Abstract
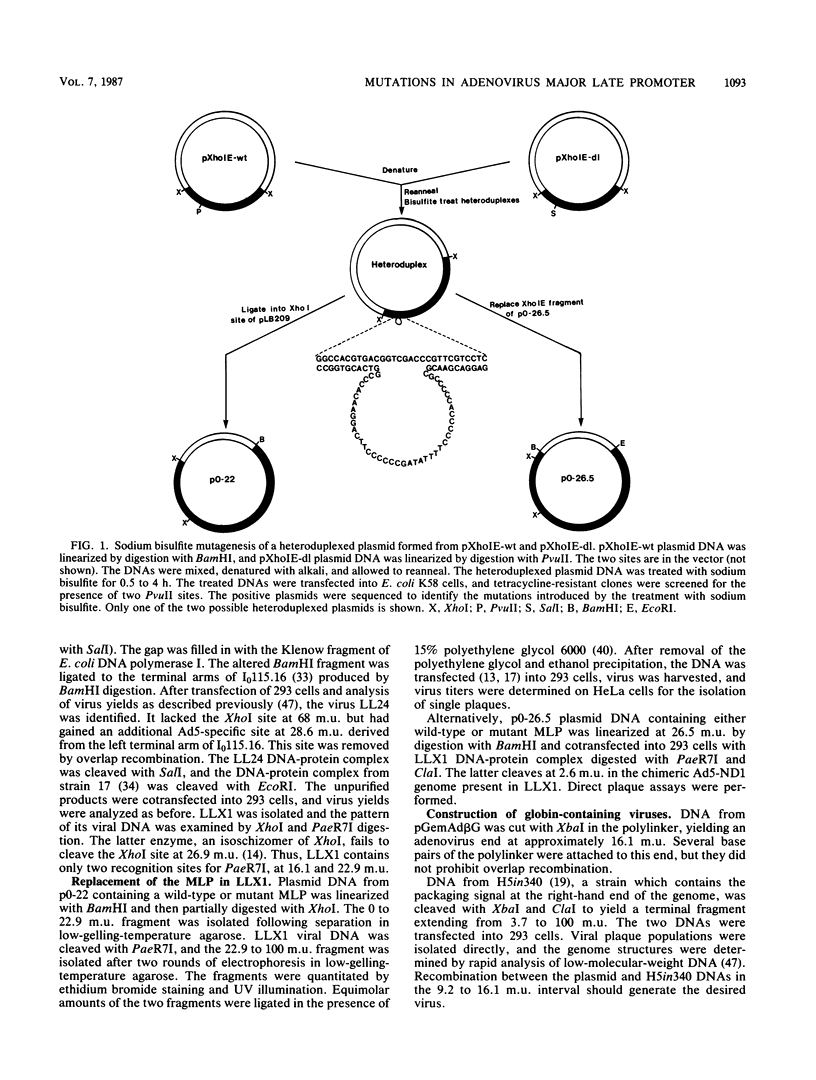
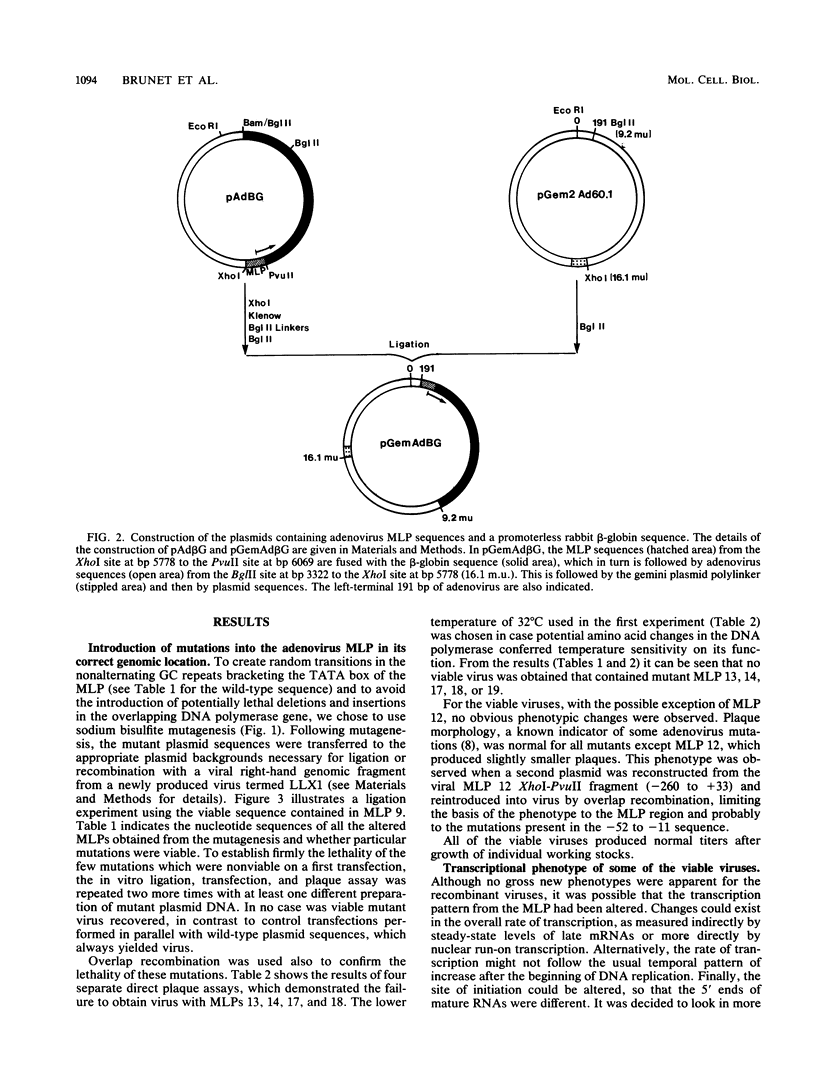
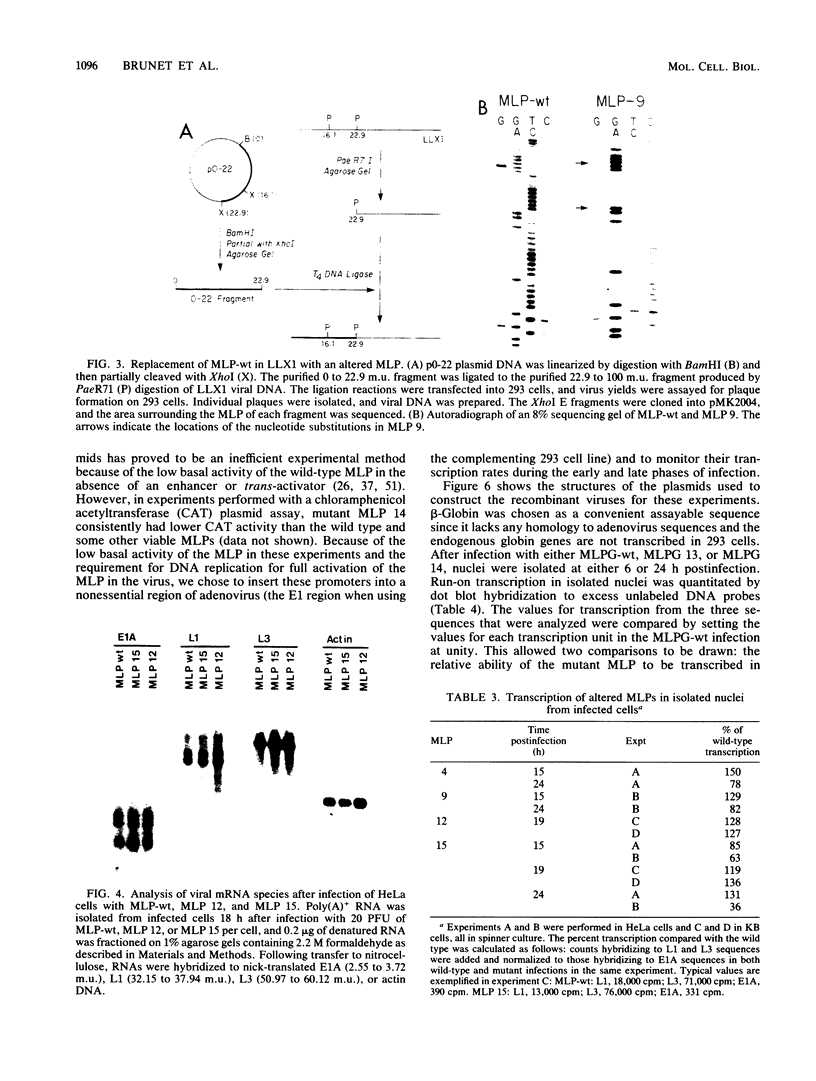
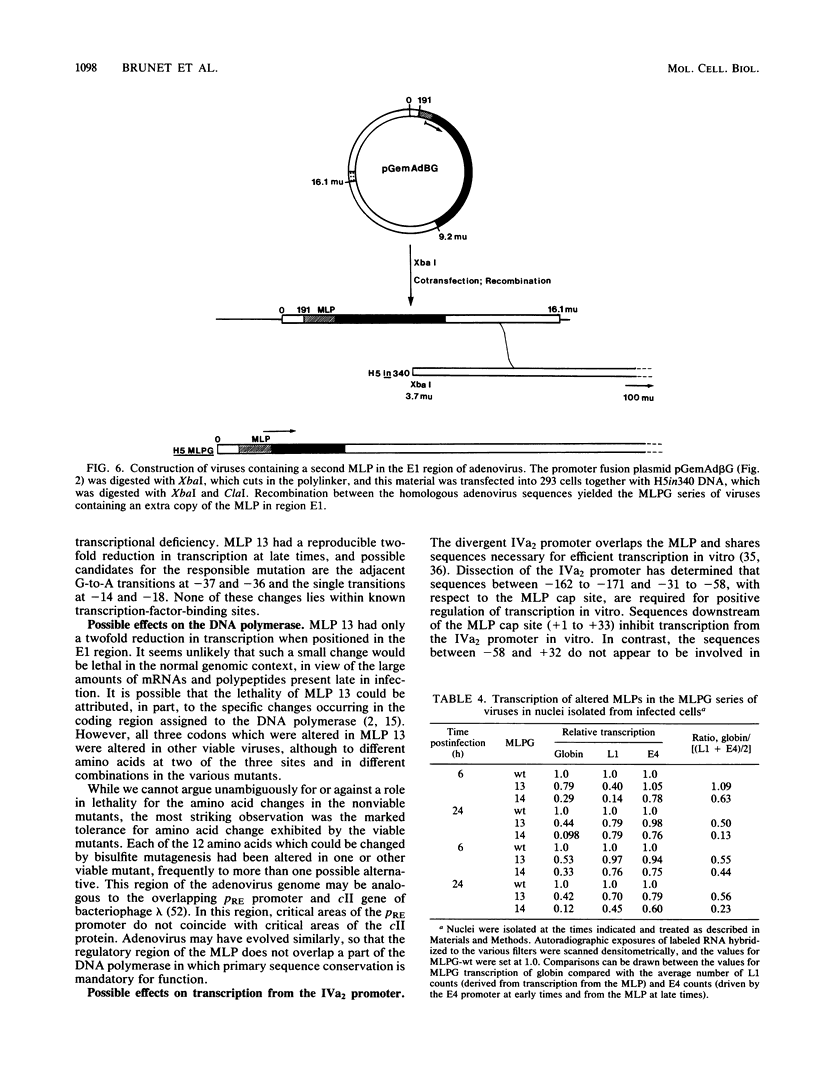
We developed an experimental system to examine the effects of mutations in the adenovirus major late promoter in its correct genomic location during a productive infection. A virus was constructed whose genome could be digested to give a rightward terminal DNA fragment extending from the XhoI site at 22.9 map units, which can be ligated or recombined with plasmid DNA containing adenovirus sequences extending from 0 to 22.9 or 26.5 map units, respectively. Mutations were made by bisulfite mutagenesis in the region between base pairs -52 and -12 with respect to the cap site at +1 and transferred to the appropriate plasmids for viral reconstruction. Of 19 mutant plasmid sequences containing single or multiple G-to-A transitions, 14 could be placed in the viral genome with no apparent change in phenotype. These mutant sequences included those which contained four transitions in the string of G residues immediately downstream of the TATA box. There were no alterations in rates of transcription from the major late promoter, sites of transcription initiation, or steady-state levels of late mRNAs. All of the five mutant sequences which could not be placed in virus contained multiple transitions both up- and downstream of the TATA box. Two of these apparently lethal mutant sequences were used in promoter fusion experiments to test their ability to promote transcription of rabbit beta-globin sequences placed in the dispensable E1 region of the virus. Both sequences showed diminished ability compared with wild-type sequences to promote transcription in this context. Comparisons between these two sequences and the viable mutant sequences suggest a role for the string of G residues located between -38 and -33 in promoting transcription from the major late promoter. The data as a whole also demonstrate that the specific nucleotide sequence of this region of the major late promoter, which overlaps transcription elements of the divergent IVa2 transcription unit and coding sequences of the adenovirus DNA polymerase, is not rigidly constrained but can mutate extensively without loss of these several functions.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akusjärvi G., Pettersson U. Nucleotide sequence at the junction between the coding region of the adenovirus 2 hexon messenger RNA and its leader sequence. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5822–5826. doi: 10.1073/pnas.75.12.5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleström P., Akusjärvi G., Pettersson M., Pettersson U. DNA sequence analysis of the region encoding the terminal protein and the hypothetical N-gene product of adenovirus type 2. J Biol Chem. 1982 Nov 25;257(22):13492–13498. [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas P. D., van Teeffelen H. A., Teertstra W. R., Jansz H. S., Veeneman G. H., van der Marel G. A., van Boom J. H. Restoration of the biological activity of in vitro synthesized phi X DNA by transfection of ung- spheroplasts or dUTPase treatment. FEBS Lett. 1980 Jan 28;110(1):15–20. doi: 10.1016/0014-5793(80)80012-3. [DOI] [PubMed] [Google Scholar]
- Babiss L. E., Fisher P. B., Ginsberg H. S. Deletion and insertion mutations in early region 1a of type 5 adenovirus that produce cold-sensitive or defective phenotypes for transformation. J Virol. 1984 Mar;49(3):731–740. doi: 10.1128/jvi.49.3.731-740.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensimhon M., Gabarro-Arpa J., Ehrlich R., Reiss C. Physical characteristics in eucaryotic promoters. Nucleic Acids Res. 1983 Jul 11;11(13):4521–4540. doi: 10.1093/nar/11.13.4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew R. W., Chodosh L. A., Sharp P. A. An RNA polymerase II transcription factor binds to an upstream element in the adenovirus major late promoter. Cell. 1985 Dec;43(2 Pt 1):439–448. doi: 10.1016/0092-8674(85)90174-6. [DOI] [PubMed] [Google Scholar]
- Chinnadurai G. Adenovirus 2 Ip+ locus codes for a 19 kd tumor antigen that plays an essential role in cell transformation. Cell. 1983 Jul;33(3):759–766. doi: 10.1016/0092-8674(83)90018-1. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Chow L. T., Broker T. R., Lewis J. B. Complex splicing patterns of RNAs from the early regions of adenovirus-2. J Mol Biol. 1979 Oct 25;134(2):265–303. doi: 10.1016/0022-2836(79)90036-6. [DOI] [PubMed] [Google Scholar]
- Concino M. F., Lee R. F., Merryweather J. P., Weinmann R. The adenovirus major late promoter TATA box and initiation site are both necessary for transcription in vitro. Nucleic Acids Res. 1984 Oct 11;12(19):7423–7433. doi: 10.1093/nar/12.19.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck-Pedersen E., Logan J., Shenk T., Darnell J. E., Jr Transcription termination within the E1A gene of adenovirus induced by insertion of the mouse beta-major globin terminator element. Cell. 1985 Apr;40(4):897–905. doi: 10.1016/0092-8674(85)90349-6. [DOI] [PubMed] [Google Scholar]
- Frost E., Williams J. Mapping temperature-sensitive and host-range mutations of adenovirus type 5 by marker rescue. Virology. 1978 Nov;91(1):39–50. doi: 10.1016/0042-6822(78)90353-7. [DOI] [PubMed] [Google Scholar]
- Gingeras T. R., Brooks J. E. Cloned restriction/modification system from Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1983 Jan;80(2):402–406. doi: 10.1073/pnas.80.2.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingeras T. R., Sciaky D., Gelinas R. E., Bing-Dong J., Yen C. E., Kelly M. M., Bullock P. A., Parsons B. L., O'Neill K. E., Roberts R. J. Nucleotide sequences from the adenovirus-2 genome. J Biol Chem. 1982 Nov 25;257(22):13475–13491. [PubMed] [Google Scholar]
- Goding C. R., Russell W. C. S1 sensitive sites in adenovirus DNA. Nucleic Acids Res. 1983 Jan 11;11(1):21–36. doi: 10.1093/nar/11.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Groudine M., Peretz M., Weintraub H. Transcriptional regulation of hemoglobin switching in chicken embryos. Mol Cell Biol. 1981 Mar;1(3):281–288. doi: 10.1128/mcb.1.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearing P., Shenk T. The adenovirus type 5 E1A transcriptional control region contains a duplicated enhancer element. Cell. 1983 Jul;33(3):695–703. doi: 10.1016/0092-8674(83)90012-0. [DOI] [PubMed] [Google Scholar]
- Hen R., Sassone-Corsi P., Corden J., Gaub M. P., Chambon P. Sequences upstream from the T-A-T-A box are required in vivo and in vitro for efficient transcription from the adenovirus serotype 2 major late promoter. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7132–7136. doi: 10.1073/pnas.79.23.7132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer E., Darnell J. E., Jr The primary transcription unit of the mouse beta-major globin gene. Cell. 1981 Feb;23(2):585–593. doi: 10.1016/0092-8674(81)90154-9. [DOI] [PubMed] [Google Scholar]
- Hu S. L., Manley J. L. DNA sequence required for initiation of transcription in vitro from the major late promoter of adenovirus 2. Proc Natl Acad Sci U S A. 1981 Feb;78(2):820–824. doi: 10.1073/pnas.78.2.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jove R., Manley J. L. In vitro transcription from the adenovirus 2 major late promoter utilizing templates truncated at promoter-proximal sites. J Biol Chem. 1984 Jul 10;259(13):8513–8521. [PubMed] [Google Scholar]
- LaFlamme S. E., Kramer F. R., Mills D. R. Comparison of pausing during transcription and replication. Nucleic Acids Res. 1985 Dec 9;13(23):8425–8440. doi: 10.1093/nar/13.23.8425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen A., Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982 Jun;29(2):609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- Lewis E. D., Manley J. L. Control of adenovirus late promoter expression in two human cell lines. Mol Cell Biol. 1985 Sep;5(9):2433–2442. doi: 10.1128/mcb.5.9.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan J., Shenk T. Adenovirus tripartite leader sequence enhances translation of mRNAs late after infection. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3655–3659. doi: 10.1073/pnas.81.12.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour S. L., Grodzicker T., Tjian R. Downstream sequences affect transcription initiation from the adenovirus major late promoter. Mol Cell Biol. 1986 Jul;6(7):2684–2694. doi: 10.1128/mcb.6.7.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982 Jul 23;217(4557):316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- Miyamoto N. G., Moncollin V., Egly J. M., Chambon P. Specific interaction between a transcription factor and the upstream element of the adenovirus-2 major late promoter. EMBO J. 1985 Dec 16;4(13A):3563–3570. doi: 10.1002/j.1460-2075.1985.tb04118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto N. G., Moncollin V., Wintzerith M., Hen R., Egly J. M., Chambon P. Stimulation of in vitro transcription by the upstream element of the adenovirus-2 major late promoter involves a specific factor. Nucleic Acids Res. 1984 Dec 11;12(23):8779–8799. doi: 10.1093/nar/12.23.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munz P. L., Young C. S. Polarity in adenovirus recombination. Virology. 1984 Jun;135(2):503–514. doi: 10.1016/0042-6822(84)90204-6. [DOI] [PubMed] [Google Scholar]
- Munz P. L., Young C., Young C. S. The genetic analysis of adenovirus recombination in triparental and superinfection crosses. Virology. 1983 Apr 30;126(2):576–586. doi: 10.1016/s0042-6822(83)80014-2. [DOI] [PubMed] [Google Scholar]
- Natarajan V., Madden M. J., Salzman N. P. Positive and negative control sequences within the distal domain of the adenovirus IVa2 promoter overlap with the major late promoter. J Virol. 1985 Jul;55(1):10–15. doi: 10.1128/jvi.55.1.10-15.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan V., Madden M. J., Salzman N. P. Proximal and distal domains that control in vitro transcription of the adenovirus IVa2 gene. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6290–6294. doi: 10.1073/pnas.81.20.6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan V., Salzman N. P. Cis and trans activation of adenovirus IVa2 gene transcription. Nucleic Acids Res. 1985 Jun 11;13(11):4067–4083. doi: 10.1093/nar/13.11.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins J. R. Mechanism of activation of early viral transcription by the adenovirus E1A gene product. Cell. 1981 Oct;26(2 Pt 2):213–220. doi: 10.1016/0092-8674(81)90304-4. [DOI] [PubMed] [Google Scholar]
- Peden K. W., Nathans D. Local mutagenesis within deletion loops of DNA heteroduplexes. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7214–7217. doi: 10.1073/pnas.79.23.7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pheiffer B. H., Zimmerman S. B. Polymer-stimulated ligation: enhanced blunt- or cohesive-end ligation of DNA or deoxyribooligonucleotides by T4 DNA ligase in polymer solutions. Nucleic Acids Res. 1983 Nov 25;11(22):7853–7871. doi: 10.1093/nar/11.22.7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawadogo M., Roeder R. G. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell. 1985 Nov;43(1):165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- Shaw A. R., Ziff E. B. Transcripts from the adenovirus-2 major late promoter yield a single early family of 3' coterminal mRNAs and five late families. Cell. 1980 Dec;22(3):905–916. doi: 10.1016/0092-8674(80)90568-1. [DOI] [PubMed] [Google Scholar]
- Shortle D., Nathans D. Local mutagenesis: a method for generating viral mutants with base substitutions in preselected regions of the viral genome. Proc Natl Acad Sci U S A. 1978 May;75(5):2170–2174. doi: 10.1073/pnas.75.5.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu L. M., Hong J. S., Wei Y. F., Engler J. A. Nucleotide sequence of the genes encoded in early region 2b of human adenovirus type 12. Gene. 1986;46(2-3):187–195. doi: 10.1016/0378-1119(86)90403-8. [DOI] [PubMed] [Google Scholar]
- Thomas G. P., Mathews M. B. DNA replication and the early to late transition in adenovirus infection. Cell. 1980 Nov;22(2 Pt 2):523–533. doi: 10.1016/0092-8674(80)90362-1. [DOI] [PubMed] [Google Scholar]
- Volkert F. C., Young C. S. The genetic analysis of recombination using adenovirus overlapping terminal DNA fragments. Virology. 1983 Feb;125(1):175–193. doi: 10.1016/0042-6822(83)90072-7. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk B., Kédinger C., Corden J., Brison O., Chambon P. Specific in vitro initiation of transcription on conalbumin and ovalbumin genes and comparison with adenovirus-2 early and late genes. Nature. 1980 Jun 5;285(5764):367–373. doi: 10.1038/285367a0. [DOI] [PubMed] [Google Scholar]
- Weintraub H. High-resolution mapping of S1- and DNase I-hypersensitive sites in chromatin. Mol Cell Biol. 1985 Jun;5(6):1538–1539. doi: 10.1128/mcb.5.6.1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyer U., Doerfler W. Species dependence of the major late promoter in adenovirus type 12 DNA. EMBO J. 1985 Nov;4(11):3015–3019. doi: 10.1002/j.1460-2075.1985.tb04037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff D. L., Mahoney M., Shatzman A., Rosenberg M. Mutational analysis of a regulatory region in bacteriophage lambda that has overlapping signals for the initiation of transcription and translation. Proc Natl Acad Sci U S A. 1984 Jan;81(2):555–559. doi: 10.1073/pnas.81.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y. T., Manley J. L. Generation and functional analyses for base-substitution mutants of the adenovirus 2 major late promoter. Nucleic Acids Res. 1984 Dec 21;12(24):9309–9321. doi: 10.1093/nar/12.24.9309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y. T., Manley J. L. Structure and function of the S1 nuclease-sensitive site in the adenovirus late promoter. Cell. 1986 Jun 6;45(5):743–751. doi: 10.1016/0092-8674(86)90788-9. [DOI] [PubMed] [Google Scholar]
- Ziff E. B., Evans R. M. Coincidence of the promoter and capped 5' terminus of RNA from the adenovirus 2 major late transcription unit. Cell. 1978 Dec;15(4):1463–1475. doi: 10.1016/0092-8674(78)90070-3. [DOI] [PubMed] [Google Scholar]
- van Ooyen A., van den Berg J., Mantei N., Weissmann C. Comparison of total sequence of a cloned rabbit beta-globin gene and its flanking regions with a homologous mouse sequence. Science. 1979 Oct 19;206(4416):337–344. doi: 10.1126/science.482942. [DOI] [PubMed] [Google Scholar]





