Abstract
This study examines the feasibility of using the adenoviral delivery of DNA for a non-native microRNA to suppress expression of a target protein (cytosolic NADP+-dependent malic-enzyme 1, ME1) in whole heart in vivo, via an isolated-heart coronary perfusion approach. Complementary DNA constructs for ME1 microRNA were inserted into adenoviral vectors. Viral gene transfer to neonatal rat cardiomyocytes yielded 65% suppression of ME1 protein. This viral package was delivered to rat hearts in vivo (Adv.miR_ME1, 1013 vp/ml PBS) via coronary perfusion, using a cardiac-specific isolation technique. ME1 mRNA was reduced by 73% at 2-6 days post-surgery in heart receiving the Adv.miR_ME1. Importantly, ME1 protein was reduced by 66% (p<0.0002) at 5-6 days relative to sham-operated control hearts. Non-target protein expression for GAPDH, calsequestrin, and mitochondrial malic enzyme, ME3, were all unchanged. The non-target isoform, ME2, was unchanged at 2-5 days and reduced at day 6. This new approach demonstrates for the first time significant and acute silencing of target RNA translation and protein content in whole heart, in vivo, via non-native microRNA expression.
Keywords: heart, RNA interference, gene therapy, microRNA, siRNA
INTRODUCTION
The recent decade has witnessed prolific advances in the methodological approaches for the overexpression of target proteins via viral-based cDNA delivery schemes to heart, in vivo [1,2]. With the recent developments in RNA interference technologies (RNAi), it is highly anticipated that these viral-based delivery approaches can be extended for the suppression of target protein expression in the heart [3]. The approach has been overwhelmingly successful in isolated cell culture models [4-6], including several studies for the suppression of malic enzyme as targeted in this study [7-9]. However, there are only a few reports for successful suppression of a target protein in heart, in vivo, based on RNAi schemes [10-14]. In those few studies, suppression was achieved via viral-based delivery of DNA code for the expression of short hairpin RNA (shRNA) [10,14], or direct injection of “naked” shRNA or short interfering RNA (siRNA) [11-13]. In this study, we will examine the potential of inhibiting protein expression, in vivo, based on an artificial microRNA scheme (miRNA), which is suggested to provide a higher level of RNA interference and less toxic side effects compared to shRNA [15,16].
Two types of small ribonucleic acid molecules are central to production of siRNA and RNA interference; the short hairpin RNA (shRNA) and microRNA (miRNA) [17-19]. The shRNA serve ideally as single-target silencing tools, whereas naturally occurring miRNA typically regulate multiple targets, and multiple miRNAs are able to target the same mRNA [20]. The miRNA structure is similar to shRNA, they include a 3′ and 5′ flanking region, and the miRNA utilize the common pathways of shRNA synthesis used to generate siRNA [17-20].
In this study, the feasibility of using viral-based delivery of DNA code for non-native miRNA to limit RNA translation in the heart was assessed. The target sequence of the miRNA construct was not designed to mimic an indigenous form of miRNA. Instead, the artificial miRNA construct was designed to include an inhibitory sequence targeted to a single protein, cytosolic NADP+ dependent malic-enzyme 1 (ME1). The DNA code for the siRNA sequence was annealed to a non-native micro-RNA template (miR_ME1), and packaged in an adenoviral vector (Adv_miR_ME1). Three separate miRNA sequences were examined, and the efficacy of inhibition was evaluated in isolated neonatal rat cardiomyocytes (see Supplemental Data). One package was selected and delivered to adult rat heart in vivo, via the open-chest isolated-heart retrograde perfusion technique as previously described by us [21-24]. Hearts were excised at 2-6 days post surgery, and the acute responses of protein and mRNA expression/suppression were assessed for target (ME1) and non-target proteins (ME2, ME3, calsequestrin, and GAPDH). Expression was compared among three separate control groups: untreated excised hearts, miR-scrambled treatment, and PBS treatment. Malic enzyme-1 was the target protein of interest based off of on-going studies in our laboratory which examine the regulatory role of ME1 on glucose oxidation and redox state in healthy and diseased heart models [25,26].
MATERIALS AND METHODS
Construction, Production, and Purification of Adenoviral Vectors
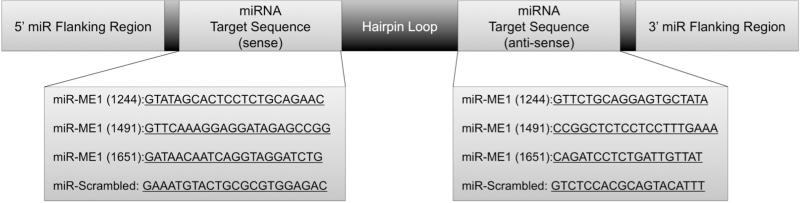
Three separate miR-RNA sequences were designed using BLOCK-iT RNAi Designer (Invitrogen, Carlsbad, California) to target rat cytoplasmic NADP(+) dependent malic enzyme mRNA (NM_012600.2). Each sequence was synthesized and annealed into Invitrogen's miR-RNA template containing an upstream / downstream adapter and hairpin loop. Figure 1 depicts the three sequences investigated. The final miR-RNA chosen for in vivo studies (#1244) included the following 22 nt siRNA sequence: GTATAGCACTCCTCTGCAGAAC. Each miR-RNA sequence was cloned into pcDNA6.2-GW/miR expression vector (Invitrogen) with a CMV promoter and verified by Invitrogen. Then each of the miR-RNA sequences were recombined into a pAd/CMV/V5-DEST Vector (Invitrogen). The adenoviral vector was transfected into HEK 293A cells using Lipofectamine (Invitrogen). The adenovirus was amplified in HEK 293 cells, harvested, and purified by cesium chloride density gradient centrifugation as previously described [21,22].
Figure 1.
The general structure of the miRNA sequences placed into the adenoviral vector include a 5′ and 3′ flanking region, hairpin loop, and the target sequence. Three separate target sequences were designed and examined in this study to target ME1 knockdown. Preliminary studies in isolated neonatal cardiomyocyte indicated sequence #1244 resulted in the greatest knockdown of ME1, and was selected for in vivo studies. A fourth sequence was designed as a scrambled control.
Vector Evaluation in Primary Cardiomyocytes
Malic enzyme-1 knockdown via the three vectors was first verified in isolated cardiomyocytes. Neonatal cardiomyocytes were isolated from 1-day-old Sprague-Dawley rats, as previously described [27]. In brief, cells were plated at a density of 240 cells/mm2 in MEM (Sigma-Aldrich, St. Louis, Missouri,) containing Hanks’ salts, 5% calf serum, vitamin B12, 1% Penicillin/Streptomycin/Amphotericin B, and 0.1mol/L bromodeoxyuridine at 37°C, in the presence of 5% CO2. After 24h, the medium was replaced with similar serum-free medium with 1% transferrin and 1% insulin. Cells were infected, with adenoviral vector (35 pfu/cell) containing one of the three miRRNA sequences. The control group was infected with an adenovirus containing DNA code for a scrambled miRRNA sequence. Cells were harvested in media 24h after viral exposure for western blot analysis of ME1 protein expression. A high MOI (35 pfu/cell) was selected in order to maximize the RNA interference process to within the timeframe of a viable cell culture preparation (ie., < 1 wk).
Vector Delivery to Rat Heart, In Vivo
Adenovirus or virus-free PBS solution was delivered to the heart in vivo by coronary perfusion as described in our previous reports [21,22]. In brief, 3 month old male Sprague-Dawley rats (350g) were anesthetized, intubated, and placed on an ice pad to cool the core body temperature to 30°C. The chest was opened at the second intercostal space. All vessels to/from the heart were cross-clamped simultaneously, and the heart was retrograde perfused in vivo for 7 min with calcium-free Tyrode solution through catheters position in the aortic root (delivery) and right ventricle (efflux). At the time of adenoviral injection, 0.2 ml of Adv.miR-ME1 or Adv.miR-scrambled (1013 vpu/ml in PBS) was first delivered through the catheter position in the aortic root. This allowed the adenovirus to circulate down the coronaries. Next, the efflux catheter positioned in the right ventricle was removed, and an additional 0.5 ml/kg of adenovirus (~0.2 ml) was delivered to the aortic root at 300 ± 100 mmHg of peak pressure. After 90 s, catheters were repositioned in the right and left ventricles, and unsequestered virus was flushed from the heart with Krebs buffer containing calcium (1.5 mM). The heart rate recovered, the cross-clamp was removed, the chest was closed, and the rats recovered. We previously reported that the percentage of cardiomyocytes transduced in vivo by this technique is 58% [21,22]. After 2-6 days, hearts were excised for the protein and mRNA analysis by western blots and PCR.
Western blots
Neonatal Cardiomyocytes
Cells were harvested in lysis buffer containing 20mM tris (hydroxymethyl) aminomethane, 100mM sodium chloride, 1mM EDTA, 0.5% Triton-X, and protease inhibitor (Sigma P8340). Following a 15 min digestion period on ice, samples were centrifuged (10 min, 3,000 rpm) and the supernatant was collected for BCA protein assays to determine protein concentration. Samples of equal protein mass were loaded onto a 4-12% Bis-Tris NuPAGE gel and transferred overnight onto a polyvinylidene difluoride membrane. Membranes were probed with a monoclonal anti-ME1 (4μg/mL, Abcam, Cambridge, UK) primary antibody, and calsequestrin was used to normalize protein loading (1:2000, Thermo Fisher Scientific, Waltham, Massachusetts). Membranes were visualized after enhanced chemiluminescence treatment (Thermo Fisher Scientific) and densitometric analysis of band intensity was used to determine changes in protein expression.
Adult Cardiac Tissue
Two to six days after Adv.miR-ME1 (n=6), PBS (n=5), or Adv.scrambled (n=5) delivery to coronaries in vivo, rat hearts were excised and retrograde perfused with a bolus of PBS to flush blood from the coronaries and ventricles [21,22]. Hearts were then freeze-clamped for Western blot analysis. Because the exogenous gene is heterogeneously expressed throughout the heart [11], whole ventricles were pulverized via a pestle and mortar, and homogenized in RIPA buffer [0.5 mM Tris-HCl, 1.5 mM NaCl, 2.5% deoxycholic acid, 10 mM EDTA, 0.1% SDS, 1% Nonidet P-40, 10% glycerol, 1% Triton X-100, and protease inhibitor]. Samples remained on ice for 15 min before being centrifuged (10 min, 3,000 rpm) and the supernatant was collected for BCA assay to determine protein concentration. Samples of equal protein mass were loaded in gels following the same general procedures described above. Primary antibodies for ME1 (3.33μg/mL, Abcam), ME2 (5μg/mL, Abcam), ME3 (6.7μg/mL, Abcam), GAPDH (1:10,000 Thermo Fisher Scientific), and calsequestrin (1:2000, Thermo Fisher Scientific) were used to assess the level of protein expression.
Measurement of RNA reduction by PCR
Total mRNAs were extracted from adult cardiac tissue using Trizol reagent (Life Technologies). The RNAs were DNase-treated and reverse transcribed using MultiScribe Reverse Transcriptase (Applied Biosystems, Life Technologies) on a Veriti Gradient PCR Thermocycler. A quantitative RT-PCR was carried out on an ABI ViiA7 using TaqMan primers and probes targeting ME1, ME2, and ME3. GAPDH served as the internal control. Primers for ME1, ME2, ME3, and GAPDH were designed by Roche (Hoffmann, Louisiana). A second primer and probe for ME1 (IDT, Coralville, Iowa) was designed to sit at the proposed cut site of the miR-RNA and had the following sequence: the forward primer 5′-TGCTTTGAGTAATCCGACCAG-3′, reverse primer: 5′-ACTGGATCAAAAGGACTGCC-3′, and probe: 5′-AGTGCTATAAAGTGACCAAGGGCCG-3′.
Statistical Analysis
Data is presented as mean ± standard error. Data set comparisons were performed with Student's unpaired, two-tailed parametric t-test, and confirmed by a non-parametric test (Mann-Whitney Rank Sum Test, or Kruskal Wallis Test). Differences in mean values were considered statistically significant at a probability level of less than 5% (p < 0.05). Reported p values are for the Student's t-test, unless otherwise stated.
Animal Use Approval
All protocols and procedures involving vertebrate animals were approved by the Animal Care Policies and Procedures Committee at the University of Illinois in Chicago (Institutional Animal Care and Use Committee accredited), and animals were maintained in accordance with the Guide for the Care and Use of laboratory Animals published by the US National Institutes of Health (NIH Publication No. 85-23, revised 1996).
RESULTS
Efficacy of miR-ME1 inhibition in primary cardiomyocytes
The protein content for the target protein, cytosolic malic enzyme-1, was assessed by western blot analysis 24 hrs after infection of isolated neonatal rat cardiomyocytes with adenoviral vectors containing the DNA templates encoded for the endogenous expression of a non-native miR-ME1 or miR-Scrambled. Three separate coding sequences for miR-ME1 and one scrambled control sequence were evaluated. All three target sequences resulted in a marked reduction of ME1 expression in the isolated cardiomyocytes relative to the scrambled miR-RNA control group. The results are presented in the Supplemental Data section. Sequence #1244 showed the greatest knockdown, at 35% of the control value. This viral package was selected for large-scale amplification and subsequent in vivo experiments.
Efficacy of miR-ME1 inhibition, in vivo
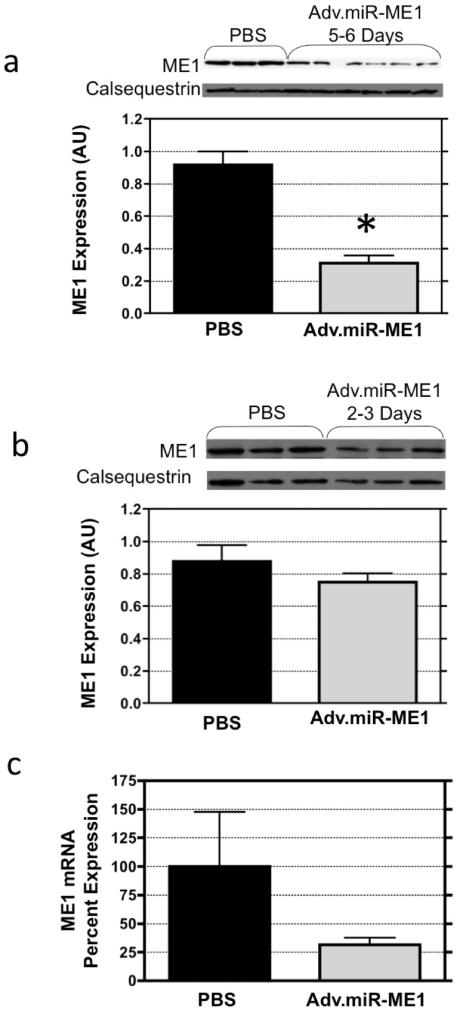
The open-chest isolated-heart perfusion technique, previously described by us [21-24], was used here to deliver adenoviral vectors (1×1013 vp/ml PBS) containing the DNA code for the endogenous expression of a non-native miR_ME1 (n=9). Sham operated controls received similar treatment with virus-free PBS (n=6). The efficacy of this strategy to inhibited ME1 expression was assessed by western blot and PCR analysis at 2-3 days and 5-6 days post-treatment. The results are illustrated in Figure 2. Consistent with our earlier reports for this open-chest adenoviral delivery procedure [21-24], survival was >80% post-surgery. Body weights were similar between groups at the time of heart excision (PBS: 348 ± 12 g; Adv.miR_ME1 348 ±9, p>0.05), as were heart weights (PBS: 2.36 ± 0.20 g; Adv.miR_ME1 2.21 ± 0.08, p>0.05). At 2-3 days post-treatment, the mRNA level for ME1 was reduced by 76% in the Adv.miR_ME1 group relative to PBS treated controls. ME1 protein content was not significantly different between groups at 2-3 days post-surgery, though the trend was lower in the Adv.miR_ME1 group (reduced 15%, p>0.05). Importantly, at 5-6 days post-treatment, ME1 mRNA was reduced by 70% (p=0.07) and protein content was significantly reduced by 66% (p<0.0002 for a Student's parametric t-test; p=0.02 for the Mann-Whitney Rank Sum non-parametric test) in the Adv.miR-ME1 group relative to the PBS treated controls. This is the first demonstration that adenoviral delivery of DNA, encoding a sequence for the endogenous expression of a non-native miRNA, results in suppression of the targeted protein in the heart, in vivo.
Figure 2.
Protein and mRNA expression of ME1 in heart (a) 5-6 days and (b) 2-3 days following delivery of Adv.miR-ME1 to rat heart via isolated-heart retrograde perfusion of the coronaries, in vivo. ME1 content was not significantly reduced at 2-3 days post-treatment (n=3) compared to PBS controls (n=3). However, at 5-6 days post-treatment, ME1 expression was reduced by 65% (n=6, p<0.0002). (c) mRNA content was reduced at both 2-3 days and 5-6 days post Adv.miR-ME1 treatment.
ME1 expression for three control group strategies
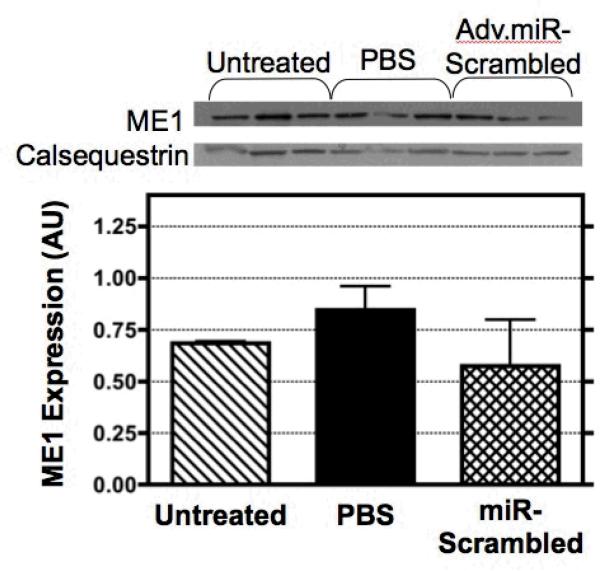
There is no ideal control group for gene transfer studies. Whether a scrambled, PBS, GFP, luciferase, or empty virus strategy is included as a control group, expression of some endogenous proteins are altered relative to untreated hearts [28,29]. Here, we assessed the content of ME1 in hearts from rats undergoing the open-chest isolated-heart perfusion procedure and receiving either a bolus of virus-free PBS (n=3) or the Adv.miR-scrambled vector (n=3). The results were compared to the level of ME1 protein measured from untreated hearts excised directly from a third group of rats (n=3). Western blots and densitometry graphs normalized to the endogenous calsequestrin content are illustrated in Figure 3. The ratio for ME1 content, relative to calsequestrin, was not significantly different between groups. However, the data suggests a non-statistical trend between groups. Compared to the untreated group, the PBS group shows an increased trend in ME1 expression, and the scrambled group shows a downward trend in ME1 expression. As described in the Discussion, this variability between the three control groups was expected based on earlier gene transfer reports from our group and others [14,23,29]. Importantly, whether we compared the Adv.miR-ME1 treated animals to the untreated excised group, the PBS group, or the Adv.miR-scrambled group, ME1 protein expression was reduced significantly by 5-6 days via this Adv.miRNA interference strategy.
Figure 3.
ME1 expression was measured by Western blot analysis for three different control conditions: untreated rat hearts, sham operated control rat hearts retrograde perfused with a bolus of virus-free PBS, and sham operated control rat hearts retrograde perfused with a bolus of PBS containing Adv.miR-scrambled. At 5-6 days post-treatment, the expression of ME1 was not significantly different between control groups.
Expression of non-target proteins
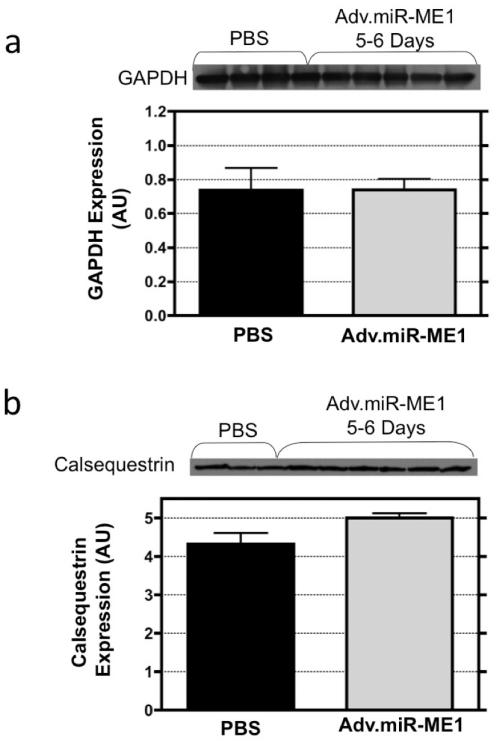
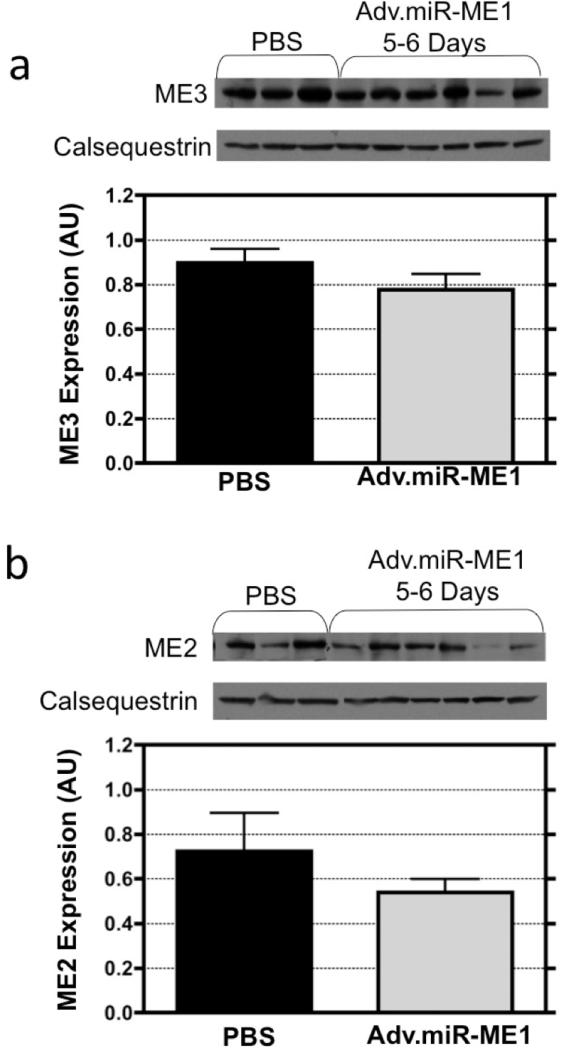
We assessed whether the strategy to inhibit ME1 affected the expression of the non-target proteins, calsequestrin and GAPDH, and the non-target mitochondrial isoforms of malic enzyme, ME2 and ME3. When selecting the code for ME1 inhibition, we performed a BLAST search to confirm that the selected 19-23 nt sequence was specific to ME1 and not these off-target proteins. The western blots and densitometry graphs are illustrated in Figure 4 and 5 for the content of the off-targets proteins from hearts excised 5-6 days following Adv.miR_ME1 (n=6) or PBS treatment (n=3). Calsequestrin and GAPDH protein expression were not different between groups (Figure 4). The non-target isoform of malic enzyme, ME3, was also not different between groups (Figure 5). ME2 expression was not statistically different between the Adv.miR-ME1 and PBS control at 5-6 days post-treatment, though the trend for ME2 expression was lower in the Adv.miR-ME1 group. Close examination of the westerns reveal that the expression at 5 days was unchanged, whereas at 6 days the protein content was reduced.
Figure 4.
The expression of the non-target proteins, calsequestrin and GAPDH, were assessed by western blot analysis from heart receiving Adv.miR-ME1 or PBS treatment. At 5-6 day post treatment, calsequestrin and GAPDH protein expression were not different between groups. (The calsequestrin blot shown in panel b is the loading control shown in Figure 2a).
Figure 5.
The expression of the non-target mitochondrial isoforms of malic enzyme (a) ME3, a mitochondrial NADP+ protein, and (b) ME2, also a mitochondrial NAD+ protein, were assessed in heart tissue by western blot analysis 5-6 days following Adv.miR-ME1 or PBS treatment. The expression of ME3 was not different between the two treatment groups. ME2 expression was also not statistically different between groups, though the expression was reduced at day 6.
DISCUSSION
As with shRNA, delivery of non-native miRNA constructs have been used to suppress target protein expression in cell cultures [4-6]. In whole animal models [30,31], pioneering studies in the cancer field have described impressive therapeutic effects with these non-native miRNAs, yet the approach had not been tested in heart, in vivo. In this study, we examined whether adenoviral delivery for gene transfer to whole heart in vivo, could inhibit the expression of the target protein, malic enzyme-1, using a strategy for induced expression of a non-native miRNA. We selected a 19-23 nt sequence for ME1, which initially resulted in a 66% reduction in ME1 expression in isolated neonatal cardiac myocytes. Subsequently, adenoviral delivery of the DNA sequence to the heart, in vivo, resulted in a 70% reduction in mRNA levels for ME1 2-6 days post-surgery. In support of the proposed strategy, the target protein, ME1, was acutely reduced by 65% at 5-6 day post-surgery relative to all control groups examined (ie., PBS, miR-scrambled, and untreated hearts).
Translational interference by non-native miRNA
Figure 6 illustrates the molecular pathway involved in the down regulation of protein expression by mRNA interference. Whereas shRNA and siRNA target RISC cleavage of the mRNA, indigenously produced miRNA can either target mRNA cleavage or mRNA translational inhibition [20,32]. Cleavage requires the miRNA sequence to fully complement the 19-23nt target of the mRNA [17,20]. An incomplete match between the two complementary sequences yield inhibition of translation rather than cleavage [17,20]. Our target sequence was fully complementary, and we anticipated mRNA degradation. However, preliminary PCR data for ME1 mRNA levels revealed no change in content at 2-6 days following Adv.miR-ME1 delivery, in vivo (data not shown). This initial PCR analysis was performed with a probe for malic enzyme-1 mRNA that was not designed to be specific to the target site of miRNA recognition. We designed a second probe, which was specific to the targeted sequence. For this second probe, PCR analysis revealed a ~73% reduction in mRNA content for ME1 at both 2-3 days and 5-6 days post-surgery. These results suggest that the mRNA was either cleaved or blocked at the target site, whereas the full strand of mRNA (as detected by the first probe) was not degraded within this acute timeframe.
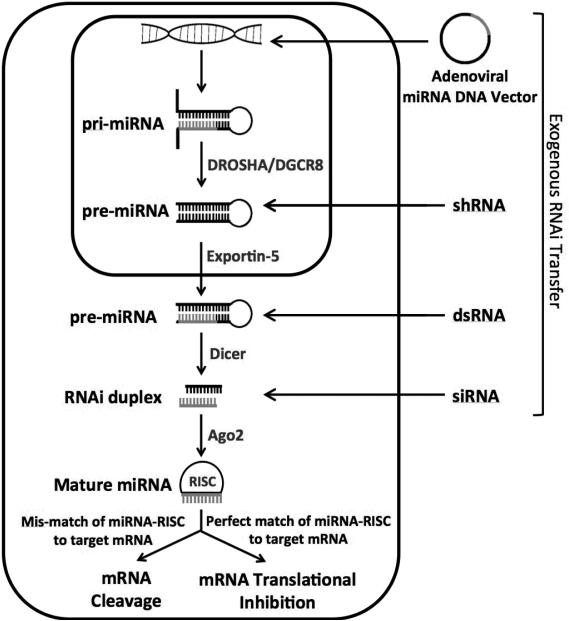
Figure 6.
While a variety of approaches can be used to introduce RNAi into the cell, all are processed similarly and incorporate into the RNAi protein machinery. microRNAs start the furthest upstream in the pathway; DNA is taken up into the nucleus and transcribed into the primary miRNA (pri-miRNA) strand which contains the target strand, hairpin loop, and a 5′ and 3′ flanking region. The flanking regions are cleaved off by DROSHA/DGCR8, into the precursor miRNA (pre-miRNA). This structure resembles shRNAs introduced into the nucleus. The pre-miRNA is then exported out of the nucleus by Exportin-5 into the cytoplasm where it is cleaved again into its characteristic 19-23nt structure. Argonaute-2 facilitates unwinding of the duplex, and the functional guide strand is incorporated into the RISC complex. Perfect complementary of the guide strand to the mRNA target leads to mRNA cleavage by RISC. Partial complementary leads to suppression of mRNA translation.
In this study, we found the mRNA level was knocked down ~70% at both 2-3 days and 5-6 days post gene transfer in vivo, whereas the protein was not knocked down until the later endpoint. This delayed response is indicative of the natural turnover rate of the endogenous ME1 pool. The delay also indicates that mRNA measures cannot be used as indirect markers of protein knockdown in siRNA based studies. Protein expression does not necessarily follow changes in mRNA content because of differences between the rates of protein turnover and mRNA translation. Similarly, changes in protein expression cannot be defined based on in vitro measurements of activity, because enzymatic flux is not solely linked to protein content [33,34].
Viral-based delivery strategies to the heart, in vivo
Our results for miRNA support proof-of-concept results (ie., RNAi in vivo) for both direct injection and intracoronary delivery of the shRNA and siRNA constructs to the heart, in vivo [10-14]. Morgan and colleagues directly injected naked siRNA to silence the expression of the Na+/H+ exchanger in the left ventricle of mouse myocardium [13]. Naked siRNA is stable in vivo for 72 hrs [12], and after 48-72 hrs the mRNA levels were reduced by 80% in the left ventricle, protein expression was down 30%, and the effects were confined to the heart. Similarly, Huang and Wu directly injected non-viral short hairpin RNA plasmid intramyocardially to inhibit prolyl hydroxylase-2 (PDH2) expression in vivo [11], and reported peak inhibition by day 14, which returned back to baseline levels by week 4. Gupta et al directly injected lentiviral vectors expressing myotropinin shRNA in the myocardium of mice, which resulted in 50% down regulation of the target gene expression in mice at 6 weeks [10]. While these direct injection approaches to RNA interference provided inhibition of target gene in heart without collateral infection of the peripheral tissue (ie., liver), silencing was restricted to the site of injection in the heart, or just proximal. Global gene transfer to the heart requires methods of intracoronary or intravenous viral vector delivery.
Poller and colleagues demonstrated the feasibility of inhibiting RNA translation in heart via viral-based delivery of code for short hairpin RNA by intracoronary and intravenous vector delivery [28]. In 2009 they examined silencing mRNA for phospholamban in whole heart of mice and rat by both an adenoviral vector (AdV_shRNA) delivered to the coronaries (acute response), and a tail vein injection of adeno-associated viral vector (AAV_shRNA) (chronic response). Phospholamban protein expression was significantly reduced by both viral-based approaches and cardiac function was enhanced. Thus, our results for the inhibition of RNA translation via the microRNA approach support these earlier results for protein knockdown via the short hairpin RNA in heart, in vivo.
One important distinction between our work, and this earlier global work by Poller, relates to the infection of peripheral tissue. In the Pollar study, tail vein injection of rAAV9_GFP in mice revealed equal gene transfer to both heart and liver [28]. Indeed, others have shown that this systemic injection of AAV vectors generally transduce to the liver more effectively than to cardiac and skeletal muscles [35]. Non-target gene transfer to the liver can be reduced by using a cardiac specific AAV serotype (such as AAV9) or a cardiac specific promoter [36]. However, higher doses of these vectors are required to achieve the same level of RNA inhibition in heart, and problems with shRNA-related toxicity, endogenous microRNA oversaturation, and liver failure are still observed [35,37]. Similarly, the conventional aortic cross-clamp method used to delivery adenoviral vectors in this Poller study was also not cardiac specific. Nevertheless, the problem with specificity is avoided by delivering the viral vectors via the fully isolated heart in vivo approach as described in this study. We previously demonstrated that the isolated-heart perfusion technique, as utilized here, is cardiac specific [21,22]. That is, we are able to flush any unsequestered virus from the heart prior to releasing the isolating cross-clamp, thereby eliminating viral accumulation or gene transfer to peripheral tissue.
Off target protein expression
While the expression of ME1 was not statistically different in heart samples from three control groups examined (ie., excised heart, PBS, and scrambled miRNA), there was some variance between groups. Western blots indicated the trend for higher ME1 expression in the PBS group relative to the content in hearts excised from non-treated animals, and lower in the group receiving scrambled miRNA. The expression of the other off target proteins, calsequestrin and GAPDH, were unchanged. This observation was not entirely unexpected. That is, in earlier adenoviral gene transfer studies performed with isolated cardiomyocytes and whole intact heart, the expression of the off-target protein, SERCA2a, was down regulated following gene transfer for both Adv.GFP (ie., GFP overexpression) and Adv.shGFP (GFP inhibition) compared to non-treated controls [28]. Whereas the expression of GAPDH, the Na+-Ca+2 exchanger, and calsequestrin were unaffected in these studies. The mechanism for this effect is unclear. Nevertheless, ME1 expression was inhibited via the miRNA treatment strategy performed under the current study relative to all three control groups.
We also examined whether the expression of two off-target isoforms of malic enzyme were affected by the ME1 miRNA treatment. Malic enzyme-1 is localized to the cytosol, and is a NADP+ dependent enzyme involved in anaplerosis in the diseased heart [25,26]. Two other isoforms of malic enzyme are localized to the mitochondria; the NAD+ dependent malic-enzyme-2 (ME2), and the NADP+ malic-enzyme-3 (ME3). While ME3 was unaffected by the treatment, western blot analysis for ME2 reveal a trend toward reduction in expression at day 6. The mechanism for this observation, in light of this new and rapidly advancing technology, is unclear, especially since ME2 is a NAD+ not NADP+ dependent isoform. Others have also observed changes in non-target isoform protein expression [18,19], and it is speculated as to whether these observations are linked to a compensatory mechanism to account for a change in the expression of the target protein, or a response via some unknown endogenous miRNA pathway.
Experimental implications
An important consideration implicates a known immune/inflammatory response to the adenovirus at 7 days following the treatment [38, 39]. The inflammatory response is not an issue if the target protein is knocked down and experimental measures are completed prior to day 7. In the case of the target enzyme examined in this study, a robust knockdown was observed at day 5-6. We could predict a moderate level of knockdown at day 4. If experimental studies are performed during the inflammatory event, a control group receiving a similar adenoviral dose of empty adenovirus or Adv.miR-scrambled should be included to account for inflammatory effects. Alternatively, immunosuppressors or immunodeficient rats could be considered in order to avoid the anti-adenoviral vector immune responses [39]. Furthermore, the AAV does not induce an inflammatory response. While AAV is ideal for long-term therapeutic treatment strategies, it is not ideal for mechanistic based studies where an acute knockdown of a single protein is desired. The adenoviral gene delivery approach provides the advantage of an acute knockdown with minimal confounding compensatory events characteristic of AAV and transgenic models.
Conclusion
While the siRNA approach has evolved as a powerful tool to down-regulate the expression of a target protein for elucidating the molecular basis of cardiovascular disease in studies of isolated cardiomyocytes, there have been far fewer attempts in whole animal models. This is a very important consideration for studies of metabolic enzymes (such as ME1), because isolated myocytes in culture, whether adult or neonatal, fail to represent the metabolic demand and mitochondrial activity of cells functioning in an intact myocardium. Therefore, extending the RNA interference approach to the intact beating heart is an important advancement and technique for molecular based studies performed under physiologically relevant conditions.
Supplementary Material
ACKNOWLEDGEMENTS
Grants. NIH grants: R37 HL49244 (Lewandowski MERIT), R01 HL62702.
LIST OF ABBREVIATIONS
- AAV
Adenoassociated virus
- Adv
Adenovirus
- BCA
Bicinchoninic acid - protein assay
- cDNA
Complementary DNA
- CMV
Cytomegalovirus
- GAPDH
Glyceraldehyde-phosphate dehydrogenase
- GFP
Green fluorescence protein
- HEK
Human embryonic kidney cells
- ME1
Malic Enzyme 1 (cytosolic isoform, NADP+ dependent)
- ME2
Malic Enzyme 2 (mitochondrial isoform, NAD+ dependent)
- ME3
Malic Enzyme 3 (mitochondrial isoform, NADP+ dependent)
- MEM
Minimum essential medium
- miR
Micro RNA
- miRNA
Micro RNA
- PBS
Phosphate buffered saline
- PDH2
Prolyl hydroxylase-2
- RIPA
Radio-immunoprecipitation assay buffer
- RISC
RNA-induced silencing complex
- RNAi
RNA interference
- RPM
revolutions per minute
- RT-PCR
Real time - polymerase chain reaction
- SERCA
Sarcoendoplasmic reticulum calcium ATPase transporter
- shRNA
Short hairpin RNA
- vpu/ml
Viral particles units per milliliter
Footnotes
Contributions. E. Douglas Lewandowski: designed research/study, interpreted results, edited manuscript. J. Michael O'Donnell: designed research/study, analyzed / interpreted results, wrote manuscript. Asha Kalichira: performed research/study, collected data, analyzed data, prepared figures. Jian Bi: performed research/study.
CONFLICTS OF INTEREST
There are no competing financial interests in relation to this work.
REFERENCES
- 1.Gray SJ, Samulski RJ. Optimizing gene delivery vectors for the treatment of heart disease. Expert Opin Biol Ther. 2008;8(7):911–22. doi: 10.1517/14712598.8.7.911. [DOI] [PubMed] [Google Scholar]
- 2.Muller OJ, Katus HA, Bekeredjian R. Targeting the heart with gene therapy-optimized gene delivery methods. Cardiovasc Res. 2007;73(3):453–62. doi: 10.1016/j.cardiores.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 3.Poller W, Fechner H. Development of novel cardiovascular therapeutics from small regulatory RNA molecules—an outline of key requirements. Curr Pharm Des. 2010;16(20):2252–68. doi: 10.2174/138161210791792813. [DOI] [PubMed] [Google Scholar]
- 4.Du J, Gao S, Lou J, Zhang G, Cong G, Shao J, Lin T, Cai X, Chang H. Effective inhibition of foot-and-mouth disease virus (FMDV) replication in vitro by vector-delivered microRNAs targeting the 3D gene. Virol J. 2011;8:292–303. doi: 10.1186/1743-422X-8-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lv Z, Xu L. Salvianolic acid B inhibits ERK and p38 MAPK signaling in TGF-b1-stumulated human hepatic stellate cell line (LX-2) via distinct pathways. Evid Based Complement Alternat Med. 2012;2012:960128. doi: 10.1155/2012/960128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wittchen ES, Aghajanian A, Burridge K. Isoform-specifc difference between Rap1A and Rap1b GTPases in the formation of endothelial cell junctions. Small GTPases. 2011;2:65–76. doi: 10.4161/sgtp.2.2.15735. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown LJ, Longacre MJ, Hasan NM, Kendrick MA, Stoker SW, Macdonald MJ. Chronic reduction of the cytosolic or mitochondrial NAD(P)-malic enzyme does not affect insulin secretion in a rat insulinoma cell line. J Biol Chem. 2009;284(51):35359–35367. doi: 10.1074/jbc.M109.040394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pongratz RL, Kibbey RG, Cline GW. Investigating the roles of mitochondrial and cytsolic malic enzyme in inulin secretion. Methods Enzymol. 2009;457:425–450. doi: 10.1016/S0076-6879(09)05024-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ronnebaum SM, Jensen MV, Hohmeier HE, Burgess SC, Zhou YP, Qian S, MacNeil D, Howard A, Thornberry N, Ilkayeva O, Lu D, Sherry AD, Newgard CB. Silencing of cytosolic or mitochondrial isoforms of malic enzyme has no effect on glucose-stimulated insulin secretion from rodent islets. J Biol Chem. 2008;283(43):28909–28917. doi: 10.1074/jbc.M804665200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta S, Maitra R, Young D, Gupta A, Sen S. Silencing the myotrophin gene by RNA interference leads to the regression of cardiac hypertrophy. Am J Physiol Heart Circ Physiol. 2009;297:H627–H636. doi: 10.1152/ajpheart.00294.2009. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Huang M, Wu JC. Molecular imaging of RNA interference therapy targeting PDH2 for treatment of myocardial ischemia. Methods Mol Biol. 2011;709:211–221. doi: 10.1007/978-1-61737-982-6_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang M, Chan DA, Jia F, Xie X, Li Z, Hoyt G, Robbins RC, Chen X, Giaccia AJ, Wu JC. Short hairpin RNA interference therapy for ischemic heart disease. Circulation. 2008;118(suppl 1):S226–S233. doi: 10.1161/CIRCULATIONAHA.107.760785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgan PE, Correa MV, Ennis IE, Diez AA, Perez NG, Cingolani HE. Silencing of sodium/hydrogen exchanger in the heart by direct injection of naked siRNA. J Appl Physiol. 2011;111:566–572. doi: 10.1152/japplphysiol.00200.2011. [DOI] [PubMed] [Google Scholar]
- 14.Poller W, Hajjar R, Schultheiss HP, Fechner H. Cardiac-targeted delivery of regulatory RNA molecules and genes for the treatment of heart failure. Cardiovasc Res. 2010;86:353–364. doi: 10.1093/cvr/cvq056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lebbink RJ, Lowe M, Chan T, Khine H, Wang X, McManus MT. Polymerase II promoter strength determines efficacy of microRNA adapted shRNAs. PLoS ONE. 2011;6(10):e26213. doi: 10.1371/journal.pone.0026213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, Gilmore BL, Burstein H, Peluso RW, Polsky B, Carter BJ, Davidson BL. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. PNAS. 2008;105(15):5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pushparaj PN, Arthi JJ, Manikandan J, Kumar SD. siRNA, miRNA, and shRNA: in vivo application. J Dent Res. 2008;87(11):992–1003. doi: 10.1177/154405910808701109. [DOI] [PubMed] [Google Scholar]
- 19.Rao DD, Vorhies JS, Senzer N, Nemunaitis J. siRNA vs shRNA” similarities and differences. Adv Drug Del Rev. 2009;61:746–759. doi: 10.1016/j.addr.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Bushati N, Cohen SM. microRNA Functions. Annu. Rev. Cell Dev. Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 21.O'Donnell JM, Lewandowski ED. Efficient, Cardiac-Specific Adenoviral Gene Transfer by Isolated Retrograde Perfusion In Vivo. Gene Therapy. 2005;12:958–964. doi: 10.1038/sj.gt.3302477. [DOI] [PubMed] [Google Scholar]
- 22.O'Donnell JM, Lewandowski ED. Controlling specificity and efficiency of adenoviral gene transfer in heart by catheter based coronary perfusion. In: Niewohner J, Tannert C, editors. Gene Therapy Prospective assessment in its societal context. Elsevier (pub); Amsterdam, Netherlands: 2006. pp. 33–46. [Google Scholar]
- 23.O'Donnell JM, Fields AD, Xu X, Chowdhury SA, Geenen DL, Bi J. Limited functional and metabolic improvements in hypertrophic and healthy hearts expressing the skeletal muscle isoform of SERCA1 by adenoviral gene transfer in vivo. Amer J Physiol (Heart and Circ.) 2008;295(6):H2483–94. doi: 10.1152/ajpheart.01023.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Donnell JM, Pound K, Xu X, Lewandowski ED. SERCA1 Expression enhances the metabolic efficiency of improved contractility in post ischemic hearts. J Mol Cell Cardio. 2009 Nov;47(5):614–21. doi: 10.1016/j.yjmcc.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pound KM, Sorokina N, Ballal K, Berkich DA, Fasano M, LaNoue KF, Taegtmeyer H, O'Donnell JM, Lewandowski ED. Substrate-enzyme competition attenuates upregulated anaplerotic flux through malic enzyme in hypertrophied hearts and restores triacylglyceride content: attenuating upregulated anaplerosis in hypertrophy. Circulation Research. 2009;104(6):805–12. doi: 10.1161/CIRCRESAHA.108.189951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sorokina N, O'Donnell JM, McKinney RD, Pound KM, Woldegiorgis G, LaNoue KF, Ballal K, Taegtmeyer H, Buttrick PM, Lewandowski ED. Recruitment of compensatory pathways to sustain oxidative flux with reduced carnitine palmitoyltransferase I activity characterizes inefficiency in energy metabolism in hypertrophied hearts. Circulation. 2007;155(15):2033–41. doi: 10.1161/CIRCULATIONAHA.106.668665. [DOI] [PubMed] [Google Scholar]
- 27.Cavagna M, O'Donnell JM, Sumbilla C, Inesi G, Klein MG. Exogenous Ca2+-ATPase isoform effects on Ca2+ transients of embryonic chicken and neonatal rat cardiac myocytes. J Physiol. 2000;528:53–63. doi: 10.1111/j.1469-7793.2000.00053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suckau L, Fechner H, Chemaly E, Krohn S, Hadri L, Kockskämper J, Westermann D, Bisping E, Ly H, Wang X, Kawase Y, Chen J, Liang L, Sipo I, Vetter R, Weger S, Kurreck J, Erdmann V, Tschope C, Pieske B, Lebeche D, Schultheiss HP, Hajjar RH, Poller WC. Long-term cardiac-targeted RNA interference for the treatment of heart failure restores cardiac function and reduces pathological hypertrophy. Circulation. 2009;119:1241–1252. doi: 10.1161/CIRCULATIONAHA.108.783852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weisser-Thomas J, Dietrich E, Janssen PML, Schmidt-Schwead S, Maier LS, Sumbilla C, Pieske B. Method-related effects of adenovirus-mediated LacZ and SERCA1 gene transfer on contractile behavior of cultured failing human cardiomyocytes. J Pharm Tox Meth. 2005;51:91–103. doi: 10.1016/j.vascn.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Lorio MV, Casalini P, Piovan C, Di Leva G, Merlo A, Triulzi T, Menard S, Croce CM, Taglisabue E. microRNA-205 regulates HER3 in human breast cancer. Cancer Res. 2009;69:2195–2200. doi: 10.1158/0008-5472.CAN-08-2920. [DOI] [PubMed] [Google Scholar]
- 31.Taulli R, Bersani F, Foglizo V, Linari A, Bigna E, Ladanyi M, Tuschl T, Ponaetto C. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotranspolanted mice by promoting myogenic differentiation. J Clin Invest. 2009;119:2366–2378. doi: 10.1172/JCI38075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 33.Griffin JL, O'Donnell JM, White LT, Hajjar RJ, Lewandowski ED. Postnatal expression and activity of the mitochondrial 2-oxoglutarate-malate carrier in intact hearts. Am J Physiol Cell Physiol. 2000 Dec;279(6):C1704–1709. doi: 10.1152/ajpcell.2000.279.6.C1704. [DOI] [PubMed] [Google Scholar]
- 34.Lewandowski ED, O'Donnell JM, Scholz TD, Sorokina N, Buttrick PM. Recruitment of NADH shuttling in pressure-overloaded and hypertrophic rat hearts. Am J Physiol Cell Physiol. 2007 May;292(5):C1880–1886. doi: 10.1152/ajpcell.00576.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayra A, Tomimitsu H, Kubodera T, Kobayashi M, Piao W, Sunaga F, Hirai Y, Shimada T, Mizusawa H, Yokota T. Intraperitoneal AAV9-shRNA inhibits target expression in neonatal skeletal and cardiac muscles. Biochem Biophys Res Commun. 2011;405(2):204–209. doi: 10.1016/j.bbrc.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 36.Prasad KMR, Xu Y, Yang Z, Acton ST, French BA. Robust cardiomyocyte-specific gene expression following systemic injection of AAV: In vivo gene delivery follows a poisson distribution. Gene Therapy. 2011;19(1):43–52. doi: 10.1038/gt.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 38.Hajjar RJ, Schmidt U, Matsui T, Guerrero JL, Lee Kyung-Han, Gwathmey JK, Dec GW, Semigran MJ, Rosenzweig A. Modulation of ventricular function through gene transfer in vivo. PNAS. 1998;95:5251–5256. doi: 10.1073/pnas.95.9.5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahi YS, Bangari DS, Mittal SK. Adenoviral Vector Immunity: Its Implications and Circumvention Strategies. Current Gene Therapy. 2011;11(4):307–320. doi: 10.2174/156652311796150372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reeti KR, Chen CY, Weaver EA, Barry MA. Advances and Future Challenges in Adenoviral Vector Pharmacology and Targeting. Current Gene Therapy. 2011;11(4):241–258. doi: 10.2174/156652311796150363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.