the University of Alabama at Birmingham Surgery Department and Dr. Erik Daniel Dohm in the University of Alabama at Birmingham Animal Resources Program for providing rat tails for isolation of collagen. DNA sequencing was performed by the University of Alabama at Birmingham Heflin Center for Genomic Sciences. The research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award CA 83679.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304855110/-/DCSupplemental.
References
- 1.de Villiers E-M, Fauquet C, Broker TR, Bernard H-U, zur Hausen H. Classification of papillomaviruses. Virology. 2004;324(1):17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 2.zur Hausen H. Papillomaviruses in the causation of human cancers—a brief historical account. Virology. 2009;384(2):260–265. doi: 10.1016/j.virol.2008.11.046. [DOI] [PubMed] [Google Scholar]
- 3.Chow LT, Broker TR, Steinberg BM. The natural history of human papillomavirus infections of the mucosal epithelia. APMIS. 2010;118(6–7):422–449. doi: 10.1111/j.1600-0463.2010.02625.x. [DOI] [PubMed] [Google Scholar]
- 4.Zhang B, Chen W, Roman A. The E7 proteins of low- and high-risk human papillomaviruses share the ability to target the pRB family member p130 for degradation. Proc Natl Acad Sci USA. 2006;103(2):437–442. doi: 10.1073/pnas.0510012103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Genovese NJ, Banerjee NS, Broker TR, Chow LT. Casein kinase II motif-dependent phosphorylation of human papillomavirus E7 protein promotes p130 degradation and S-phase induction in differentiated human keratinocytes. J Virol. 2008;82(10):4862–4873. doi: 10.1128/JVI.01202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McLaughlin-Drubin ME, Münger K. The human papillomavirus E7 oncoprotein. Virology. 2009;384(2):335–344. doi: 10.1016/j.virol.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimmermann H, Degenkolbe R, Bernard H-U, O’Connor MJ. The human papillomavirus type 16 E6 oncoprotein can down-regulate p53 activity by targeting the transcriptional coactivator CBP/p300. J Virol. 1999;73(8):6209–6219. doi: 10.1128/jvi.73.8.6209-6219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar A, et al. Human papillomavirus oncoprotein E6 inactivates the transcriptional coactivator human ADA3. Mol Cell Biol. 2002;22(16):5801–5812. doi: 10.1128/MCB.22.16.5801-5812.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas MC, Chiang CM. E6 oncoprotein represses p53-dependent gene activation via inhibition of protein acetylation independently of inducing p53 degradation. Mol Cell. 2005;17(2):251–264. doi: 10.1016/j.molcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 10.Patel D, Huang SM, Baglia LA, McCance DJ. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 1999;18(18):5061–5072. doi: 10.1093/emboj/18.18.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137(4):609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howie HL, Katzenellenbogen RA, Galloway DA. Papillomavirus E6 proteins. Virology. 2009;384(2):324–334. doi: 10.1016/j.virol.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pim D, Tomaic V, Banks L. The human papillomavirus (HPV) E6* proteins from high-risk, mucosal HPVs can direct degradation of cellular proteins in the absence of full-length E6 protein. J Virol. 2009;83(19):9863–9874. doi: 10.1128/JVI.00539-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lepik D, Ilves I, Kristjuhan A, Maimets T, Ustav M. p53 protein is a suppressor of papillomavirus DNA amplificational replication. J Virol. 1998;72(8):6822–6831. doi: 10.1128/jvi.72.8.6822-6831.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Massimi P, Pim D, Bertoli C, Bouvard V, Banks L. Interaction between the HPV-16 E2 transcriptional activator and p53. Oncogene. 1999;18(54):7748–7754. doi: 10.1038/sj.onc.1203208. [DOI] [PubMed] [Google Scholar]
- 16.Brown C, Kowalczyk AM, Taylor ER, Morgan IM, Gaston K. P53 represses human papillomavirus type 16 DNA replication via the viral E2 protein. Virol J. 2008;5:5. doi: 10.1186/1743-422X-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jian Y, et al. Post-transcriptional induction of p21cip1 protein by human papillomavirus E7 inhibits unscheduled DNA synthesis reactivated in differentiated keratinocytes. Oncogene. 1998;17(16):2027–2038. doi: 10.1038/sj.onc.1202142. [DOI] [PubMed] [Google Scholar]
- 18.Demers GW, Halbert CL, Galloway DA. Elevated wild-type p53 protein levels in human epithelial cell lines immortalized by the human papillomavirus type 16 E7 gene. Virology. 1994;198(1):169–174. doi: 10.1006/viro.1994.1019. [DOI] [PubMed] [Google Scholar]
- 19.Eichten A, Westfall M, Pietenpol JA, Münger K. Stabilization and functional impairment of the tumor suppressor p53 by the human papillomavirus type 16 E7 oncoprotein. Virology. 2002;295(1):74–85. doi: 10.1006/viro.2002.1375. [DOI] [PubMed] [Google Scholar]
- 20.Thomas JT, Hubert WG, Ruesch MN, Laimins LA. Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc Natl Acad Sci USA. 1999;96(15):8449–8454. doi: 10.1073/pnas.96.15.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee C, Wooldridge TR, Laimins LA. Analysis of the roles of E6 binding to E6TP1 and nuclear localization in the human papillomavirus type 31 life cycle. Virology. 2007;358(1):201–210. doi: 10.1016/j.virol.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 22.Wang HK, Duffy AA, Broker TR, Chow LT. Robust production and passaging of infectious HPV in squamous epithelium of primary human keratinocytes. Genes Dev. 2009;23(2):181–194. doi: 10.1101/gad.1735109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pim D, Banks L. HPV-18 E6*I protein modulates the E6-directed degradation of p53 by binding to full-length HPV-18 E6. Oncogene. 1999;18(52):7403–7408. doi: 10.1038/sj.onc.1203134. [DOI] [PubMed] [Google Scholar]
- 24.Crook T, Tidy JA, Vousden KH. Degradation of p53 can be targeted by HPV E6 sequences distinct from those required for p53 binding and trans-activation. Cell. 1991;67(3):547–556. doi: 10.1016/0092-8674(91)90529-8. [DOI] [PubMed] [Google Scholar]
- 25.Foster SA, Demers GW, Etscheid BG, Galloway DA. The ability of human papillomavirus E6 proteins to target p53 for degradation in vivo correlates with their ability to abrogate actinomycin D-induced growth arrest. J Virol. 1994;68(9):5698–5705. doi: 10.1128/jvi.68.9.5698-5705.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, et al. Multiple functions of human papillomavirus type 16 E6 contribute to the immortalization of mammary epithelial cells. J Virol. 1999;73(9):7297–7307. doi: 10.1128/jvi.73.9.7297-7307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noya F, Chien WM, Broker TR, Chow LT. p21cip1 Degradation in differentiated keratinocytes is abrogated by costabilization with cyclin E induced by human papillomavirus E7. J Virol. 2001;75(13):6121–6134. doi: 10.1128/JVI.75.13.6121-6134.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan W, Chen X. GPX2, a direct target of p63, inhibits oxidative stress-induced apoptosis in a p53-dependent manner. J Biol Chem. 2006;281(12):7856–7862. doi: 10.1074/jbc.M512655200. [DOI] [PubMed] [Google Scholar]
- 29.Chow LT, Broker TR. Human papillomavirus RNA transcription. In: Garcea RL, DiMaio D, editors. The Papillomaviruses. New York: Springer; 2006. pp. 109–144. [Google Scholar]
- 30.Wang X, Meyers C, Wang HK, Chow LT, Zheng ZM. Construction of a full transcription map of human papillomavirus type 18 during productive viral infection. J Virol. 2011;85(16):8080–8092. doi: 10.1128/JVI.00670-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiang CM, Broker TR, Chow LT. An E1M—E2C fusion protein encoded by human papillomavirus type 11 is asequence-specific transcription repressor. J Virol. 1991;65(6):3317–3329. doi: 10.1128/jvi.65.6.3317-3329.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banerjee NS, Wang HK, Broker TR, Chow LT. Human papillomavirus (HPV) E7 induces prolonged G2 following S phase reentry in differentiated human keratinocytes. J Biol Chem. 2011;286(17):15473–15482. doi: 10.1074/jbc.M110.197574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park RB, Androphy EJ. Genetic analysis of high-risk e6 in episomal maintenance of human papillomavirus genomes in primary human keratinocytes. J Virol. 2002;76(22):11359–11364. doi: 10.1128/JVI.76.22.11359-11364.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hubert WG, Laimins LA. Human papillomavirus type 31 replication modes during the early phases of the viral life cycle depend on transcriptional and posttranscriptional regulation of E1 and E2 expression. J Virol. 2002;76(5):2263–2273. doi: 10.1128/jvi.76.5.2263-2273.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spangle JM, Münger K. The human papillomavirus type 16 E6 oncoprotein activates mTORC1 signaling and increases protein synthesis. J Virol. 2010;84(18):9398–9407. doi: 10.1128/JVI.00974-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu Z, et al. Human papillomavirus 16 E6 oncoprotein interferences with insulin signaling pathway by binding to tuberin. J Biol Chem. 2004;279(34):35664–35670. doi: 10.1074/jbc.M403385200. [DOI] [PubMed] [Google Scholar]
- 37.Shahbazian D, Parsyan A, Petroulakis E, Hershey J, Sonenberg N. eIF4B controls survival and proliferation and is regulated by proto-oncogenic signaling pathways. Cell Cycle. 2010;9(20):4106–4109. doi: 10.4161/cc.9.20.13630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Shea C, et al. Adenoviral proteins mimic nutrient/growth signals to activate the mTOR pathway for viral replication. EMBO J. 2005;24(6):1211–1221. doi: 10.1038/sj.emboj.7600597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pim D, Massimi P, Dilworth SM, Banks L. Activation of the protein kinase B pathway by the HPV-16 E7 oncoprotein occurs through a mechanism involving interaction with PP2A. Oncogene. 2005;24(53):7830–7838. doi: 10.1038/sj.onc.1208935. [DOI] [PubMed] [Google Scholar]
- 40.Menges CW, Baglia LA, Lapoint R, McCance DJ. Human papillomavirus type 16 E7 up-regulates AKT activity through the retinoblastoma protein. Cancer Res. 2006;66(11):5555–5559. doi: 10.1158/0008-5472.CAN-06-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suprynowicz FA, et al. The human papillomavirus type 16 E5 oncoprotein inhibits epidermal growth factor trafficking independently of endosome acidification. J Virol. 2010;84(20):10619–10629. doi: 10.1128/JVI.00831-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khan J, et al. Role of calpain in the formation of human papillomavirus type 16 E1^E4 amyloid fibers and reorganization of the keratin network. J Virol. 2011;85(19):9984–9997. doi: 10.1128/JVI.02158-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burnett S, Jareborg N, DiMaio D. Localization of bovine papillomavirus type 1 E5 protein to transformed basal keratinocytes and permissive differentiated cells in fibropapilloma tissue. Proc Natl Acad Sci USA. 1992;89(12):5665–5669. doi: 10.1073/pnas.89.12.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morgenstern JP, Land H. Advanced mammalian gene transfer: High titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18(12):3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hardouin N, Nagy A. Gene-trap-based target site for cre-mediated transgenic insertion. Genesis. 2000;26(4):245–252. doi: 10.1002/(sici)1526-968x(200004)26:4<245::aid-gene50>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 46.Banerjee NS, Chow LT, Broker TR. Retrovirus-mediated gene transfer to analyze HPV gene regulation and protein functions in organotypic “raft” cultures. Methods Mol Med. 2005;119:187–202. doi: 10.1385/1-59259-982-6:187. [DOI] [PubMed] [Google Scholar]
- 47.Van Tine BA, Broker TR, Chow LT. Simultaneous in situ detection of RNA, DNA, and protein using tyramide-coupled immunofluorescence. Methods Mol Biol. 2005;292:215–230. doi: 10.1385/1-59259-848-x:215. [DOI] [PubMed] [Google Scholar]
- 48.Jha S, et al. Destabilization of TIP60 by human papillomavirus E6 results in attenuation of TIP60-dependent transcriptional regulation and apoptotic pathway. Mol Cell. 2010;38(5):700–711. doi: 10.1016/j.molcel.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.