SUMMARY
Adenosine deaminases acting on RNA (ADARs) are involved in RNA editing that converts adenosine residues to inosine specifically in double-stranded RNAs. In this study, we investigated the interaction of the RNA editing mechanism with the RNA interference (RNAi) machinery and found that ADAR1 forms a complex with Dicer through direct protein-protein interaction. Most importantly, ADAR1 increases the maximum rate (Vmax) of pre-microRNA (miRNA) cleavage by Dicer and facilitates loading of miRNA onto RNA-induced silencing complexes, identifying a new role of ADAR1 in miRNA processing and RNAi mechanisms. ADAR1 differentiates its functions in RNA editing and RNAi by formation of either ADAR1/ADAR1 homodimer or Dicer/ADAR1 heterodimer complexes, respectively. As expected, expression of miRNAs is globally inhibited in ADAR1−/− mouse embryos, which in turn alters expression of their target genes and might contribute to their embryonic lethal phenotype.
INTRODUCTION
Adenosine deaminases acting on RNA (ADARs) are the enzymes involved in one type of RNA editing that converts adenosine residues into inosine (A-to-I RNA editing) in double-stranded RNAs (dsRNAs). The translation machinery reads inosine as guanosine, leading to recoding of a relatively small number of neurotransmitter and ion channel genes and diversification of their functions (Hood and Emeson, 2012). However, A-to-I editing occurs most frequently in non-coding regions that contain repetitive elements such as Alu and LINE (Ramaswami et al., 2012). The biological significance of repetitive RNA editing is largely unknown. Three ADAR gene family members (ADAR1-3) are known in vertebrates. Members of the ADAR gene family share common structural features, such as the presence of multiple dsRNA binding domains (dsRBDs) and a separate deaminase domain. Three repeats of dsRBD are present in ADAR1, while only two repeats are found in ADAR2 and ADAR3 (Nishikura, 2010). The inactivation of ADAR1 leads to an embryonic lethal phenotype due to widespread apoptosis (Hartner et al., 2004; Wang et al., 2004), indicating the absolute requirement of ADAR1 for life. However, the mechanism that underlies the ADAR1−/− mouse phenotype is unknown.
The RNase III gene family members Drosha and Dicer are key components of the microRNA (miRNA) processing machinery (Krol et al., 2010). In the nucleus, Drosha with a dsRNA-binding protein DGCR8 cleaves primary transcripts of miRNA genes (pri-miRNAs) to produce intermediate precursors, pre-miRNAs. Once exported to the cytoplasm, pre-miRNAs are cleaved by Dicer, releasing 21–24 nt mature miRNAs (Krol et al., 2010). A separate dsRNA-binding protein, HIV-1 TAR RNA-binding protein (TRBP), binds to Dicer and enhances the Dicer activity (Chakravarthy et al., 2010). The Dicer/TRBP complex also acts as the platform for RNA-induced silencing complex (RISC) assembly by recruiting Ago2 (Chendrimada et al., 2005), which is the catalytic engine to execute the RISC-mediated silencing either by translational suppression or degradation of target mRNAs (Liu et al., 2004; Meister et al., 2004). The RISC consisting of Dicer, TRBP, and Ago2 is the core miRNA-mediated silencing machinery that couples the miRNA processing and silencing steps (Gregory et al., 2005).
RNAi-dependent endogenous short RNAs (endo-siRNA) derived from loci enriched for inverted repeats and transposons are dramatically upregulated in ADAR null mutant worms, indicating that A-to-I RNA editing of dsRNA regions of transcripts derived from these loci inhibits their entry into the RNAi silencing pathway (Wu et al., 2011). A-to-I RNA editing mechanism appears to compete for shared dsRNA substrates with the RNAi machinery and inhibit synthesis of endo-siRNAs (Wu et al., 2011). Furthermore, certain pri-miRNAs are subject to A-to-I RNA editing. Editing of pri-miRNAs could inhibit their processing (Kawahara et al., 2008; Kawahara et al., 2007a; Yang et al., 2006), suppress RISC loading (Iizasa et al., 2010), or lead to silencing of a different set of target genes by the edited miRNAs (Kawahara et al., 2007b).
With these recent studies implicating antagonistic interaction of A-to-I RNA editing and RNAi pathways, we investigated the interaction of ADAR1 with the RISC component proteins. We found that ADAR1 interacts directly with Dicer in an RNA binding-independent manner and promotes processing of siRNA and miRNA, RISC loading of miRNAs, and consequently silencing of target RNAs, revealing a stimulative, instead of antagonistic, role of ADAR1 in RNAi. In contrast to the requirement of homodimerization for the A-to-I RNA editing activities of ADAR1 (Cho et al., 2003), one monomer of ADAR1 binds to one molecule of Dicer. ADAR1 thus acts as an RNA editing enzyme or as a modulator of the RNAi machinery by choosing its complex partner, i.e., ADAR1 itself or Dicer.
RESULTS
Interaction of ADAR1 with RISC Member Proteins
Two isoforms of ADAR1, a full-length interferon-inducible ADAR1p150 and a shorter and constitutive ADAR1p110 truncated at the N-terminus, are known. A recent study indicates that both ADAR1p150 and ADAR1p110 shuttle between nucleus and cytoplasm (Fritz et al., 2009). We confirmed that both ADAR1p150 and p110 are located in the cytoplasm and nucleus of human cell lines (Figure S1A). ADAR2, known to localize mainly in the nucleus (Nishikura, 2010), and Dicer, known to localize in the cytoplasm (Krol et al., 2010), served as controls for the quality of extraction (Figure S1A).
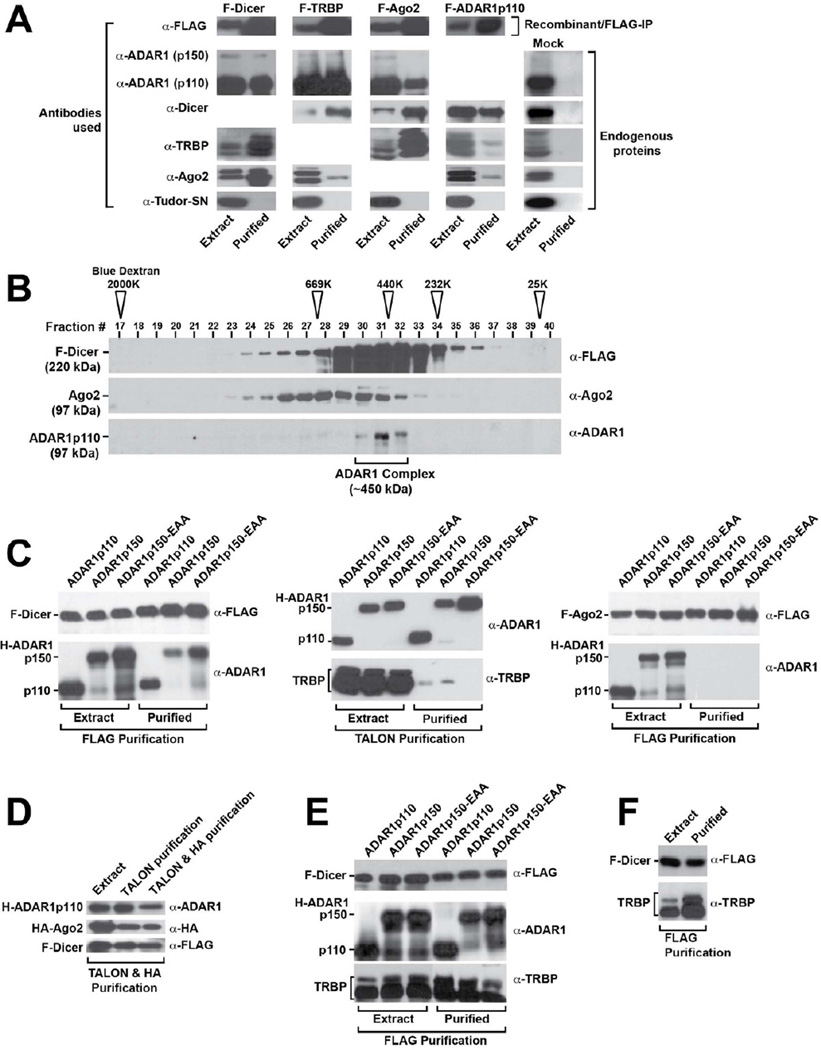
A-to-I editing is considered to take place mainly in the nucleus, and thus the cytoplasmic distribution of ADAR1 suggests that it may have functions separate from RNA editing, possibly in the RNAi mechanism (Nishikura, 2006). We investigated the interaction of the cytoplasmic ADAR1 with RISC component proteins. FLAG-epitope tagged RISC proteins; F-Dicer, F-TRBP, and F-Ago2, were all individually purified from permanently transformed HEK293 cell lines (Figure S1B). As expected, endogenous RISC partner proteins were detected in F-Dicer, F-TRBP, or F-Ago2 IP products (Figure 1A, left three columns). Most significantly, we found that each FLAG-tagged RISC member also pulled down endogenous ADAR1p110 (Figure 1A). ADAR1p150 showed lesser interaction, probably due to its low expression in HEK293 cells (Figure S1A). The reciprocal F-ADAR1p110 IP experiment pulled down endogenous Dicer as well as TRBP and Ago2 to a lesser extent (Figure 1A, F-ADAR1p110 column). As a negative control, all FLAG IP products were checked for interaction with endogenous Tudor-SN, a protein reported to promote cleavage of hyper-edited inosine containing dsRNAs (Scadden, 2005). We found no interaction with Tudor-SN, confirming that the interaction observed between ADAR1 and RISC proteins was specific (Figure 1A, bottom panels). Mock FLAG-IP experiments done with untransfected HEK293 cells indicated that none of endogenous Dicer, TRBP, Ago2, or ADAR1 contaminated the mock purification products (Figure 1A, Mock column), confirming that the endogenous ADAR1 was specifically pulled down with RISC member proteins. Similar F-ADAR2 IP experiments revealed no interaction of ADAR2 with RISC proteins (data not shown). Size exclusion chromatography of F-Dicer IP products revealed complexes of ∼450 kDa size consisting of endogenous ADAR1p110 proteins (Figure 1B). The expected size of a complex consisting of Dicer, ADAR1p110, and Ago2 is ∼450 kDa.
Figure 1. ADAR1 interacts with Dicer in an RNA binding-independent manner.
(A) Pulldown products of FLAG-tagged RISC member proteins and reciprocal pulldown of FLAG-tagged ADAR1 (F-ADAR1p110) purified from HEK293 cell extracts. Cytoplasmic extract (20 μg) and FLAG-IP peak eluate (15 μl) were examined by immunoblotting analysis. A mock FLAG-IP conducted with untransfected HEK293 cells was used to monitor the background levels of protein that may associate with the FLAG Ab resin (right panels). (B) F-Dicer IP products fractionated by Superose 6 gel filtration column chromatography were analyzed by immunoblotting. The positions of the molecular mass size markers are indicated by open arrowheads. (C) Analysis of recombinant protein interactions in the Sf9 cell co-infection/co-purification system. Various H-ADAR1 expression baculoviruses were co-infected with F-Dicer, untagged TRBP, or F-Ago2 expression viruses. FLAG IP isolated F-Dicer or F-Ago2, and immunoblotting determined their interaction with ADAR1. TALON purifications isolated each H-ADAR1s, and immunoblotting determined their interaction with TRBP. (D) Analysis of recombinant proteins purified from Sf9 cells triple-infected with F-Dicer, H-ADAR1, and HA-Ago2. Immunoblotting determined ADAR1 indirect interaction with Ago2 mediated by Dicer. (E) Analysis of recombinant proteins purified from Sf9 cells triple-infected with F-Dicer, H-ADAR1, and untagged TRBP. FLAG purifications isolated F-Dicer, and immunoblotting determined its relative interaction with ADAR1 and TRBP. (F) Analysis of recombinant proteins purified from Sf9 cells co-infected with F-Dicer and TRBP. FLAG purifications isolated F-Dicer, and immunoblotting determined its relative interaction with TRBP in the absence of ADAR1.
RNA-binding Independent ADAR1 Interaction with Dicer
While the above experiments showed strong interaction of ADAR1 with Dicer, they also showed lesser interaction with the other RISC components. We therefore used a recombinant baculovirus co-infection/co-purification system to more precisely characterize the interactions of ADAR1. A HAT (histidine affinity tag)-tagged ADAR1 (H-ADAR1) expression virus was co-infected in Sf9 cells with an F-Dicer, F-Ago2, or untagged TRBP expression virus, and the resulting complexes were purified by FLAG IP or His-based TALON affinity chromatography. Each type of affinity purification is completely specific for its cognate epitope tag, as demonstrated by mock purification control experiments (Figure S1C). We examined both ADAR1p110 and ADAR1p150 as well as a mutant ADAR1p150-EAA, which is devoid of dsRNA-binding due to point mutations (KKxxK®EAxxA) introduced in each of three dsRBDs (Valente and Nishikura, 2007). Co-purification of F-Dicer revealed its interaction with all three forms of ADAR1, including the ADAR1p150-EAA RNA-binding defective mutant (Figure 1C, left panels). The results established an RNA-binding independent interaction of ADAR1 with Dicer. In contrast, the amount of TRBP associated with H-ADAR1p110 or H-ADAR1p150 was substantially lower, and was virtually absent for H-ADAR1p150-EAA (Figure 1C, center panels). Finally, attempted co-purification of F-Ago2 and H-ADAR1 from co-infected Sf9 cells failed, indicating that they do not interact directly with each other (Figure 1C, right panels). These results demonstrate that the association of ADAR1 with TRBP and Ago2 is indirect, i.e., bridged by Dicer and/or dsRNA. We then purified ADAR1 associated complexes from Sf9 cells triple-infected with F-Dicer, H-ADAR1, and human influenza hemagglutinin-tagged Ago2 (HA-Ago2) expression viruses by first TALON column and then HA IP. The FLAG IP procedure was not used for purification of these complexes. Nevertheless, the purified complex contained not only H-ADAR1 and HA-Ago2 but also F-Dicer (Figure 1D), indicating that ADAR1 does form a triple complex with Dicer and also with Ago2, through its interaction with Dicer.
Finally, we examined whether the interaction of ADAR1 with Dicer would preclude interaction of TRBP with Dicer, or vice versa by analyzing FLAG IP products co-purified from Sf9 cells triple-infected with 1:1:1 ratio of F-Dicer, H-ADAR1, and untagged TRBP expression baculoviruses. ADAR1, including ADAR1p150-EAA RNA-binding defective mutant, and TRBP were co-purified with Dicer (Figure 1E). Since ADAR1 does not interact with TRBP (Figure 1C, center panels), ADAR1 and TRBP detected here in the F-Dicer pulldown products most likely represent proteins present in two separate Dicer complexes, one Dicer/ADAR1 and another Dicer/TRBP complexes. The results suggest that the presence (co-expression) of ADAR1 did not exclude formation of Dicer/TRBP complexes, and the presence (co-expression) of TRBP did not exclude formation of Dicer/ADAR1 complexes. FLAG-Dicer pulldown of TRBP conducted in the absence of ADAR1 (Figure 1F, co-infection) indicates that perhaps slightly more (but not significantly more) TRBP proteins can be complexed with Dicer if ADAR1 is not co-expressed as compared to TRBP proteins pulled down in the presence of ADAR1 (Figure 1E, triple infection). These results suggest that there is no extreme affinity difference between ADAR1 and TRBP for binding to Dicer.
The Domains Involved in the Dicer/ADAR1 Interaction
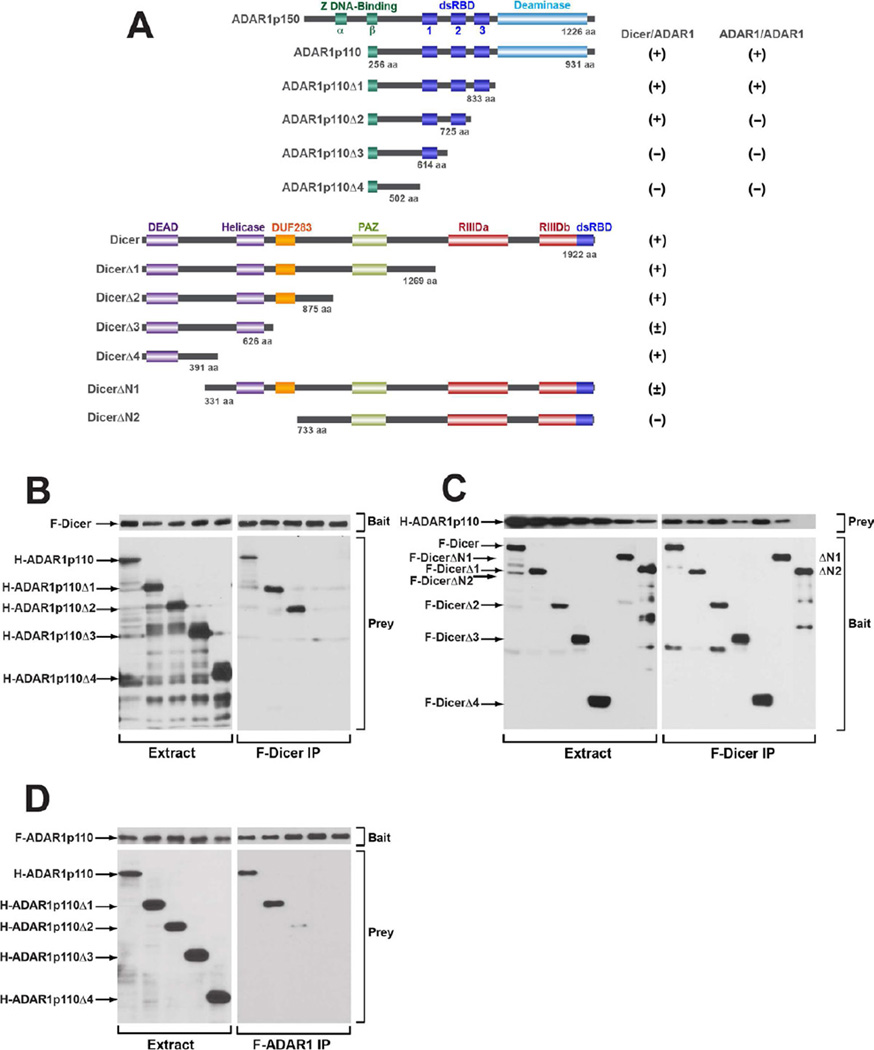
We examined the domains of ADAR1 and Dicer that are involved in their protein-protein interaction. ADAR1p150 contains two Z-DNA binding domains (α and β), three dsRBDs (dsRBD1, 2, 3), and a C-terminal deaminase domain. The N-terminus region containing the Z-DNA binding domain a is missing in ADAR1p110 (Figure 2A). Dicer contains a DEAD-box RNA helicase domain at the N-terminus region followed by DUF283 and PAZ domains, two catalytic RNase III domains, and a dsRNA-binding domain at the C-terminus (Figure 2A). The PAZ domain of Dicer likely binds to the 2-nt 3′ overhangs of miRNAs and siRNAs (Krol et al., 2010), whereas the DEAD-box RNA helicase domain may be involved in an auto-inhibition mechanism (Ma et al., 2008).
Figure 2. Domains involved in the Dicer/ADAR1 interaction.
(A) The domain structures of human ADAR1, Dicer, and their deletion mutants are shown. The extent of Dicer/ADAR1 or ADAR1 homodimer interaction is indicated; positive (+), weakly positive (±), or negative (−). (B) Mapping of the ADAR1 domain involved in the Dicer interaction. The full-length F-Dicer (bait) and full-length or one of the H-ADAR1p110 deletion mutants (prey) were co-infected in Sf9 cells. (C) Mapping of the Dicer domain involved in the ADAR1 interaction. The full-length or one of the F-Dicer deletion mutants (bait) and the full-length H-ADAR1 (prey) were co-infected in Sf9 cells. (D) Mapping of the ADAR1 domain involved in the ADAR1 homodimer interaction. The full-length F-ADAR1p110 (bait) and full-length or one of the H-ADAR1p110 deletion mutants (prey) were co-infected in Sf9 cells. (B, C) FLAG-IP purification was done for isolation of F-Dicer/H-ADAR1 interacting complexes. (D) FLAG-IP purification isolated F-ADAR1/H-ADAR1 interacting complexes. (B, C, D) FLAG-IP products were analyzed by immunoblotting. Left panel, input cell extracts. Right panel, FLAG-IP purified proteins.
Both ADAR1p150 and ADAR1p110 interacted with Dicer (Figure 1C), so ADAR1p110 was used for domain mapping analysis. C-terminal deletion constructs of H-ADAR1p110 and C-terminal deletion and N-terminal deletion constructs of F-Dicer were prepared (Figure 2A). These ADAR1 and Dicer deletion constructs were co-infected with the full-length partner construct in Sf9 cells. The FLAG-IP purification is in general cleaner than that of TALON purification. Therefore, we used the FLAG-IP procedure both for identification of H-ADAR1 deletion mutants interacting with F-Dicer (Figure 2B) and also for identification of F-Dicer deletion mutants interacting with H-ADAR1 (Figure 2C). The full length F-Dicer (bait) was purified efficiently (Figure 2B, right upper panel), and the full-length H-ADAR1, and H-ADAR1 Δ1 and H-ADAR1 Δ2 but not H-ADAR1 Δ3 and H-ADAR1 Δ4 mutants (prey) were pulled down by F-Dicer (Figure 2B, right lower panel), indicating that the second dsRBD (dsRBD2) of ADAR1 is involved in the interaction with Dicer. All F-Dicer mutants (baits) were purified efficiently via FLAG-IP (Figure 2C, right lower panel). While the full-length H-ADAR1 was co-purified and thus interacted robustly with the full length F-Dicer, and F-DicerΔ1, F-DicerΔ2, and F-DicerΔ4 mutants, the F-DicerΔ3 mutant lacking the DUF283 domain and F-DicerΔN1 lacking a part of the DEAD-box RNA helicase domain displayed substantially reduced (<4 fold) interaction with the H-ADAR1p110 (prey) (Figure 2C, right upper panel). Finally, the F-DicerΔN2 mutant lacking both DUF283 and DEAD-box RNA helicase domains lost its interaction with ADAR1 completely (Figure 2C, right upper panel), indicating that both domains are involved in the interaction with ADAR1.
We also determined the ADAR1 domains involved in the ADAR1 homodimer interaction by using the full-length F-ADAR1p110 (bait) and H-ADAR1p110 deletion mutants (prey), which revealed that the third dsRBD (dsRBD3) is required for the interaction (Figure 2D, right lower panel). Together, our results indicate that ADAR1 uses dsRBD2 and dsRBD3 differentially for formation of the Dicer/ADAR1 heterodimer and the ADAR1 homodimer complexes, respectively.
ADAR1 Increases the Rate of miRNA and siRNA Processing by Dicer
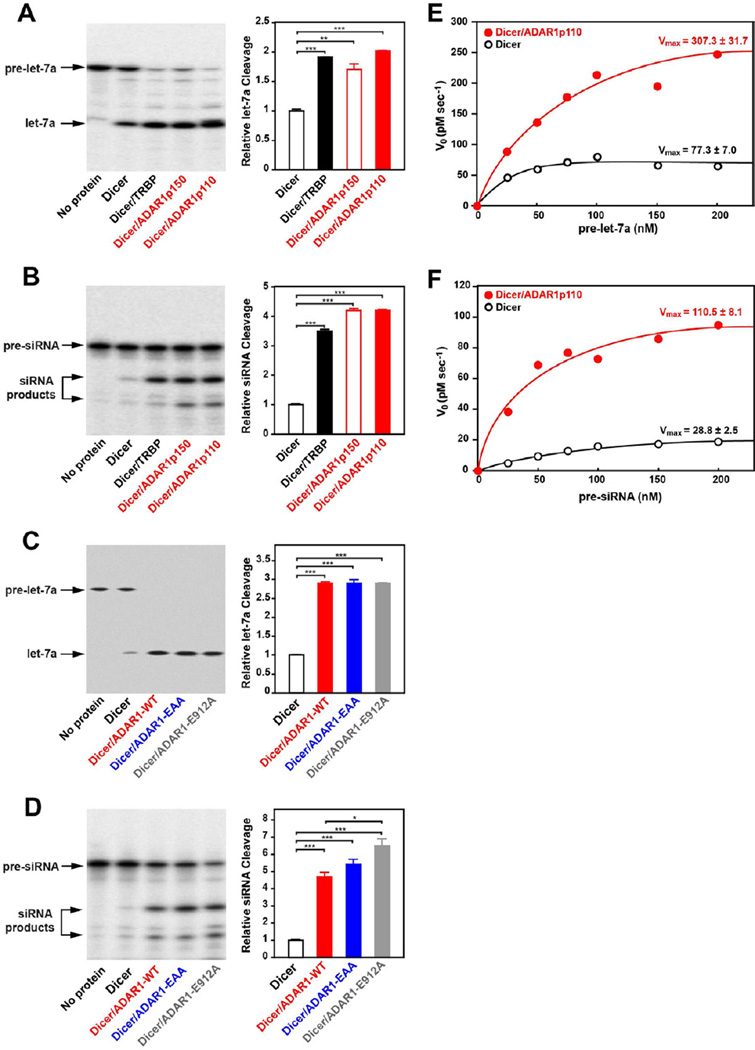
Although Dicer alone can cleave pre-miRNA to mature miRNA (pre-miRNA dicing reaction), its catalytic activity is known to be modulated by TRBP and PACT (Chendrimada et al., 2005; Haase et al., 2005; Lee et al., 2006). For instance, TRBP binds to Dicer and increases the maximum rate (Vmax) of the pre-miRNA dicing reaction (Chakravarthy et al., 2010). Accordingly, we investigated the effect of ADAR1 interaction with Dicer on the miRNA processing mechanism. We co-expressed F-Dicer with H-ADAR1p110 or H-ADAR1p150 in Sf9 cells, and the F-Dicer/H-ADAR1 complex was purified by consecutive TALON and FLAG affinity chromatography (Figure S2A), which resulted in isolation of the Dicer/ADAR1 complex from free F-Dicer or H-ADAR1 forms (Figure S2B). We also prepared F-Dicer alone and the F-Dicer/H-TRBP complex (Figure S2C). Dicer alone, Dicer/TRBP, and Dicer/ADAR1 complexes were subjected to in vitro Dicer cleavage assay using pre-let7a as substrate (Figure S3A). We also examined a 35-bp completely complementary dsRNA containing 3’ 2 nt overhangs as a different type of Dicer substrate, which can be processed into ∼22 bp small interfering RNAs (siRNAs) and used as an siRNA precursor (pre-siRNA) (Figure S4A). We found that the Dicer complexed with ADAR1p150 or ADAR1p110 did cleave both pre-let-7a and pre-siRNA into mature let-7a (Figure 3A) and siRNA (Figure 3B). Furthermore, cleavage of pre-let-7a and pre-siRNA was substantially increased, 2 and 4 fold, by both ADAR1p150 and ADAR1p110. This ADAR1 stimulation of Dicer cleavage activities was equivalent to that of TRBP (Figure 3A, 3B). In order to understand how ADAR1 augments the Dicer cleavage, we prepared Dicer/ADAR1 complexes consisting of either ADAR1p110-EAA or ADAR1p110-E912A mutant. As already mentioned, ADAR1p110-EAA cannot bind any (long and short) dsRNA (Valente and Nishikura, 2007). ADAR1p110-E912A has no deaminase activity because of a point mutation of Glu912 to Ala (Lai et al., 1995). The glutamate residue is involved in the proton transfer function of the enzyme. The Dicer/ADAR1-E912A complex, just as the Dicer/ADAR1-WT complex, diced pre-let-7a (3 fold) and also pre-siRNA (6 fold) more efficiently as compared to the Dicer alone control (Figure 3C, 3D), indicating that the ADAR1 catalytic activity is not required for augmentation of the Dicer cleavage reaction. To our surprise, however, we found that the complex consisting of ADAR1-EAA mutant also increased Dicer cleavage of pre-let-7a and pre-siRNA (Figure 3C, 3D). The result indicates that promotion of the Dicer cleavage reaction by ADAR1 is not mediated via direct ADAR1 binding to the substrate RNAs (pre-miRNA or pre-siRNA), but more likely results from conformational changes induced in Dicer by ADAR1.
Figure 3. ADAR1 augments the Dicer cleavage reaction rate and increases miRNA/siRNA production.
(A, C) In vitro miRNA processing analysis. (B, D) In vitro siRNA processing analysis. (A, B, C, D) The Dicer cleavage reaction was done under single-turnover conditions. (E) Michaelis-Menten analysis of the pre-let-7a cleavage reaction. (F) Michaelis-Menten analysis of pre-siRNA cleavage reaction. (E, F) The Dicer cleavage reaction was conducted under multiple-turnover conditions. V0 values were plotted against substrate concentrations, leading to estimation of Vmax values.
To assess more quantitatively the facilitative effects of ADAR1, steady-state enzyme kinetics analysis was conducted using Dicer alone and Dicer/ADAR1p110. The Dicer cleavage reaction was carried out under multiple turnover conditions, in which the substrate concentration is in molar excess over the enzyme, i.e., Dicer. First order reaction rate (V0) values were obtained through time course analysis of Dicer cleavage products of pre-let-7a (Figure S3B) and pre-siRNA (Figure S4B). Analysis of Michaelis-Menten plots made at various substrate concentrations gave estimates of the maximum rate (Vmax) values of the pre-let-7a cleavage reaction; 77.3 and 307.3 pM/sec (Figure 3E) and of the pre-siRNA cleavage reaction; 28.8 and 110.5 pM/sec (Figure 3F) for Dicer alone and Dicer/ADAR1, respectively, indicating that ADAR1 increases the Vmax of the Dicer cleavage of both pre-let-7a and pre-siRNA by 4-fold in comparison to the reaction conducted with Dicer alone. Our results suggest that the RNA editing enzyme ADAR1 affects the turnover rate of Dicer and increases substantially the overall efficacy of miRNA and siRNA processing.
ADAR1 Augments RISC Loading and Target RNA Silencing Efficacy
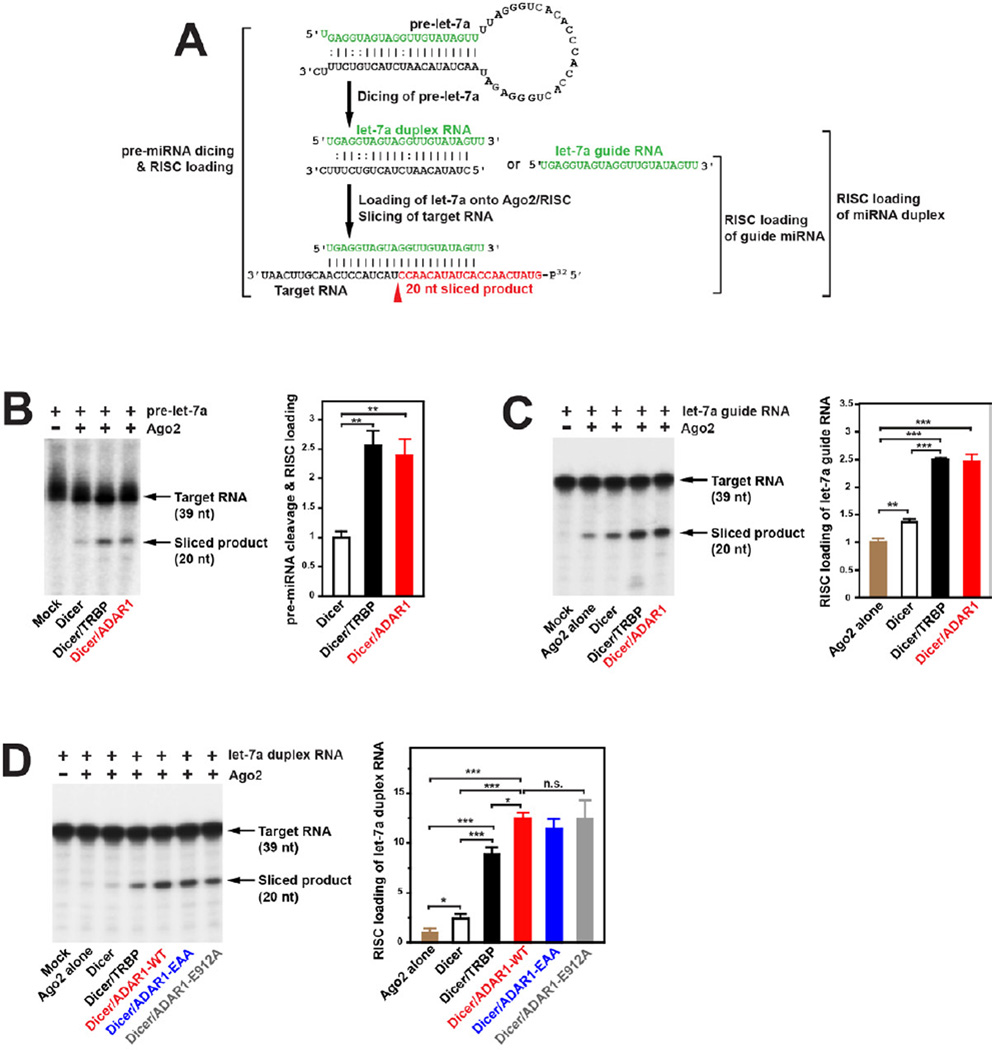
We next investigated whether ADAR1 would facilitate the overall target RNA silencing. It is possible that miRNA or siRNA cleaved by the Dicer/ADAR1 complex may not be loaded onto a functional RISC, resulting in accumulation of abortive miRNA or siRNA. We, therefore, first examined the effects of ADAR1 on loading of let-7a RNAs cleaved from pre-let-7a by the Dicer/ADAR1 complex and subsequent silencing of its target RNA (Figure 4A, pre-miRNA dicing & RISC loading assay). Dicer, Dicer/TRBP, or Dicer/ADAR1 was pre-incubated with F-Ago2 followed by addition of pre-let-7a for RISC assembly and further incubated with the cognate target RNA for slicing. We found a 2.4 fold increase in the sequence specific Ago2 slicing of the target RNA by ADAR1p110 (stimulation equivalent to TRBP) (Figure 4B). Having established that let-7a RNAs diced by the Dicer/ADAR1 are functional, we next examined the effects of ADAR1 on two separate RISC activities previously reported (Gregory et al., 2005): loading of the single-stranded guide miRNA or loading of the already diced miRNA duplex (Figure 4A). We found that ADAR1 facilitates the guide RNA loading by 1.7–2.5 fold (Figure 4C) and also the miRNA duplex loading by 5–12.5 fold (Figure 4D) as compared to Ago2 alone or Dicer/Ago2. The magnitude of the ADAR1 effects was again very similar to that observed with TRBP (Figure 4C, 4D). For the miRNA duplex loading assay, we tested also two ADAR1 mutants, ADAR1-EAA and ADAR1-E912A. These mutants exhibited activities equivalent to those of wildtype ADAR1 (Figure 4D), indicating again that neither the dsRNA-binding nor deaminase activities of ADAR1 are required for miRNA duplex loading. Together with the fact that ADAR1 forms the Dicer/ADAR1/Ago2 triple complex (Figure 1D), ADAR1 appears to function in augmentation of Dicer cleavage of pre-miRNA/pre-siRNA, RISC assembly, and loading of miRNA, as does TRBP (Chendrimada et al., 2005; Gregory et al., 2005).
Figure 4. ADAR1 augments RISC loading and target RNA silencing efficacy.
(A) RISC loading and assays using pre-let-7a, let-7a guide, or let-7a duplex RNA are schematically shown. The let-7a-5p target RNA (39 nt) was 5’ 32P-labeled. Ago2 slicing of the target RNA guided by let-7a-5p would produce a 20-nt product (highlighted in red). (B) Pre-let-7a dicing-coupled RISC loading assay: 10 nM Ago2 used. (C) RISC loading of let-7a guide RNA: 5 nM Ago2 used. (D) RISC loading of let-7a duplex RNA: 5 nM Ago2 used. (B, C, D) Sliced target RNA products were fractionated by 15% Urea-PAGE (left panel). A quantitative summary is also shown (right panel). The target RNA slicing efficiency was estimated by the ratio of the radioactivity of the correctly cleaved band over that of the uncut control band. Significant differences were identified by Student’s t-tests: *, p<0.05; **, p<0.01; ***, p<0.001. Error bars, S.E. (n = 3).
Differentiation of ADAR1 Functions by Selective Formation of Homo- and Hetero-dimer Complexes
How does ADAR1 differentiate its two roles, one as an RNA editing enzyme and the other as an RNAi machinery component? SDS-PAGE and colloidal blue staining of the F-Dicer/H-ADAR1p110 complex, which was purified through consecutive affinity columns (Figure S2A), revealed only two bands of Dicer and ADAR1, in approximately a 1:1 ratio (Figure S2B). We then examined the molecular mass of the complex by FPLC size fractionation chromatography. The Dicer/ADAR1 complex eluted around ∼330 kDa (Figure S2D). This is consistent with a complex containing one molecule each of Dicer (219 kDa) and ADAR1 (110 kDa), but not with one containing Dicer plus an ADAR1 homodimer. As we already shown, the ADAR1-E912A deamination defective mutant can form the Dicer complex and function as efficient as the wildtype ADAR1 (Figure 3C, 3D). This raises the question of whether ADAR1 complexed with Dicer remains active as an editor.
To test this, we examined processing of pre-miR-151 by the Dicer/ADAR1 heterodimer. Prior to processing of pre-miR-151 to mature miRNA, ADAR1 edits a so called +3 site of pre-miR-151 with ∼90% efficiency and the −1 site with lesser efficiency (Figure S5A) (Kawahara et al., 2007a). We confirmed that the Dicer/ADAR1 complex efficiently diced pre-miR-151 and processes it to mature miR-151 RNAs, whereas the ADAR1 homodimer complex did not have the miRNA processing activity (Figure S5B). Unprocessed pre-miR-151 and mature miR-151 RNAs generated by the miRNA processing assay were then separated by gel purification and examined for A-to-I RNA editing by sequencing of the cDNA clones derived from RT-PCR products. Neither any pre-miR-151 nor mature miR-151 RNAs was edited (Figure S5C). The results indicate that ADAR1 complexed with Dicer acts as an RNAi machinery component but has no A-to-I RNA editing activity, because the latter requires homodimerization of ADAR1 (Cho et al., 2003). Our results revealed how ADAR1 differentiates its RNA editor and RNAi functions: forming two different complexes, ADAR1 homodimers and Dicer/ADAR1 heterodimers, respectively (Figure S6).
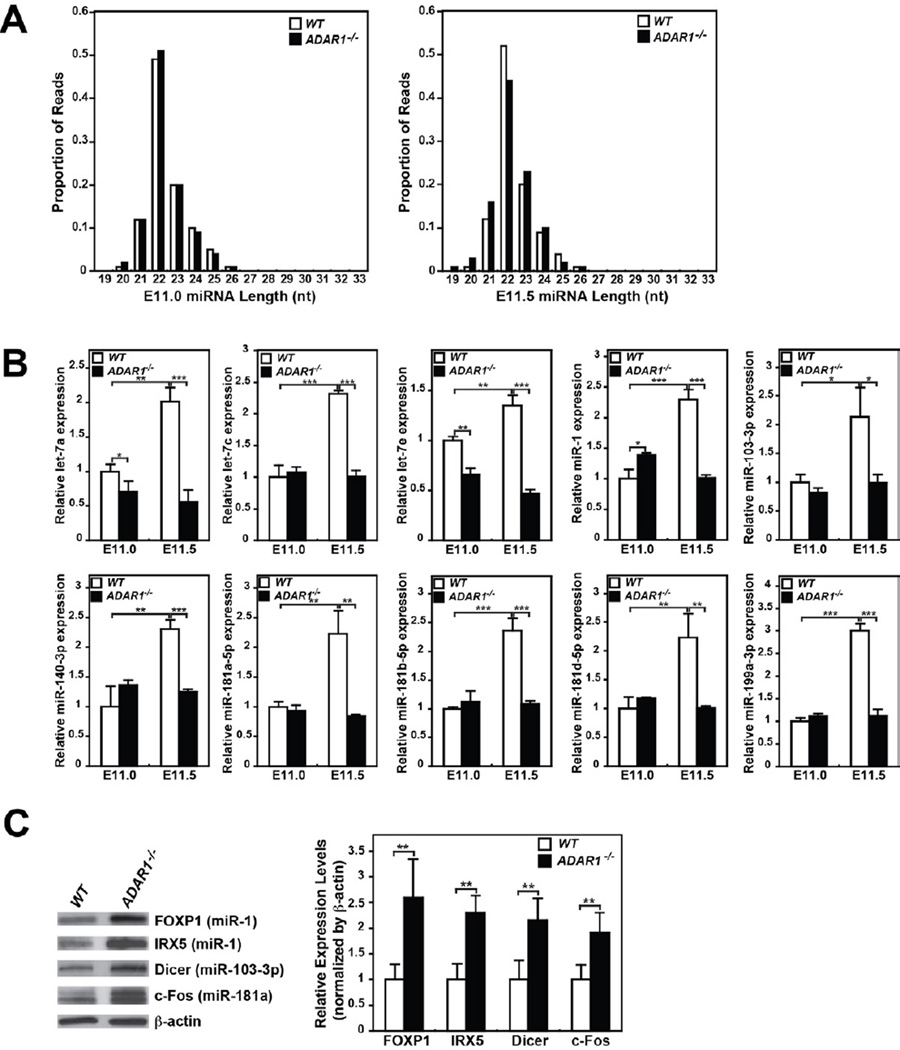
Global Suppression of miRNA Production in ADAR1−/− Mouse Embryos
To assess the in vivo effects of ADAR1 on miRNA processing, we investigated global miRNA expression in ADAR1−/− mouse embryos, which die around E12 (Wang et al., 2004). Total RNA samples were prepared from ADAR1−/− and control wildtype mouse embryos collected live at E11.0 and E11.5. Small RNAs in the miRNA size range were examined by deep sequencing (Table S1, Table S2A). Size distribution of miRNA reads, peaking at 22 nt, did not differ among wildtype and ADAR1−/− embryos at E11.0 and E11.5 (Figure 5A). At E11.0, the ADAR1 knockout had no major effect on miRNA read numbers (Table S1), but a significant decrease of miRNA reads was observed in ADAR1−/− embryos at E11.5 as compared to control wildtype embryos (Table S1, Table S2B). An increased number of reads was observed for other class small RNAs of E11.5 ADAR1−/− embryos, i.e. rRNA as well as snoRNA and snRNA related small RNA, which could have affected the normalized miRNA read numbers (Table S1). To confirm the reduced miRNA expression in ADAR1−/− embryos independently and more quantitatively, we determined the expression of representative miRNAs highly expressed in wildtype embryos by quantitative RT-PCR. We spiked a control RNA in RNA samples, which allowed us to obtain absolute miRNA concentrations. We found that concentrations of the miRNAs examined were indeed reduced by 2 to 3 fold in E11.5 ADAR1−/− embryos (Figure 5B), confirming the trend predicted by deep sequencing and reflecting more accurately the changes in miRNA. Furthermore, qRT-PCR analysis revealed that the expression of miRNAs dramatically increased in wildtype embryos during the half-day period from E11.0 to E11.5 (e.g. let-7a, miR-1, and miR-181a-5p), whereas their expression remained at the E11.0 level in E11.5 ADAR1−/− embryos (Figure 5B). Together, our findings suggest that ADAR1 interacts with Dicer and promotes rapid increase of miRNA production globally around E11–12, which is likely essential for embryo development.
Figure 5. Reduced expression of miRNAs and upregulation of their target genes inADAR1−/− mouse embryos at E11.5.
(A) Distribution of miRNA deep sequencing read lengths was compared between wildtype and ADAR1−/− mouse embryos at E11.0 and E11.5 stages. (B) Quantitation of select miRNAs expressed in E11.0 and E11.5 embryos. A spike control, 2 × 1010 copies of a chemically synthesized RNA, was added to the RT reaction mixture. Quantitative PCR was conducted in quadruplicate. The relative miRNA expression level standardized by the spiked RNA is presented as the expression level relative to that of the wildtype embryo at E11.0. Significant differences were identified by Student’s t-tests: *, p<0.05; **, p<0.01; ***, p<0.001 Error bars, S.D. (n = 5). (C) Increased protein expression of the genes targeted by the miRNAs reduced in ADAR1−/− embryos. Immunoblotting analysis of select target gene levels in E11.5 embryos (left) and a quantitative summary (right). Significant differences were identified by Student’s t-tests: **, p<0.01. Error bars, S.D. (n = 5).
Finally, we determined the protein expression level of genes already validated as targets of the miRNAs, which had reduced expression (Figure 5B). We expect that reduction in the synthesis of miRNAs should result in increased protein expression level of their target genes. We tested FOXP1 and IRX5 as miR-1 targets (Nasser et al., 2008; Zhao et al., 2007), Dicer as a miR-103-3p target (Martello et al., 2010), and c-Fos as a miR-181a target (Wu et al., 2012). As expected, increased target gene protein levels (2–2.5 fold) were indeed noted in E11.5 ADAR1−/− embryos (Figure 5C). Globally reduced miRNA synthesis is thus likely to have significant effects on expression of their target genes in ADAR1−/− embryos.
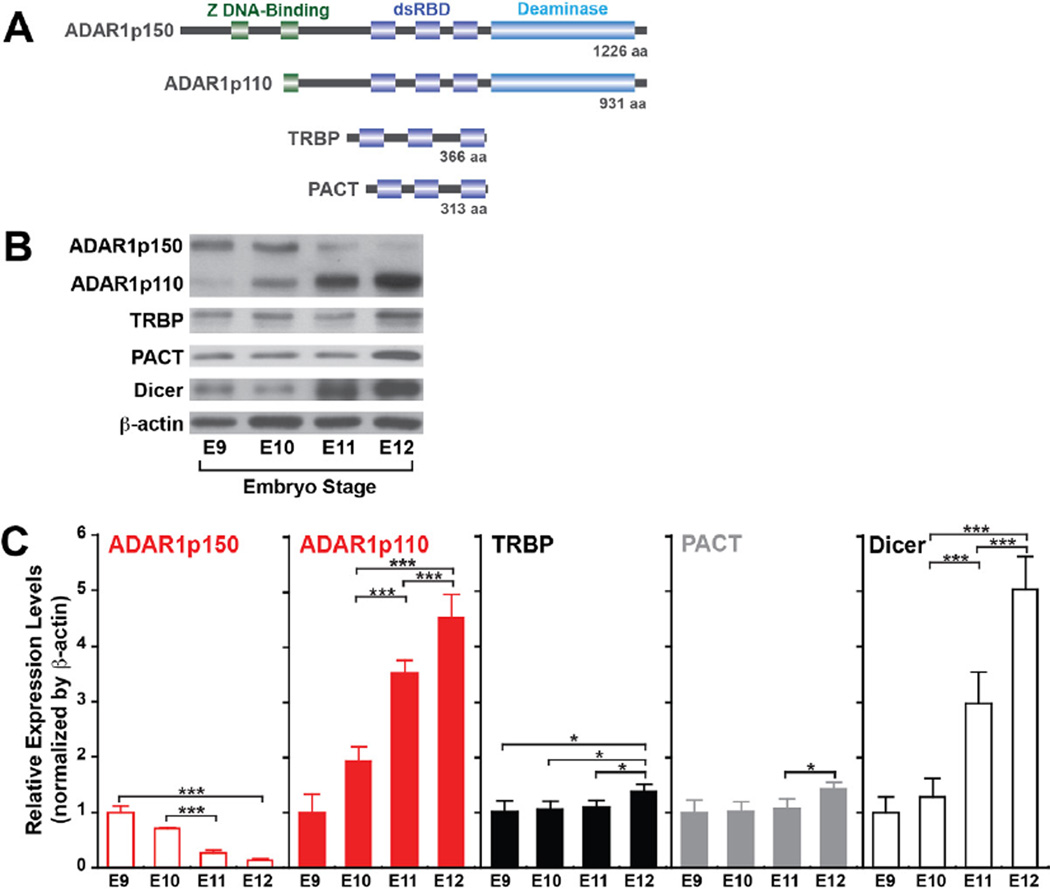
Concomitant Upregulation of Dicer and ADAR1 in E11-E12 Mouse Embryos
The dramatic increase of miRNA expression detected in E11.5 wildtype embryos (Figure 5B) suggests a possible upregulation of the pre-miRNA processing machinery during this particular period of embryo development. We therefore investigated the expression levels of Dicer, ADAR1, and TRBP and PACT, two dsRNA binding proteins known to enhance Dicer activity (Figure 6A) (Chakravarthy et al., 2010; Lee et al., 2006). We found that Dicer levels indeed increased significantly at E11–12 as compared to those at E9–10 (3–5 fold). Most importantly, ADAR1p110 expression also increased (4–5 fold), concomitantly with Dicer (Figure 6B, 6C). Interestingly, the ADAR1p150 inversely decreased around this time, possibly indicating a special function of this ADAR1 isoform in even earlier embryos, such as maintenance of hematopoietic stem cells as reported previously (Hartner et al., 2009). In contrast to the dramatic upregulation of Dicer and ADAR1p110, only a very modest increase in the TRBP and PACT expression was noted at E12 (Figure 6B, 6C). Our findings suggest that a dramatic increase in the pre-miRNA processing capability occurs in E11–12 embryos, caused by concomitant upregulation of Dicer and ADAR1p110, which is likely to be required at these stages of rapidly growing embryos. Interestingly, we found that the Dicer protein level increased by ∼2 fold in E11.5 ADAR1−/− embryos (Figure 5C), possibly reflecting the presence of a feedback mechanism to attempt to compensate the loss of ADAR1. Alternatively, the increased expression may reflect the reduced expression of miRNAs, which usually target Dicer, e.g., miR-103-3p (Figure 5C) (Martello et al., 2010). In any case, the results suggest that the loss of ADAR1 apparently cannot be compensated by the increased Dicer alone in upregulation of miRNA synthesis. ADAR1 perhaps plays a major role in the miRNA synthesis and miRNA-mediated gene silencing mechanisms that may exceed that of TRBP or PACT, at least during this period of embryonic development.
Figure 6. Dicer and ADAR1 are rapidly induced in E11–12 stage embryos.
(A) The domain structures of two ADAR1 isoforms, TRBP, and PACT. (B) Immunoblotting analysis of proteins involved in the pre-miRNA processing mechanism. Wildtype mouse embryos collected live at various embryonic stages (E9.0–12.0), and the extracted protein was fractionated by 4–15% SDS-PAGE and tested for immunoblotting analysis. (C) Quantitative summary of immunoblotting analysis results. The relative expression level was normalized by β-actin and presented as the expression level relative to that of the E9.0 embryo. Significant differences were identified by Student’s t-tests: *, p<0.05; ***, p<0.001. Error bars, S.D. (n = 4).
Upregulation of Genes Predicted as Targets of miRNAs Suppressed in ADAR1−/− Mouse Embryos
Having established a new function of ADAR1 in the miRNA biogenesis mechanism, we anticipated that suppression of many miRNAs is likely to affect their target gene expression. We therefore next examined the global gene expression patterns in E11.5 ADAR1−/− embryos by deep sequencing analysis of the transcriptome (Table S3A). A dynamic change in the gene expression pattern, i.e., some up and others down, not necessarily a systemic shut down of all genes, was detected in E11.5 ADAR1−/− embryos compared to wildtype embryos (Figure S7A, Table S3B). We asked whether upregulation of some genes could be the result of downregulation of their relevant miRNAs. We first identified the miRNAs for which read numbers were reduced more than 4-fold (Table S2B) and the gene transcripts upregulated more than 2-fold in E11.5 ADAR1−/− embryos (Figure S7B, S7C, Table S3B). In silico target prediction analysis revealed that a large fraction of the upregulated genes were indeed targets of the downregulated miRNAs (miRNA-target pairs) (Figure S7A, Table S3C). We found that these genes are enriched in many different functions and pathways such as cell death, cell proliferation, hematopoiesis, and cardiovascular system development (Table S3C). We already showed upregulation of select target gene protein expression (Figure 5D). Together, our results indicate that ADAR1 plays an important role in facilitating Dicer cleavage and increasing the expression of miRNA, which in turn controls coordinated gene expression involved in many different biological functions and pathways during development.
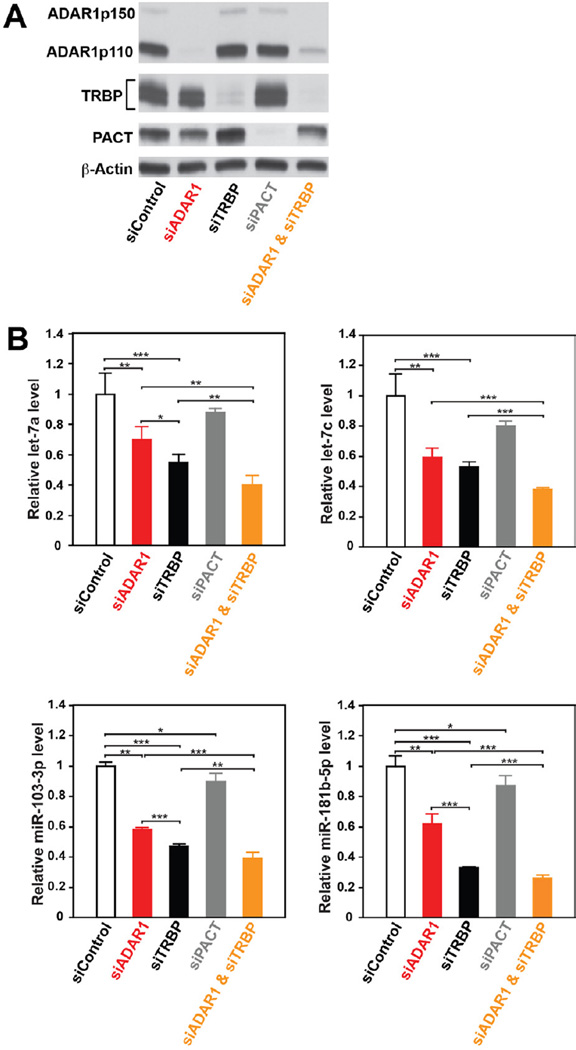
Effects of ADAR1 on Synthesis of miRNAs in Cultured Cells
Having established ADAR1 RNAi functions in developing mouse embryos, we next examined the ADAR1 effects on miRNA synthesis in cultured cells. We carried out single or combination knockdowns of ADAR1, TRBP, and PACT (Figure 7A) and quantitated select miRNA expression levels in HeLa cells (Figure 7B). The four miRNAs monitored have no A-to-I editing sites (Kawahara et al., 2008), excluding the possibility of their expression being affected by the ADAR1 editing function. We found that PACT knockdown had very little effects on the monitored miRNA expression, whereas TRBP knockdown reduced miRNA levels by 40–70%, as reported previously (Koscianska et al., 2011). ADAR1 knockdown caused a significant reduction (30–40 % reduction), slightly less than TRBP. Knockdown of both ADAR1 and TRBP generated an additional 10–20 % suppression effect as compared to TRBP single knockdown (Figure 7B). We investigated the effects of ADAR1 on miRNA synthesis also by deep sequencing of miRNAs expressed in HeLa cells treated for ADAR1 knockdown (Table S4). A global reduction of miRNA synthesis was noted in ADAR1 knockdown HeLa cells as compared to the control siRNA treated HeLa cells. The extent of the reduction was similar to that expected from the qRT-PCR analysis done for selected miRNAs. Our results indicate that ADAR1 does have an impact on miRNA synthesis also in cultured cells.
Figure 7. Effects of ADAR1 on RNAi efficacy in HeLa cells.
(A) Immunoblotting analysis of ADAR1, TRBP, and PACT protein levels in HeLa cells 72 hrs after transfection with ADAR1, TRBP, PACT, and ADAR1/TRBP siRNAs. (B) Quantification of selected miRNA expression levels by qRT-PCR. The relative expression level was normalized by β-actin and presented as the expression level relative to that of HeLa cells treated with control siRNA. Significant differences were identified by Student’s t-tests: *, p<0.05; **, p<0.01***, p<0.001 Error bars, S.D. (n = 4).
DISCUSSION
The antagonistic interaction of A-to-I editing with RNAi, i.e., competitive inhibition of the RNAi pathway by A-to-I editing of dsRNA substrates, has been reported previously. A-to-I editing of these dsRNAs results in their structural alterations (reduction in double-strandedness) and reduced production of end-siRNAs (Wu et al., 2011) or functional miRNAs (Iizasa et al., 2010; Kawahara et al., 2008; Kawahara et al., 2007a; Kawahara et al., 2007b; Yang et al., 2006), leading to suppression of the RNAi efficacy (Figure S6, left panel).
In this study, we demonstrated the presence of a completely different type of interaction (stimulative interaction) between RNA editing and RNAi mechanisms through formation of Dicer/ADAR1 complexes (Figure S6, right panel). Mass spectrometric analyses of the proteins associated with Dicer have previously identified ADAR1 as one of putative Dicer interacting-proteins, although the mode and biological significance of their association was not investigated (Landthaler et al., 2008). Size fractionation column chromatography analysis of the Dicer/ADAR1 complex indicated that monomeric ADAR1 associates with Dicer in a 1:1 ratio. Most importantly, our studies have demonstrated that ADAR1 in the complex positively controls the activity of Dicer by increasing Vmax of pre-miRNA or pre-siRNA cleavage reaction, thereby generating significantly more mature miRNA or siRNA. We have also demonstrated that miRNAs generated by the Dicer/ADAR1 complex are functional, i.e., they are loaded onto Ago2/RISC and capable of silencing the target RNA (Figure S6, right panel).
Involvement of certain dsRBDs in RNA binding-independent protein-proetin interaction has been reprted (Haase et al., 2005; Hitti et al., 2004). We previously showed that the second dsRBD of ADAR1, but not the first or third, is dispensable for A-to-I editing activity of ADAR1 (Lai et al., 1995). Here we showed that the second dsRBD of ADAR1 is required for its RNA binding-independent interaction with Dicer. Both the DUF283 domain and the N-terminal half of the DEAD-box RNA helicase domain of Dicer seem to be involved in the interaction with ADAR1. The very similar N-terminal region of Dicer has been reported to be involved in the RNA binding-independent Dicer/TRBP interaction via the third dsRBD of TRBP (Daniels et al., 2009; Haase et al., 2005), revealing that ADAR1 and TRBP utilize the same DEAD-box RNA helicase domain for their interaction with Dicer. Recent computational studies revealed a significant structural similarity of the DUF283 domain to the dsRBD domain αβββα folding) (Dlakic, 2006). The involvement of the DUF283 domain in the Dicer/ADAR1 complex but not in the Dicer/TRBP complex may be related to the fact that the second dsRBD (dsRBD2) of ADAR1 is utilized instead of the third dsRBD (dsRBD3) being used by TRBP (Figure 6A), leading to a different conformational change induced in Dicer by ADAR1. The exact function of the DUF283 domain uniquely involved in the Dicer/ADAR1 complex formation remains to be established.
The pre-miRNA cleavage and RISC loading experiments using ADAR1 mutants defective in dsRNA binding or A-to-I deamination revealed that dsRNA binding and deaminase activities of ADAR1 are not required for the mechanism that increases the rate of pre-miRNA cleavage or for the mechanism that facilitates RISC loading. We demonstrated that ADAR1 in the Dicer complex is not competent for A-to-I editing. Thus, augmentation of the Dicer activity by the ADAR1 deamination defective mutant was anticipated. However, it was surprising to find that the dsRNA-binding defective ADAR1 mutant facilitated both pre-miRNA cleavage and RISC loading steps just as well as wildtype ADAR1, indicating that no direct binding of ADAR1 to pre-miRNAs or already diced miRNA duplexes is involved in the Dicer activity augmentation mechanisms. TRBP complexes with Dicer via its third dsRNA binding domain (dsRBD3) and increases the Dicer cleavage activity (Daniels et al., 2009; Haase et al., 2005), but deletion of the other two dsRBDs (dsRBD1 and dsRBD2) abolished its Dicer augmentation activity, leading to proposal of a model in which direct binding of TRBP to pre-miRNA substrates via dsRBD1 and dsRBD2 increases the initial substrate recognition rate (Chakravarthy et al., 2010). However, the direct binding of TRBP within the Dicer/TRBP complex to pre-miRNAs has never been demonstrated (Chakravarthy et al., 2010). In view of the dsRNA-binding independent ADAR1 effects, conformational changes induced in Dicer by ADAR1 are likely to underlie the mechanism that upregulates the Dicer function. TRBP has been reported to increase significantly the stability of Dicer (Chendrimada et al., 2005). We found increased Dicer protein level in E11.5 ADAR1−/− embryos as compared to wildtype embryos (Figure 5C), which rules out any role for ADAR1 in the Dicer stabilization. Interestingly, the DEAD-box RNA helicase domain has been reported to autoinhibit the catalytic efficiency of Dicer (Ma et al., 2008), suggesting the possibility that ADAR1 stimulates the Dicer activity (pre-miRNA cleavage and RISC loading) through its binding to this region, releasing the auto-inhibitory effects of the DEAD-box RNA helicase domain.
It was surprising to find that ADAR1 with its well known function in the A-to-I editing mechanism has a completely different persona as an RNAi stimulating regulator like TRBP and PACT. The formation of a homodimer complex of ADAR1 mediated via the third dsRBD (Figure 2D) is essential for its enzymatic activity (Cho et al., 2003), and the monomeric ADAR1 in the Dicer/ADAR1 complex thus lacks A-to-I deamination activities as we demonstrated in this study. Thus, ADAR1 seems to differentiate its two roles in the A-to-I RNA editing and RNAi mechanisms by forming two different types of complexes, ADAR1 homodimers and Dicer/ADAR1 heterodimers (Figure S6). However, we do not know the exact mechanism that determines the balance of the two functions. Since the RNAi function of ADAR1 is exerted in the cytoplasm, cytoplasmic vs nuclear distribution and concentration of ADAR1 as well as concentrations of TRBP or PACT in developing embryos, various tissues, and cell lines may be parameters that determine the ratio of ADAR1 homodimer and Dicer/ADAR1 heterodimer formation. We previously reported that editing occurs in approximately 20% of pri-miRNAs in human brains and often inhibits their processing to mature miRNAs (Kawahara et al., 2008). Despite a significant increase in the ADAR1 expression level (Figure 6B, 6C), we found that A-to-I editing of pri-miRNAs is very limited in E11–12 wildtype mouse embryos (Y. Kawahara and K. Nishikura, unpublished studies). Thus, the ADAR1 function may be more skewed toward an RNAi regulator than an RNA editor in growing embryos (Figure S6).
Inactivation of the mouse ADAR1 gene results in widespread apoptosis and death of embryos at around E12 (Wang et al., 2004). Aberrant activation of interferon signaling pathways has been suggested as the underlying mechanism (Hartner et al., 2009). In contrast to the severe ADAR1−/− mouse phenotype, only mild phenotypic alterations, i.e., growth retardation and male sterility for TRBP−/− mutant mice (Zhong et al., 1999) and defective ear development and hearing loss for PACT−/− mice (Rowe et al., 2006) have been reported. These results may indicate a more important contribution of ADAR1 than TRBP and PACT in the miRNA-mediated RNAi, especially in E11–12 embryos. The expression of ADAR1 (p110 isoform) is rapidly induced around this time, concomitantly with Dicer, in contrast to the delayed and modest increase of TRBP and PACT expression revealed in this study (Figure 6B, 6C). In ADAR1−/− embryos, the rapid and significant upregulation of miRNA production can not occur because of the lack of formation of the Dicer/ADAR1 complex. This seems to result in dysregulated expression of many genes, which are otherwise repressed by these miRNAs during normal development of wildtype embeyos (Figure S7). It is tempting to speculate that deficiency in the RNAi function rather than RNA editing function of ADAR1 underlies the ADAR1−/− mouse phenotype.
EXPERIMENTAL PROCEDURES
Mice
The ADAR1−/− mouse line (Wang et al., 2004), back crossed more than 10 times to C57BL/6J mice, was used for Small RNA-seq, RNA-seq, and qRT-PCR analyses as well as protein expression analyses. Animal protocols for breeding and preparation of embryonic RNA and protein samples were approved by the Institutional Animal Care and Use Committee of The Wistar Institute and performed in accordance to the US National Institutes of Health Guidelines.
Dicer Cleavage Assay
The pre-let-7a RNA substrate for Dicer cleavage assay was synthesized by in vitro transcription of a plasmid template containing a double ribozyme system (plasmid, a gift from Dr. J. Doudna) (Ferre-D'Amare and Doudna, 1996). The 35-bp duplex RNA used as a pre-siRNA substrate was made by annealing pre-siRNA sense and antisense RNAs, which were synthesized by IDT. Pre-let-7a and the sense strand of pre-siRNA were 5’ end-labeled using T4 polynucleotide kinase (New England Biolabs) and (γ-32P)-ATP. The sequences of the substrates: pre-let-7a; 5′-UGAGGUAGUAGGU UGUAUAGUUUUAGGGUCACACCCACCACUGGGAGAUAACUAUACAAUCUACUGUCUUAC C-3′, pre-siRNA sense; 5′-UGAGGUAGUAGGUUGUAUAGUUUGAAAGUUCACGAUU-3’, pre-siRNA antisense; 5’-UCGUGAACUUUCAAACUAUACAACCUACUACCUCAAA-3’.
In vitro Dicer cleavage assays were done in a 40 µl reaction mixture containing 20 mM PIPES pH6.5, 1.5 mM MgCl2, 80 mM NaCl, 1 mM DTT, 10% glycerol, 5 nM Dicer, and 5’-end 32P-labeled substrate (pre-let-7a or pre-siRNA) at concentrations of 25, 50, 75, 100, 150 and 200 nM. Reactions were incubated at 37 °C, and 5 µl aliquots were taken after 0, 2, 5, 10, 20 and 40 minutes. The reactions were stopped by adding 1.2 volumes of Gel Loading Buffer II (Ambion). After heating at 75°C for 10 min, the samples were analyzed by 15% Urea-PAGE, and quantified using MolecularDynamics and ImageQuant (GE Healthcare). Initial velocities (vo) at each substrate concentration (S) were determined by linear regression, using Kaleidagraph (Synergy Software). Vmax along with standard errors were calculated by fitting to the Michaelis-Menten equation, vo = (Vmax × S)/(Km + S), using Graphpad Prism (Graphpad Software).
Pre-miRNA Cleavage-coupled RISC Loading Assay
A 7 μl reaction mixture in 20 mM PIPES, pH6.5, 1.5 mM MgCl2, 80 mM NaCl, 1 mM DTT, 10% glycerol, containing 10 fmol F-Dicer, F-Dicer/H-TRBP or F-Dicer/H-ADAR1 and 100 fmol F-Ago2 was pre-incubated at 4°C for 30 min. Subsequently, 100 fmol human pre-let-7a-1 was added to the reactions, and further incubated for dicing of pre-let-7a-1 and loading of let-7a-1 to F-Ago2/RISC at 37 °C for 10 minutes. The target RNA cleavage reaction in a 10 μl reaction mixture was mixed with 10 fmol of 5’-end 32P-labeled let-7a-1 target RNA and 2 units/μl RNasin (Promega) and incubated at 37 °C for 10 minutes. The reactions were stopped by adding 1.2 volumes of Gel Loading Buffer II (Ambion). RISC loading products (32P-labeled sliced target RNAs) were analyzed by 15% Urea-PAGE, and quantified using MolecularDynamics. The sequences of the substrates: human pre-let-7a-1; 5’-UGAGGUAGUAGGUUGUAUAGUUUUAGGGUCACACCCACCACUGGGAGAUAACUA UACAAUCUACUGUCUUUC-3’ (Dharmacon), let-7a-1 target RNA; 5’-GUAUCAACCACUAUACA ACCUACUACCUCAACGUUCAAU-3’ (IDT).
Assay for RISC Loading of Single-stranded Guide miRNA and miRNA Duplexes
RISC loading of single-stranded guide let-7a or let-7a RNA duplex was done exactly as described for the pre-miRNA dicing-coupled RISC loading assay, except as follows. Human let-7a-1 or let-7a-1 duplex (100 fmol) was used for the reaction. An additional incubation for loading of let-7a-1 or let-7a-1 duplex to F-Ago2/RISC was done at 30°C for 30 minutes. The target RNA slicing reaction (10 μl) was incubated with 10 fmol of 5’-end 32P-labeled let-7a-1 target RNA and 4 units/ml RNasin (Promega) at 30 °C for 90 minutes. The reaction was stopped by adding Proteinase K (Roche) to 1 mg/ml followed by incubation at 37° for 30 min prior to Phenol/Chloroform/Isopropanol extraction. Let-7a-1 RNA duplex was made by annealing let-7a S22: 5’-UGAGGUAGUAGGUUGUAUAGUU-3’ and let-7a A21: 5’-CUAUACAAUCUACUGUCUUUC-3’ synthesized at IDT.
Small RNA-seq and RNA-seq and Bioinformatics Data Analysis
Procedures for small RNA-seq and RNA-seq and bioinformatics data analysis are described in the Extended Experimental Procedures (Supplemental Information).
Supplementary Material
HIGHLIGHTS.
ADAR1 forms a complex with Dicer and enhances cleavage of miRNA and siRNA.
The ADAR1-Dicer heterodimer facilitates RISC loading and RNA silencing.
Global expression of miRNAs and target genes depends on ADAR1 in developing embryos.
Although ADAR1 homodimers antagonize RNAi, ADAR1-Dicer heterodimers stimulate it.
ACKNOWLEDGEMENTS
We thank Jennifer A. Doudna for the pre-let-7a template plasmid, Zissimos Mourelatos for discussion on RISC loading analysis, and John M. Murray for discussion on enzyme kinetics analysis and also critical reading of the manuscript. The work was supported by grants from the National Institutes of Health (GM040536, HL070045, and HL099342), the Ellison Medical Foundation (AG-55-2281-09), and the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health to K.N. B-E.W. is a recipient of a University of Pennsylvania, Vagelos scholarship. M.S. is supported in part by a fellowship from Japan Society for the Promotion of Science (JSPS 2010–22). We thank also for services provided by the Protein Expression, Genomics, and Bioinformatics Shared Facilities of The Wistar Cancer Center, which are supported by the National Cancer Institute (P30 CA010815).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Statistical Analysis
Experiments were run in triplicate or quadruplicate and repeated in a minimum of three independent trials. Image quantitation was done using ImageQuant image analysis software (GE Healthcare). Data are presented as means ± standard deviation (SD). Two-tailed t tests were conducted where the minimum level of significance was p < 0.05.
ACCESSION NUMBERS
The small RNA-seq and RNA-seq data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO) under series accession no. GSE43192 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE43192)
SUPPLEMENTAL INFORMATION
Supplemental information includes Extended Experimental Procedures, eleven figures, and four tables, which can be found with this article online at doi:xxxx.
REFERENCES
- Chakravarthy S, Sternberg SH, Kellenberger CA, Doudna JA. Substrate-specific kinetics of Dicer-catalyzed RNA processing. J. Mol. Biol. 2010;404:392–402. doi: 10.1016/j.jmb.2010.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho DS, Yang W, Lee JT, Shiekhattar R, Murray JM, Nishikura K. Requirement of dimerization for RNA editing activity of adenosine deaminases acting on RNA. J. Biol. Chem. 2003;278:17093–17102. doi: 10.1074/jbc.M213127200. [DOI] [PubMed] [Google Scholar]
- Daniels SM, Melendez-Pena CE, Scarborough RJ, Daher A, Christensen HS, El Far M, Purcell DF, Laine S, Gatignol A. Characterization of the TRBP domain required for dicer interaction and function in RNA interference. BMC Mol. Biol. 2009;10:38. doi: 10.1186/1471-2199-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlakic M. DUF283 domain of Dicer proteins has a double-stranded RNA-binding fold. Bioinformatics. 2006;22:2711–2714. doi: 10.1093/bioinformatics/btl468. [DOI] [PubMed] [Google Scholar]
- Ferre-D'Amare AR, Doudna JA. Use of cis- and trans-ribozymes to remove 5' and 3' heterogeneities from milligrams of in vitro transcribed RNA. Nucleic Acids Res. 1996;24:977–978. doi: 10.1093/nar/24.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz J, Strehblow A, Taschner A, Schopoff S, Pasierbek P, Jantsch MF. RNA-regulated interaction of transportin-1 and exportin-5 with the double-stranded RNA-binding domain regulates nucleocytoplasmic shuttling of ADAR1. Mol. Cell Biol. 2009;29:1487–1497. doi: 10.1128/MCB.01519-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartner JC, Schmittwolf C, Kispert A, Muller AM, Higuchi M, Seeburg PH. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J. Biol. Chem. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- Hartner JC, Walkley CR, Lu J, Orkin SH. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat. Immunol. 2009;10:109–115. doi: 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitti EG, Sallacz NB, Schoft VK, Jantsch MF. Oligomerization activity of a double-stranded RNA-binding domain. FEBS Lett. 2004;574:25–30. doi: 10.1016/j.febslet.2004.07.080. [DOI] [PubMed] [Google Scholar]
- Hood JL, Emeson RB. Editing of neurotransmitter receptor and ion channel RNAs in the nervous system. Curr. Top. Microbiol. Immunol. 2012;353:61–90. doi: 10.1007/82_2011_157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizasa H, Wulff BE, Alla NR, Maragkakis M, Megraw M, Hatzigeorgiou A, Iwakiri D, Takada K, Wiedmer A, Showe L, et al. Editing of Epstein-Barr virus-encoded BART6 microRNAs controls their dicer targeting and consequently affects viral latency. J. Biol. Chem. 2010;285:33358–33370. doi: 10.1074/jbc.M110.138362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Megraw M, Kreider E, Iizasa H, Valente L, Hatzigeorgiou AG, Nishikura K. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 2008;36:5270–5280. doi: 10.1093/nar/gkn479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007a;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007b;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koscianska E, Starega-Roslan J, Krzyzosiak WJ. The role of Dicer protein partners in the processing of microRNA precursors. PLoS One. 2011;6:e28548. doi: 10.1371/journal.pone.0028548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Lai F, Drakas R, Nishikura K. Mutagenic analysis of double-stranded RNA adenosine deaminase, a candidate enzyme for RNA editing of glutamate-gated ion channel transcripts. J. Biol. Chem. 1995;270:17098–17105. doi: 10.1074/jbc.270.29.17098. [DOI] [PubMed] [Google Scholar]
- Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Ma E, MacRae IJ, Kirsch JF, Doudna JA. Autoinhibition of human dicer by its internal helicase domain. J. Mol. Biol. 2008;380:237–243. doi: 10.1016/j.jmb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, Enzo E, Guzzardo V, Rondina M, Spruce T, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Nasser MW, Datta J, Nuovo G, Kutay H, Motiwala T, Majumder S, Wang B, Suster S, Jacob ST, Ghoshal K. Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J Biol Chem. 2008;283:33394–33405. doi: 10.1074/jbc.M804788200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Nishikura K. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat. Rev. Mol. Cell Biol. 2006;7:919–931. doi: 10.1038/nrm2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G, Lin W, Piskol R, Tan MH, Davis C, Li JB. Accurate identification of human Alu and non-Alu RNA editing sites. Nat. Methods. 2012 doi: 10.1038/nmeth.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe TM, Rizzi M, Hirose K, Peters GA, Sen GC. A role of the double-stranded RNA-binding protein PACT in mouse ear development and hearing. Proc. Natl. Acad. Sci. U S A. 2006;103:5823–5828. doi: 10.1073/pnas.0601287103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scadden AD. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat. Struct. Mol. Biol. 2005;12:489–496. doi: 10.1038/nsmb936. [DOI] [PubMed] [Google Scholar]
- Valente L, Nishikura K. RNA binding-independent dimerization of adenosine deaminases acting on RNA and dominant negative effects of nonfunctional subunits on dimer functions. J. Biol. Chem. 2007;282:16054–16061. doi: 10.1074/jbc.M611392200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Miyakoda M, Yang W, Khillan J, Stachura DL, Weiss MJ, Nishikura K. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J. Biol. Chem. 2004;279:4952–4961. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- Wu C, Gong Y, Yuan J, Zhang W, Zhao G, Li H, Sun A, Zou Y, Ge J. microRNA-181a represses ox-LDL-stimulated inflammatory response in dendritic cell by targeting c-Fos. J Lipid Res. 2012;53:2355–2363. doi: 10.1194/jlr.M028878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Lamm AT, Fire AZ. Competition between ADAR and RNAi pathways for an extensive class of RNA targets. Nat. Struct. Mol. Biol. 2011;18:1094–1101. doi: 10.1038/nsmb.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- Zhong J, Peters AH, Lee K, Braun RE. A double-stranded RNA binding protein required for activation of repressed messages in mammalian germ cells. Nat. Genet. 1999;22:171–174. doi: 10.1038/9684. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.