Abstract
Niemann-Pick C disease (NPC) is a neurodegenerative lysosomal disorder characterized by storage of cholesterol and other lipids caused by defects in NPC1, a transmembrane protein involved in cholesterol export from the lysosome, or NPC2, an intralysosomal cholesterol transport protein. Alterations in lysosomal activities have been implicated in NPC pathogenesis therefore the aim of this study was to conduct a proteomic analysis of lysosomal proteins in mice deficient in either NPC1 or NPC2 to identify secondary changes that might be associated with disease. Lysosomal proteins containing the specific mannose 6-phosphate modification were purified from wild-type and Npc1−/− and Npc2−/− mutant mouse brains at different stages of disease progression and identified by bottom-up LC-MS/MS and quantified by spectral counting. Levels of a number of lysosomal proteins involved in lipid catabolism including prosaposin and the two subunits of β-hexosaminidase were increased in both forms of NPC, possibly representing a compensatory cellular response to the accumulation of glycosphingolipids. Several other lysosomal proteins were significantly altered, including proteases and glycosidases. Changes in lysosomal protein levels corresponded with similar alterations in activities and transcript levels. Understanding the rationale for such changes may provide insights into the pathophysiology of NPC.
Keywords: Niemann-Pick C, lysosome, mouse, spectral counting, mannose 6-phosphate
1. Introduction
The lysosome is a cellular organelle responsible for the degradation and recycling of macromolecules and to this end, it contains numerous hydrolytic enzymes (“acid hydrolases”) and accessory proteins that have evolved to function in its acidic pH as well as transporters to export catabolic products [1]. Defects in lysosome proteins are associated with lysosomal storage disorders (LSDs) [2] which are monogenic diseases with a cellular phenotype that typically reflects the function of the defective protein. For example, the loss of an acid hydrolase may result in an accumulation of its substrate(s) within the lysosome. Alternatively, loss of a lysosomal transporter will result in a lysosomal accumulation of the molecules that it normally exports from this organelle. In LSDs, disease typically results from cell death or dysfunction as a consequence of the primary defect but for one, Niemann-Pick C disease (NPC), there is evidence (see below) that secondary alterations in the function of the lysosomal system may play a role in pathogenesis.
NPC is a fatal, neurovisceral LSD that exists in two clinically similar but genetically distinct forms that result from mutations in genes encoding proteins involved in the intracellular transport of cholesterol (reviewed in [3, 4]). NPC type 1 accounts for the majority of cases (~95%) and results from mutations in the gene encoding NPC1 [5], an endolysosomal transmembrane protein with a sterol sensing domain that binds cholesterol and oxysterols [6–8]. NPC type 2 results from mutations in the gene that encodes a small soluble glycoprotein, NPC2 [9], that resides within the lumen of the lysosome [9, 10] and binds cholesterol and related sterols [11–14]. While the precise interactions between NPC1 and NPC2 that result in the transport of LDL-derived cholesterol from the lysosome are not completely clear, there is evidence consistent with the hypothesis that NPC2 shuttles cholesterol from inner lysosomal membrane structures or microaggregates to the sterol binding site of NPC1, which then facilitates the egress of cholesterol from the lysosome to other intracellular sites [15–17].
NPC is characterized by the storage of cholesterol within the lysosomes of affected individuals and this is consistent with the known functional properties of NPC1 and NPC2. However, other lipids also accumulate, including sphingomyelin, glycosphingolipids (gangliosides GA2, GM2 and GM3), glycosylceramides (glucosylceramide and lactosylceramide), and bis(monoacylglycerol)phosphate (reviewed in [18]). The basis for the accumulation of these lipids is not clear but several studies suggest that in the absence of NPC1, a secondary alteration in a lysosomal activity might be involved. Along with changes in other lysosomal enzyme activities, the activity of acid sphingomyelinase (SMPD1) is reduced in Npc1−/− mice [19] and there is evidence from cell-culture experiments that this is at a posttranslational level [20]. While the mechanism underlying this reduction remains unclear, it does provide a rational explanation for the increased levels of sphingomyelin. Restoration of SMPD1 activity in NPC1-deficient cells greatly reduced cholesterol storage [21] and improved other biochemical features of disease, suggesting a complex interplay between SMPD1, cholesterol and sphingomyelin. Altered glycosylation of select lysosomal proteins is another secondary phenotype observed in both Npc1−/− and Npc2−/− mutant mice [22] although its relevance towards pathogenesis, if any, is unclear.
The fact that a secondary change in a lysosomal activity may play an important role in the cellular pathways of pathogenesis in NPC led us to speculate whether other lysosomal changes might also be involved. To address this question, we have conducted a proteomic comparison of the levels of soluble lysosomal proteins in the brains of mutant mouse strains that lack either NPC1 [23] or NPC2 [24] and which both recapitulate NPC disease.
Materials and Methods
2.1 Animals
Experiments and procedures involving live animals were conducted in compliance with protocols approved by the Robert Wood Johnson Medical School Institutional Animal Care and Use Committee (Protocol number I10–010). Npc1−/− andNpc2−/− mutant mice have been described previously [23, 24] and were in an isogenic BALB/c genetic background [22]. Mice were killed by injection of a sodium pentobarbital/sodium phenytoin mixture (Euthasol, Delmarva Laboratories) and perfusion with saline by intracardiac puncture. Brains were dissected, frozen on dry ice and stored at −80°C prior to use. Animal information is given in Supplementary Table 1.
2.2 Purification of lysosomal proteins
Lysosomal proteins containing mannose 6-phosphate (Man-6-P) were purified on immobilized bovine soluble cation-independent Man-6-P receptor (sCI-MPR) columns using a modified version of published procedures [25]. In brief, each mouse brain was powdered using a Bessmann tissue pulverizer then homogenized on ice using a Brinkmann Polytron in 10 ml of PBS containing 0.2% Tween-20, 2.5 mM EDTA, 5 mM β-glycerophosphate, 1 µg/ml pepstatin, 1 µg/ml Leupeptin and 0.5 mM Pefabloc. Homogenates were centrifuged at 6,000 × g for 30 min at 4°C then supernatants gravity-loaded onto 1 ml bed volume immobilized sCI-MPR. Columns were washed twice with 10 ml PBS / 0.2 % Tween-20 and twice with 10 ml PBS. A mock elution with 4 ml of PBS containing 5 mM glucose 6-phosphate / 5 mM mannose was conducted to release proteins that were bound to the column or immobilized sCI-MPR via interactions that were independent of the presence of Man-6-P [25]. Man-6-P glycoproteins were eluted with 4 ml PBS containing 5 mM Man-6-P. Eluates were concentrated using Amicon Ultra-4 Ultracel-10k filtration devices.
2.3 Tandem mass spectrometry
Tryptic digests of samples of purified Man-6-P glycoproteins were prepared as described previously [25]. Each sample was analyzed in duplicate using an LTQ Orbitrap Velos tandem mass spectrometer (Thermo Scientific) coupled to a Ultimate 3000 RLSCnano System. Peptides were solubilized in 0.1% TFA and 0.5 µg loaded on to an in-house generated fused silica trap-column of 100 µm × 2 cm packed with Magic C18 AQ (5um bead size, 200 Å pore size Michrom Bioresources, Inc.) and washed with 0.2% formic acid at a flow-rate of 10 µl/min for 5 min. Retained peptides were separated on an in-house generated fused silica column of 75 µm × 50 cm packed with Magic C18 AQ (3 µm bead size, 200 Å pore size, Michrom Bioresources, Inc.) using a segmented linear gradient from 4 to 90% B (A: 0.1% formic acid, B: 0.08% formic acid, 80% ACN): 5 min, 4–10% B; 60 min, 10–40% B; 15 min, 40–55% B; 10 min, 55–90% B. For each cycle, one full MS was scanned in the Orbitrap with resolution of 60000 from 300–2000 m/z and the 20 most intense peaks fragmented by CID using a normalized collision energy of 35% and products scanned in the ion trap. Data dependent acquisition was set for a repeat count of 2 and exclusion of 60 sec.
2.4 Protein identification and quantification
Peak lists were generated using Proteome Discoverer 1.2. No constraints were used with respect to retention time, charge state or peak count. Minimum precursor mass was 350 Da and maximum was 6000. Data were searched against the Ensembl release 64 of the NCBIM37 mouse genome assembly using a local Global Proteome Machine (GPM) XE Manager version 2.2.1 (Beavis Informatics Ltd., Winnipeg, Canada) with X!Tandem version Cyclone2011.12.01 to assign spectral data [26, 27]. Parameters were: fragment mass error, 0.4 Da; parent mass error, 10 ppm; maximum charge, +8; minimum 5 peaks assigned; one missed cleavage allowed; carbamidomethylation was a constant modification; methionine oxidation was a variable modification during initial modeling; methionine and tryptophan oxidation and asparagine and glutamine deamidation were variable modifications during model refinement; and point mutations were allowed. All MS data files were analyzed together in a MudPit analysis and individual data extracted to ensure that peptides that could be assigned to more than one protein were assigned consistently for all samples. To avoid problems of redundancy that may arise from multiple protein identifiers that can be assigned to the same gene, Ensembl protein identifiers were converted to associated gene names using the BioMart tool. Protein assignments were filtered for a log GPM expectation score of −10 or better and a minimum assignment of two unique peptides based on amino acid sequence.
2.5 Statistical analysis
Relative protein levels in preparations of purified Man-6-P glycoproteins were determined by spectral counting [28, 29]. Duplicate LC-MS/MS runs were performed on each sample and spectral counts for individual gene products were summed. Our analysis was conducted on known lysosomal proteins and probable lysosomal candidates based on similarity to known lysosomal proteins, demonstrated presence of Man-6-P or specificity of purification on immobilized sCI-MPR [25, 30]. As counting error for Poisson data is inversely proportional to the square of the spectral counts, group comparisons of proteins with very low spectral counts have very low statistical power [31]. Thus, data were filtered to only include proteins with an average of ≥ 10 spectral counts per animal, yielding a final dataset of 62 lysosomal or potential lysosomal proteins. For each of these proteins and for each animal, we measured the number of spectral counts. There were three genotypes (wild type, Npc1−/−, and Npc2−/−), each analyzed at two ages corresponding to different stages of disease progression (early and late), for a total of six groups with five animals per group. To adjust for experimental variation from animal to animal, we computed the total number of spectral counts for each animal. This total served as a reference denominator for comparing protein counts. To construct a statistical model for the counts, we used Poisson regression with “count” as the response variable, “group” as a predictor variable, and the log (base e) of the total spectral count as an offset [31]. We fitted this Poisson model separately for each of the 62 proteins using the “glm” package in the R statistical system (Version 2.15) (R Development Core Team R: A Language and Environment for Statistical Computing, http://www.R-project.org) with “family” specified as “Poisson”. We used a Bonferroni correction to account for multiple comparisons of 62 proteins. Given that there were few significant differences between the wild-type control at the two different age groups, the effect of the NPC mutations for each age group was compared to the average of all wild-type controls (see Results).
2.6 Enzyme assays
Most lysosomal enzyme activities were measured as described previously [32–34]. Hexosaminidase A (HEXA/HEXB heterodimer) activity was measured using 4-methylumbelliferyl-2-acetamido-2-deoxy-6-sulfo-β-D-glucopyranoside [35]. Note that this assay also measures hexosaminidase S (HEXA/HEXA homodimer) activity but this is a minor isoenzyme.
2.7 scriptional profiling data
Gene expression data Series GSE5944 for P49 Npc1−/− mouse cerebellum and corresponding normal controls (n=3 per group) was extracted from the NCBI GEO database [36] (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE5944). Signal intensities for multiple probes corresponding to a single gene were summed.
3. Results and Discussion
3.1 Experimental design
The aim of this study was to perform a quantitative analysis of lysosomal proteins in the brains of Npc1−/− and Npc2−/− mice to determine whether secondary changes in levels of lysosomal proteins could be related to disease phenotype. In addition, a comparison of secondary alterations between Npc1−/− and Npc2−/− mice could potentially provide valuable insights into the respective functions of these proteins. Our approach was to isolate Man-6-P glycoproteins from NPC mutant mice by affinity purification on immobilized sCI-MPR, identify individual components of the mixture by LC-MS/MS and use spectral counting as a semi-quantitative approach to compare the relative levels of each protein in the mutant and wild-type mice. For each NPC mouse model, we analyzed both presymptomatic early-stage (ages 30–44 days) and symptomatic late-stage (ages 64–88 days) disease (n=5 per group) (Supplementary Table 1). Our rationale was that this approach would help to distinguish fundamental lysosomal responses to the defects in cholesterol trafficking from differences reflecting alterations in the distribution of cell type within the CNS as disease progresses (e.g., a diminishing number of neurons and increasing number of activated microglia as a response to disease [37, 38]).
3.2 Expression of lysosomal proteins in Balb/c mouse brain
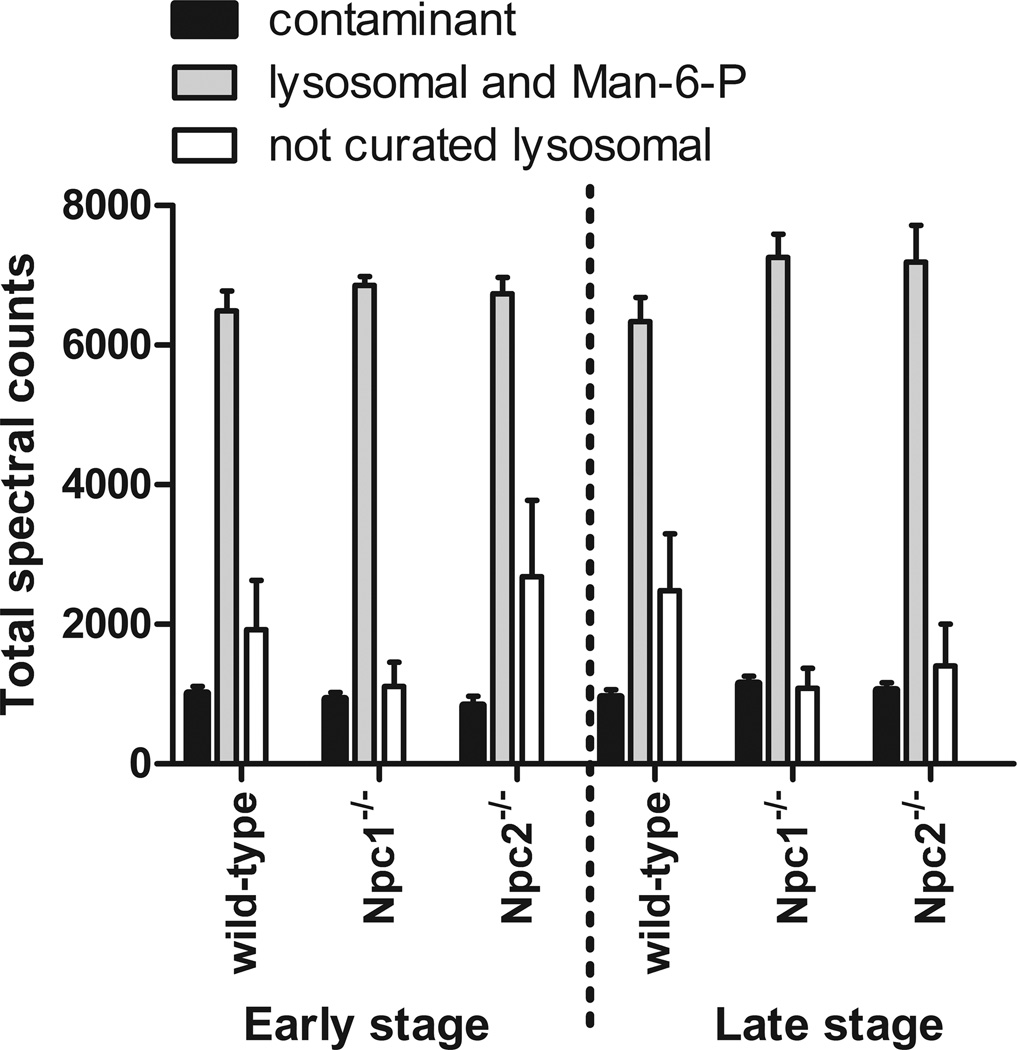
The protein composition of the Man-6-P glycoprotein preparation isolated from the brains of wild-type control and NPC-mutant mice was analyzed by LC-MS/MS. Data supporting protein assignments are shown in Supplementary Tables 2 (protein assignments) and 3 (peptide identifications and classification). After mapping protein assignments to the corresponding genes, in total, 912 gene products met our inclusion criteria (GPM log expectancy score of −10 or better with 2 or more unique peptides). Of these, 15 were known contaminants of non-murine origin, 827 were proteins that are not assigned to the lysosome and 69 were classified as lysosomal (62 known lysosomal proteins and 7 potential lysosomal candidates [25, 30]). We also identified myeloperoxidase, a neutrophil alpha granule protein known to contain Man-6-P. However, while the majority of assignments were made to non-lysosomal proteins, the majority of spectra were assigned to lysosomal proteins (Fig. 1).
Figure 1. Classification of protein assignments.
“Contaminant” represent assignments that are not of murine origin (e.g., trypsin and keratins); “lysosomal and Man-6-P” represents known lysosomal proteins, select lysosomal candidates and myeloperoxidase, a neutrophil granule protein that contains Man-6-P; “not curated lysosomal” represents murine proteins that are not assigned to the lysosome.
3.3 Differential expression of lysosomal proteins in the brains of NPC mutant animals
Levels of lysosomal proteins identified in purified samples from the wild-type control, Npc1−/− and Npc2−/− mice were estimated by spectral counting. Data were filtered for proteins expressed with an average of ≥ 10 spectral counts per animal to eliminate non-informative assignments (see Methods), thus we focus on 62 of the original 69 known or candidate lysosomal proteins.
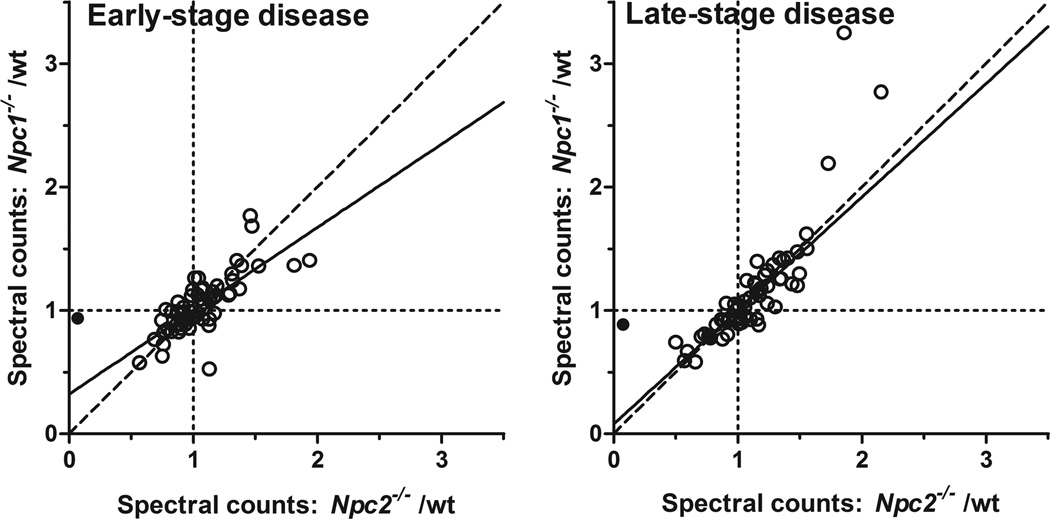
Relative levels of the majority of lysosomal proteins were unchanged in the absence of either NPC1 or NPC2 at either early or late stage disease (regions where ratios of both mutants/wild type is ~1, indicated by intersection of dotted lines in Fig. 2). However, the levels of some lysosomal proteins were altered and there is a close correlation between changes in the two different NPC mutants (Fig. 2) compared to wild-type, presumably reflecting similarities in cellular phenotypes despite the difference in genetic basis.
Figure 2. Spectral counts for NPC1 and NPC2 mutants normalized to controls in early and late-stage disease.
Data represent known lysosomal proteins and potential candidates with an average of at least 10 spectral counts per animal. The filled symbol represents NPC2 which we expect to be essentially absent from the Npc2−/− animals. Data are fit to a first order polynomial using the robust fit option in Graphpad Prism 5 (solid line). Dashed line represents x=y; dotted lines, x=1 and y=1.
The primary goal of this study was to identify lysosomal changes that are common to both NPC mutants and which were observed in both symptomatic and presymptomatic animals as these may indicate a direct cellular response to the disruption in cholesterol trafficking. As age does not appear to affect levels of the vast majority of all lysosomal proteins for the wild type controls (Table 1 and Supplementary Fig. 1), to increase statistical power, we have combined data from all ten of these animals as a single control group. There are a number of proteins that are clearly increased in disease at the expense of others when considering both the fold-change compared to wild-type levels and the statistical significance of the change (Table 1).
Table 1. Relative level of lysosomal proteins by MS analysis.
Wild type control column shows the ratio of spectral counts for the two age groups after normalizing to total spectral counts per animal. The Npc1−/− and Npc2−/− mutants at early and late stage disease are compared to the average of all wild type controls. Parentheses indicate the estimate of the error ranging from −/+ one standard error of the mean. A number of proteins were increased in the mutants at early and late stage and these are indicated with a bold protein identifier.
| Wild-type control | Early-stage disease | Late-stage disease | |||
|---|---|---|---|---|---|
| Protein | young/old | Npc1/wild-typeav | Npc2/wild-typeav | Npc1/wild-typeav | Npc2/wild-typeav |
| Aga | 0.97 (0.90 – 1.04) | 0.95 (0.89 – 1.01) | 0.97 (0.91 – 1.03) | 0.78 (0.73 – 0.83) ** | 0.88 (0.82 – 0.93) |
| Arsa | 1.08 (1.02 – 1.14) | 0.78 (0.74 – 0.82) *** | 0.84 (0.80 – 0.88) * | 0.85 (0.81 – 0.89) | 0.85 (0.81 – 0.89) |
| Arsb | 1.03 (0.98 – 1.08) | 1.05 (1.01 – 1.10) | 1.13 (1.08 – 1.17) | 0.82 (0.78 – 0.85) *** | 0.84 (0.80 – 0.87) *** |
| Arsg | 1.03 (0.89 – 1.18) | 0.59 (0.51 – 0.68) * | 0.71 (0.62 – 0.82) | 0.53 (0.45 – 0.61) *** | 0.51 (0.44 – 0.59) *** |
| Arsk | 1.08 (0.96 – 1.22) | 0.91 (0.81 – 1.01) | 0.90 (0.81 – 1.00) | 0.74 (0.66 – 0.83) | 0.68 (0.60 – 0.76) |
| Asah1 | 1.12 (1.08 – 1.17) | 0.99 (0.96 – 1.03) | 1.06 (1.03 – 1.10) | 1.13 (1.10 – 1.17) ** | 1.35 (1.30 – 1.39) *** |
| Cln5 | 0.85 (0.78 – 0.93) | 1.05 (0.97 – 1.13) | 0.96 (0.88 – 1.04) | 1.02 (0.94 – 1.10) | 1.09 (1.01 – 1.17) |
| Creg1 | 0.87 (0.79 – 0.97) | 0.95 (0.87 – 1.04) | 1.02 (0.94 – 1.12) | 0.99 (0.91 – 1.07) | 1.22 (1.13 – 1.32) |
| Ctbs | 0.95 (0.85 – 1.05) | 0.86 (0.78 – 0.94) | 1.12 (1.02 – 1.22) | 0.80 (0.73 – 0.88) | 0.87 (0.80 – 0.96) |
| Ctsa | 1.05 (1.01 – 1.10) | 1.07 (1.03 – 1.11) | 1.09 (1.05 – 1.13) | 1.05 (1.01 – 1.09) | 1.06 (1.02 – 1.10) |
| Ctsb | 1.07 (1.02 – 1.11) | 1.00 (0.97 – 1.04) | 0.99 (0.95 – 1.02) | 0.98 (0.94 – 1.01) | 0.97 (0.93 – 1.00) |
| Ctsc | 0.89 (0.81 – 0.97) | 1.13 (1.05 – 1.21) | 1.01 (0.93 – 1.09) | 1.16 (1.08 – 1.24) | 0.96 (0.89 – 1.04) |
| Ctsd | 0.94 (0.91 – 0.98) | 0.91 (0.88 – 0.94) | 1.07 (1.04 – 1.10) | 0.96 (0.93 – 0.99) | 1.01 (0.98 – 1.04) |
| Ctsf | 0.96 (0.89 – 1.04) | 1.14 (1.07 – 1.21) | 1.35 (1.27 – 1.43) *** | 1.12 (1.05 – 1.19) | 1.30 (1.23 – 1.38) *** |
| Ctsh | 0.57 (0.44 – 0.72) | 1.70 (1.43 – 2.01) | 2.38 (2.03 – 2.78) *** | 1.76 (1.49 – 2.08) * | 1.38 (1.15 – 1.64) |
| Ctsl | 1.16 (1.10 – 1.23) | 0.83 (0.79 – 0.87) ** | 0.90 (0.86 – 0.94) | 0.84 (0.80 – 0.88) ** | 0.78 (0.75 – 0.82) *** |
| Ctso | 1.05 (0.92 – 1.21) | 0.92 (0.81 – 1.03) | 0.82 (0.73 – 0.93) | 0.60 (0.53 – 0.69) * | 0.54 (0.47 – 0.62) *** |
| Ctss | 0.76 (0.63 – 0.90) | 1.82 (1.61 – 2.06) *** | 1.62 (1.42 – 1.84) * | 2.46 (2.19 – 2.76) *** | 1.41 (1.24 – 1.61) |
| Ctsz | 1.10 (1.03 – 1.17) | 1.12 (1.07 – 1.18) | 1.21 (1.15 – 1.27) ** | 1.22 (1.16 – 1.28) *** | 1.15 (1.09 – 1.21) |
| Dnase2a | 0.89 (0.76 – 1.04) | 0.96 (0.84 – 1.10) | 1.04 (0.92 – 1.19) | 0.77 (0.67 – 0.88) | 0.92 (0.80 – 1.05) |
| Dpp7 | 0.93 (0.87 – 0.99) | 0.87 (0.82 – 0.92) | 0.90 (0.85 – 0.95) | 0.88 (0.83 – 0.93) | 0.86 (0.81 – 0.91) |
| Epdr1 | 1.16 (1.12 – 1.20) *** | 0.82 (0.79 – 0.84) *** | 1.01 (0.98 – 1.04) | 0.97 (0.94 – 1.00) | 1.01 (0.98 – 1.03) |
| Fuca1 | 0.87 (0.79 – 0.96) | 0.95 (0.87 – 1.03) | 0.98 (0.90 – 1.07) | 0.96 (0.89 – 1.05) | 0.97 (0.90 – 1.06) |
| Fuca2 | 0.91 (0.85 – 0.97) | 1.12 (1.06 – 1.18) | 1.05 (0.99 – 1.11) | 1.00 (0.95 – 1.06) | 1.04 (0.99 – 1.10) |
| Gaa | 0.89 (0.86 – 0.92) * | 1.07 (1.05 – 1.10) | 0.89 (0.87 – 0.92) *** | 1.04 (1.02 – 1.07) | 1.12 (1.10 – 1.15) *** |
| Galc | 1.05 (0.96 – 1.15) | 0.77 (0.70 – 0.83) | 0.83 (0.77 – 0.90) | 0.95 (0.88 – 1.03) | 1.12 (1.04 – 1.21) |
| Galns | 0.87 (0.80 – 0.95) | 0.83 (0.77 – 0.90) | 0.79 (0.72 – 0.85) | 0.66 (0.60 – 0.71) *** | 0.76 (0.70 – 0.82) * |
| Ggh | 0.92 (0.82 – 1.02) | 0.96 (0.87 – 1.05) | 0.91 (0.83 – 1.00) | 0.81 (0.73 – 0.89) | 0.89 (0.81 – 0.98) |
| Gla | 0.90 (0.84 – 0.97) | 1.01 (0.95 – 1.07) | 0.79 (0.74 – 0.84) * | 0.92 (0.86 – 0.98) | 0.92 (0.87 – 0.98) |
| Glb1 | 1.00 (0.95 – 1.04) | 0.87 (0.84 – 0.91) * | 0.72 (0.69 – 0.75) *** | 0.82 (0.78 – 0.85) *** | 0.77 (0.74 – 0.80) *** |
| Gns | 1.10 (1.05 – 1.16) | 1.08 (1.04 – 1.13) | 1.09 (1.05 – 1.14) | 1.19 (1.14 – 1.24) *** | 1.13 (1.09 – 1.18) |
| Gusb | 0.96 (0.91 – 1.02) | 1.05 (1.00 – 1.10) | 1.11 (1.06 – 1.17) | 0.98 (0.94 – 1.03) | 1.00 (0.95 – 1.05) |
| Hexa | 0.97 (0.93 – 1.02) | 1.14 (1.10 – 1.18) * | 1.05 (1.01 – 1.09) | 1.19 (1.15 – 1.23) *** | 1.12 (1.08 – 1.16) |
| Hexb | 1.03 (0.99 – 1.08) | 1.65 (1.60 – 1.70) *** | 1.38 (1.34 – 1.43) *** | 1.96 (1.90 – 2.02) *** | 1.56 (1.51 – 1.61) *** |
| Ids | 1.09 (1.02 – 1.16) | 1.07 (1.01 – 1.13) | 0.99 (0.93 – 1.04) | 0.96 (0.91 – 1.02) | 0.90 (0.85 – 0.95) |
| Idua | 1.07 (0.97 – 1.19) | 0.81 (0.74 – 0.89) | 0.81 (0.73 – 0.89) | 0.72 (0.65 – 0.79) * | 0.70 (0.63 – 0.77) * |
| Lgmn | 0.99 (0.93 – 1.06) | 1.08 (1.03 – 1.15) | 1.25 (1.19 – 1.32) *** | 1.23 (1.17 – 1.30) *** | 1.21 (1.14 – 1.27) * |
| Lipa | 1.01 (0.90 – 1.13) | 1.29 (1.18 – 1.41) | 1.33 (1.22 – 1.45) | 1.44 (1.32 – 1.56) *** | 1.39 (1.27 – 1.51) *** |
| Man2b1 | 0.88 (0.84 – 0.93) | 1.27 (1.22 – 1.32) *** | 1.04 (0.99 – 1.08) | 1.18 (1.13 – 1.22) *** | 1.16 (1.12 – 1.21) * |
| Man2b2 | 0.97 (0.92 – 1.02) | 0.96 (0.92 – 1.01) | 0.81 (0.77 – 0.85) *** | 0.88 (0.84 – 0.93) | 0.87 (0.83 – 0.92) |
| Manba | 1.19 (1.13 – 1.25) * | 0.92 (0.88 – 0.96) | 0.96 (0.91 – 1.00) | 0.76 (0.72 – 0.79) *** | 0.68 (0.65 – 0.71) *** |
| Naaa | 1.06 (0.98 – 1.15) | 1.17 (1.09 – 1.25) | 0.98 (0.91 – 1.05) | 1.10 (1.03 – 1.17) | 1.04 (0.97 – 1.11) |
| Naga | 0.93 (0.89 – 0.97) | 0.91 (0.88 – 0.95) | 0.93 (0.90 – 0.96) | 1.05 (1.02 – 1.09) | 0.91 (0.88 – 0.95) |
| Naglu | 0.96 (0.90 – 1.03) | 1.13 (1.07 – 1.19) | 0.98 (0.92 – 1.04) | 1.05 (1.00 – 1.11) | 0.98 (0.93 – 1.04) |
| Neu1 | 1.05 (0.95 – 1.16) | 1.30 (1.21 – 1.41) * | 1.27 (1.18 – 1.38) | 1.33 (1.24 – 1.43) *** | 1.34 (1.25 – 1.45) *** |
| Npc2 | 1.41 (1.30 – 1.54) *** | 0.74 (0.68 – 0.80) ** | 0.06 (0.04 – 0.07) *** | 0.91 (0.85 – 0.98) | 0.08 (0.06 – 0.10) *** |
| Pgcp | 0.86 (0.81 – 0.92) | 1.01 (0.96 – 1.06) | 0.90 (0.85 – 0.95) | 0.73 (0.69 – 0.78) *** | 0.84 (0.80 – 0.89) |
| Pla2g15 | 0.91 (0.85 – 0.97) | 0.82 (0.78 – 0.87) | 0.83 (0.79 – 0.88) | 0.77 (0.73 – 0.82) *** | 0.75 (0.70 – 0.79) *** |
| Plbd2 | 0.88 (0.85 – 0.91) * | 0.90 (0.87 – 0.93) | 0.96 (0.93 – 0.99) | 0.78 (0.75 – 0.81) *** | 0.89 (0.86 – 0.91) * |
| Pld3 | 0.97 (0.85 – 1.11) | 0.82 (0.73 – 0.93) | 0.94 (0.84 – 1.06) | 0.78 (0.69 – 0.87) | 0.88 (0.79 – 0.99) |
| Pofut2 | 0.87 (0.73 – 1.03) | 0.53 (0.45 – 0.64) * | 1.16 (1.02 – 1.33) | 0.61 (0.51 – 0.72) | 0.41 (0.34 – 0.50) *** |
| Ppt1 | 1.12 (1.07 – 1.17) | 1.01 (0.97 – 1.05) | 1.17 (1.13 – 1.22) *** | 0.95 (0.92 – 0.99) | 1.22 (1.17 – 1.26) *** |
| Ppt2 | 0.81 (0.72 – 0.91) | 0.61 (0.54 – 0.68) *** | 0.60 (0.54 – 0.68) *** | 0.46 (0.40 – 0.52) *** | 0.52 (0.46 – 0.59) *** |
| Prcp | 0.79 (0.74 – 0.84) * | 0.81 (0.76 – 0.86) * | 0.74 (0.70 – 0.78) *** | 0.72 (0.68 – 0.77) *** | 0.77 (0.73 – 0.82) *** |
| Psap | 1.29 (1.21 – 1.38) ** | 1.14 (1.08 – 1.20) | 1.53 (1.46 – 1.61) *** | 1.41 (1.35 – 1.49) *** | 1.33 (1.27 – 1.40) *** |
| Rnaset2b | 1.17 (1.02 – 1.35) | 1.19 (1.07 – 1.33) | 1.36 (1.22 – 1.51) | 0.88 (0.78 – 1.00) | 1.10 (0.99 – 1.23) |
| Scpep1 | 1.16 (1.07 – 1.24) | 0.86 (0.81 – 0.92) | 1.05 (0.99 – 1.11) | 0.95 (0.89 – 1.01) | 0.94 (0.88 – 1.00) |
| Sgsh | 0.96 (0.85 – 1.08) | 1.07 (0.97 – 1.18) | 1.12 (1.02 – 1.23) | 0.76 (0.68 – 0.85) | 1.01 (0.92 – 1.11) |
| Siae | 0.88 (0.82 – 0.95) | 0.73 (0.68 – 0.79) *** | 0.78 (0.72 – 0.83) * | 0.87 (0.82 – 0.93) | 0.75 (0.70 – 0.81) *** |
| Smpd1 | 1.13 (1.06 – 1.21) | 0.74 (0.70 – 0.79) *** | 0.70 (0.65 – 0.74) *** | 0.72 (0.68 – 0.76) *** | 0.82 (0.78 – 0.87) |
| Smpdl3a | 0.95 (0.88 – 1.02) | 0.83 (0.77 – 0.88) | 0.79 (0.73 – 0.84) * | 0.66 (0.62 – 0.71) *** | 0.67 (0.62 – 0.72) *** |
| Tpp1 | 0.94 (0.88 – 1.01) | 1.27 (1.20 – 1.34) *** | 1.30 (1.23 – 1.37) *** | 1.28 (1.22 – 1.35) *** | 1.34 (1.27 – 1.41) *** |
P values with Bonferonni correction: p<0.005/62;
p<0.01/62;
p<0.05/62.
3.4 Lysosomal enzyme activities in NPC-mutant mouse brain
To extend the MS results and to determine whether observed alterations in the levels in the Man-6-Phosphorylated forms of lysosomal proteins reflect total activity, regardless of phosphorylation state, we measured the activity of select lysosomal enzymes in the brains of the Npc1−/− and Npc2−/− animals (Fig. 3, Panel A). We find that the activity of most lysosomal enzymes is elevated in both mutants at both time points, although relatively more in late-stage disease. On first examination, these results would appear to contrast with the determinations from purified Man-6-P glycoproteins by MS analysis, in which few lysosomal proteins were determined to be elevated and to a lesser degree. However, this is a function of the respective experimental designs. Enzyme activity measurements were normalized to the total protein concentration of each brain homogenate, thus a generalized increase in lysosomal activity can be identified. For the MS analysis, the same quantity of purified Man-6-P glycoproteins was analyzed. Here, if the levels of lysosomal proteins were generally increased but the amounts of non-lysosomal proteins remained invariant, then we would observe some increase in amounts of lysosomal proteins measured by MS at the expense of the non-lysosomal constituents (or lysosomal proteins that are not elevated, e.g., MANBA). Consistent with this hypothesis, we find that the relative proportion of non-lysosomal proteins is decreased in the groups of animals where lysosomal activities were most elevated (early stage Npc1−/− and late stage Npc1−/− and Npc2−/−) (Fig. 1). Thus, in the MS analysis, in the face of a generalized increase in levels of Man-6-P-containing lysosomal proteins, an increase in the spectral counts of any given protein would indicate that it was elevated greater than average. In practical terms, this means that the fold increases relative to average wild-type control reported in Table 1 are likely underestimates in absolute terms. Conversely, proteins that are apparently decreased may not be when considering absolute amounts.
Figure 3. Activity of lysosomal enzymes in NPC-mutant and control mice.
Panel A: enzyme activities normalized to the average of all wild-type controls are shown at ages corresponding to early-stage (E) and late-stage (L) disease. There are three animals per group and error bars represent the standard error of the mean. The following enzyme assays reflect the activity of the respective lysosomal proteins: β-glucosidase, GBA; β-mannosidase, MANBA; dipeptidyl-peptidase 7, DPP7; α-fucosidase, FUCA1; legumain, LGMN; cathepsin B, CTSB; α-mannosidase, MAN2B1; dipeptidyl-peptidase I, CTSC; total β-hexosaminidase, HEXA/HEXB heterodimer (HexA) and HEXB/HEXB homodimer (HexB); β-hexosaminidase A, HEXA/HEXB heterodimer; tripeptidyl-peptidase I, TPP1. Panels B and C: average enzyme activities for NPC1 and NPC2 mutants normalized to appropriate age controls for early and late-stage disease, respectively. Data are fit to a first order polynomial using the robust fit option in Graphpad Prism 5 (solid line). Dashed line represents x=y. Panel D. Enzyme activities in serum from wild-type and mutant animals. Data are normalized to the average of all control animals, error bars represent standard error of the mean. Statistical significance (*) at the p ≤ 0.05 level was determined using the T.TEST function in Microsoft Excel.
Changes in lysosomal activities corresponded well in both mutants (Fig. 3, Panels B and C) as noted earlier for the MS analysis (Fig. 2), reflecting the linked functional roles of NPC1 and NPC2 in cholesterol transport. Activity of β-glucocerebrosidase (GBA, β-glucosidase), which is targeted via a Man-6-P-independent pathway [39], was unaffected by the loss of either NPC1 or NPC2.
We also measured lysosomal enzyme activities in liver and brain in a separate cohort of animals (Supplementary Fig. 2, Panels A and B). Increases in lysosomal enzyme activities observed in the brain of the two NPC mutants were typically paralleled in liver but observed to a higher degree (Supplementary Fig. 2, Panel C). Given that the activity of β-hexosaminidase altered quite dramatically in response to the loss of either NPC1 or NPC2 in both liver and brain, we measured total β-hexosaminidase and β-hexosaminidase A activity in serum from control and NPC mutant animals (Fig. 3, Panel D). Both activities were elevated in the serum of the NPC mutants although to a slightly lower degree (up to 2.5 fold that of average control) than found in liver or brain.
3.5 Transcriptional profiling of lysosomal proteins in Npc1−/− mutant mouse brain
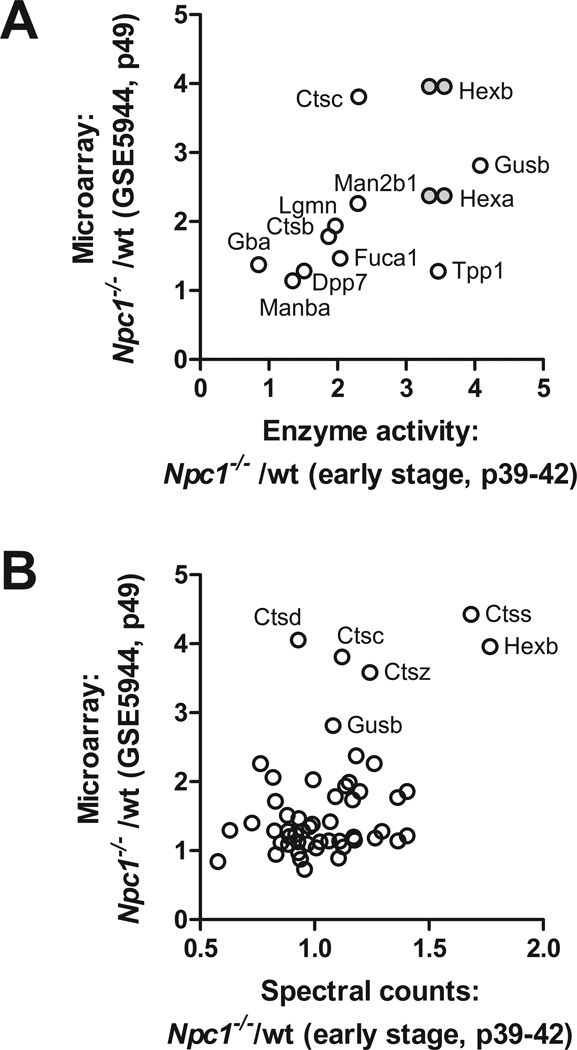
We compared enzyme activity measurements and spectral count data with an analysis of gene expression in Npc1−/− mutant animals. We extracted transcriptional profiling data for lysosomal proteins from an analysis of Npc1−/− cerebellum at P49 (GEO data series GSE5944), an age that approximated to our early stage group. Changes in lysosomal enzyme activity in the early stage Npc1−/− mutant animals compared to wild-type control correlate well with changes in the same lysosomal proteins measured by transcriptional profiling (Fig. 4, Panel A). This indicates that alterations in the expression of lysosomal proteins appear to reflect transcriptional changes. When expression measured by transcriptional profiling is compared with spectral count analysis (Fig.4, Panel B), there is less correlation. In part, this may be due to the fact that increases in absolute expression levels are underestimated using the MS approach.
Figure 4. Relationship between lysosomal gene expression profiles and protein levels and activities in Npc1−/− mice.
Fold-change of Npc1−/− compared to wild-type as measured by transcription profiling was compared with fold-change of enzyme activity (Panel A) and relative protein levels estimated by spectral counting (Panel B). Note that in Panel A, transcription profile data for HexA and HexB (grey filled symbols) are plotted against both total β-hexosaminidase activity (i.e., HEXA/HEXB heterodimer and HEXB homodimer) and β-hexosaminidase A activity (HEXA/HEXB heterodimer).
3.6 Biological significance of lysosomal changes
In this study, secondary changes in the expression of lysosomal proteins in mouse models of Niemann-Pick C type 1 and 2 disease were examined using proteomic methods, enzyme activity measurements and transcriptional profiling. Our goal was to identify lysosomal responses to the primary cholesterol storage defect that might provide useful cellular insights into these two similar yet genetically distinct diseases. There are two broad conclusions to be drawn from this study.
First, from enzyme activity measurements, there is a generalized increase in numerous (10 of the 11 measured) lysosomal enzyme activities in NPC disease that became more pronounced as disease progressed. Increases in activity were similar in both Npc1−/− and Npc2−/− animals although they tended to be greater in the former, presumably reflecting the more severe phenotype of disease. These increases appear to be specific to NPC. For example, in a previous study of lysosomal enzymes in brain autopsy material from patients with two other genetically distinct lysosomal storage diseases (late-infantile and juvenile neuronal ceroid lipofuscinosis), most of the activities were similar in patients and controls [40]. This includes β-hexosaminidase A which we find to be markedly elevated in NPC. The molecular basis for increased lysosomal activities is not clear but it may be relevant that the only activity that was not increased (i.e., GBA) was the only lysosomal enzyme assayed that is not transported to the lysosome via the Man-6-P targeting pathway.
Second, the proteomic analysis highlighted several lysosomal proteins that were elevated to a higher degree than the generalized increase (Table 1), suggesting a more specific response to disease. Several of the lysosomal alterations may represent cellular responses to the accumulation of various undigested materials within the cell, including lipids that may accumulate in response to a disruption in normal lysosomal function as a result of altered lipid composition of internal membrane structures. For example, degradation of sphingolipids is enhanced by a component of internal lysosomal membranes, bis(monoacylglycero)phosphate, and the concentration of this lipid increases during endocytosis with a concomitant decrease in cholesterol concentration (reviewed in [41]). The accumulation of cholesterol in the lysosomes of NPC mutants could quite conceivably interfere with this process.
β-Hexosaminidase
β-Hexosaminidase is a dimeric lysosomal enzyme with several isoenzymes that consist of different combinations of two distinct gene products, HEXA and HEXB. β-Hexosaminidase A is a heterodimer of HEXA and HEXB while β-hexosaminidase B is a HEXB homodimer. Levels of both HEXA and HEXB were increased in both NPC mutants. β-hexosaminidase A, together with GM2 activator protein (GM2A), plays a key role in the degradation of ganglioside GM2 (GM2). Lysosomal accumulation of GM2 is a well characterized cellular hallmark of human NPC1 and NPC2 disease [18] and was previously demonstrated in the respective mouse models at age 50 days [24]. Results presented here are consistent with an up-regulation of β-hexosaminidase as a compensatory cellular response to the elevated levels of GM2 and while not detected here, it is possible that levels of GM2A would also be increased. It is worth noting that β-hexosaminidase activity was previously found to be elevated 2- to 3-fold in the brain of Niemann-Pick A/B (acid sphingomyelinase (Smpd1) mutant) mice [42]. β-Hexosaminidase also functions in the degradation of proteoglycans (see below).
Prosaposin
PSAP was elevated in both mutants at early- and late-stage disease. PSAP encodes a lysosomal glycoprotein that is proteolytically processed into four small glycoproteins, saposins A-D. While the individual saposins are highly homologous, each has a distinct lysosomal function in the degradation of different glycosphingolipids (reviewed in [43]): saposin A functions with galactosylceramidase (GALC) in the degradation of galactosylceramide; saposin B acts with arylsulfatase A (ARSA) and α-galactosidase A (GLA), in the degradation of lactosylceramide and globotriaosylceramide, respectively; saposin C functions with GBA in the degradation of glucosylceramide; saposin D is thought to act with acid ceramidase (ASAH1) in the degradation of ceramide. Peptides are assigned here that correspond to all four of the saposins, and some of these peptides extend across the sites of proteolytic processing, indicating that it is the precursor prosaposin that is elevated rather than an individual mature saposin. PSAP accumulates within many lysosomal storage diseases [44] thus it is possible that its elevation in NPC is relatively non-specific. However, there is a significant accumulation of glycosphingolipids in both NPC1 and NPC2 disease and, as observed for β-hexosaminidase, the elevation in PSAP levels may be a compensatory response to this. It is interesting to note that, with the exception of ASAH1, catalytic proteins involved in glycosphingolipid degradation (e.g., ARSA, GLA and GBA) were not elevated in the NPC mutants. Given that the saposins do not serve a catalytic function but are instead thought to increase accessibility of the lipid substrates to the respective catabolic enzymes, these results may suggest that the lysosomal accumulation of cholesterol in the NPC mutants interferes with the accessibility of glycosphingolipids within the lysosomal membranes, rather than directly with their degradation per se.
Acid ceramidase (ASAH1)
ASAH1 was elevated in the NPC mutant animals in late-stage disease and is the only enzyme directly involved in the degradation of glycosphingolipids that was increased (see above).
Lysosomal acid lipase (LIPA)
LIPA plays a key role in lysosomal cholesterol processing in hydrolyzing lipoprotein-derived cholesterol esters to free cholesterol which is subsequently transported from the lysosome by the concerted action of NPC1 and NPC2. LIPA was elevated in the late- but not early-stage NPC animals. This is consistent with earlier observations of increased levels of Lipa transcript in the cerebellum of Npc1−/− mice, where this increase was interpreted to be a compensatory cellular response to the defect in cholesterol transport [45]. The latter study also identified an increase in Npc2 transcript levels in the cerebellum of Npc1−/− mice but in terms of the whole brain, we observe no specific elevation of NPC2 relative to that of other Man-6-phosphorylated proteins.
Other
Increased levels were also observed for a number of lysosomal proteins that are not involved in the degradation of known storage material in NPC disease. These include a number of lysosomal proteins involved in the degradation of proteins and peptides (Table 1) including tripeptidyl peptidase 1, cathepsins F, H, S and Z and these were elevated at both early and late stage. Lysosomal alpha mannosidase (MAN2B1) was also elevated, as was observed previously in the NPC mutant mice [22]. The physiological relevance of these changes is difficult to evaluate but they may suggest the accumulation of material within the lysosome that has been uncharacterized to date. Thus, understanding such changes from a mechanistic standpoint may provide valuable insights into NPC and potentially novel targets for therapeutic intervention.
3.7 Concluding remarks
As therapies for more LSDs enter the clinic, there is an increasing need for surrogates to follow disease progression, benchmark efficacy and titrate therapy. Given that the most immediate effects of a lysosomal defect may be reflected by compensatory changes in lysosomal activities, identification of secondary lysosomal changes may provide a rational and targeted approach to the discovery of useful biomarkers. In this study, mass spectrometric methods provided a platform for the discovery that β-hexosaminidase activity is significantly elevated in serum in response to NPC disease. This provides a proof-of-principle for proteomic approaches to identify secondary alterations in lysosomal activities that could potentially provide information of clinical value in LSDs.
Supplementary Material
Acknowledgements
This study was supported by The Ara Parseghian Medical Research Foundation and grants R01DK054317 and S10 RR024584 from the National Institutes of Health. We thank Caifeng Zhao for her excellent assistance with the mass spectrometry.
Abbreviations
- GPM
Global Proteome Machine
- LSD
lysosomal storage disease
- Man-6-P
mannose 6-phosphate
- sCI-MPR
soluble cation-independent mannose 6-phosphate receptor
- NPC
Niemann-Pick C disease; NPC1
Footnotes
The authors have no financial or commercial conflicts of interests to disclose.
References
- 1.Holtzman E. Lysosomes. New York: Plenum Press; 1989. [Google Scholar]
- 2.Scriver CR. The metabolic & molecular bases of inherited disease. New York: McGraw-Hill; 2001. [Google Scholar]
- 3.Rosenbaum AI, Maxfield FR. Niemann-Pick type C disease: molecular mechanisms and potential therapeutic approaches. J. Neurochem. 2011;116:789–795. doi: 10.1111/j.1471-4159.2010.06976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vanier MT. Niemann-Pick disease type C. Orphanet J. Rare. Dis. 2010;5:16. doi: 10.1186/1750-1172-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carstea ED, Morris JA, Coleman KG, Loftus SK, et al. Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science. 1997;277:228–231. doi: 10.1126/science.277.5323.228. [DOI] [PubMed] [Google Scholar]
- 6.Infante RE, Abi-Mosleh L, Radhakrishnan A, Dale JD, et al. Purified NPC1 protein. I. Binding of cholesterol and oxysterols to a 1278-amino acid membrane protein. J. Biol. Chem. 2008;283:1052–1063. doi: 10.1074/jbc.M707943200. [DOI] [PubMed] [Google Scholar]
- 7.Infante RE, Radhakrishnan A, Abi-Mosleh L, Kinch LN, et al. Purified NPC1 protein: II. Localization of sterol binding to a 240-amino acid soluble luminal loop. J. Biol. Chem. 2008;283:1064–1075. doi: 10.1074/jbc.M707944200. [DOI] [PubMed] [Google Scholar]
- 8.Ohgami N, Ko DC, Thomas M, Scott MP, et al. Binding between the Niemann-Pick C1 protein and a photoactivatable cholesterol analog requires a functional sterol-sensing domain. Proc. Natl. Acad. Sci. USA. 2004;101:12473–12478. doi: 10.1073/pnas.0405255101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naureckiene S, Sleat DE, Lackland H, Fensom A, et al. Identification of HE1 as the second gene of Niemann-Pick C disease. Science. 2000;290:2298–2301. doi: 10.1126/science.290.5500.2298. [DOI] [PubMed] [Google Scholar]
- 10.Chikh K, Vey S, Simonot C, Vanier MT, Millat G. Niemann-Pick type C disease: importance of N-glycosylation sites for function and cellular location of the NPC2 protein. Mol. Genet. Metab. 2004;83:220–230. doi: 10.1016/j.ymgme.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 11.Baker CS, Magargee SF, Hammerstedt RH. Cholesterol transfer proteins from ram cauda epidydimal and seminal plasma. Biol. Reprod. Suppl. 1993;48:86. [Google Scholar]
- 12.Liou HL, Dixit SS, Xu S, Tint GS, et al. NPC2, the protein deficient in Niemann-Pick C2 disease, consists of multiple glycoforms that bind a variety of sterols. J. Biol. Chem. 2006;281:36710–36723. doi: 10.1074/jbc.M608743200. [DOI] [PubMed] [Google Scholar]
- 13.Okamura N, Kiuchi S, Tamba M, Kashima T, et al. A porcine homolog of the major secretory protein of human epididymis, HE1, specifically binds cholesterol. Biochim. Biophys. Acta. 1999;1438:377–387. doi: 10.1016/s1388-1981(99)00070-0. [DOI] [PubMed] [Google Scholar]
- 14.Xu S, Benoff B, Liou HL, Lobel P, Stock AM. Structural basis of sterol binding by NPC2, a lysosomal protein deficient in Niemann-Pick type C2 disease. J. Biol. Chem. 2007;282:23525–23531. doi: 10.1074/jbc.M703848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Infante RE, Wang ML, Radhakrishnan A, Kwon HJ, et al. NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc. Natl. Acad. Sci. USA. 2008;105:15287–15292. doi: 10.1073/pnas.0807328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon HJ, Abi-Mosleh L, Wang ML, Deisenhofer J, et al. Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell. 2009;137:1213–1224. doi: 10.1016/j.cell.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang ML, Motamed M, Infante RE, Abi-Mosleh L, et al. Identification of surface residues on Niemann-Pick C2 essential for hydrophobic handoff of cholesterol to NPC1 in lysosomes. Cell. Metab. 2010;12:166–173. doi: 10.1016/j.cmet.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vanier MT. Lipid changes in Niemann-Pick disease type C brain: personal experience and review of the literature. Neurochem Res. 1999;24:481–489. doi: 10.1023/a:1022575511354. [DOI] [PubMed] [Google Scholar]
- 19.Pentchev PG, Gal AE, Booth AD, Omodeo-Sale F, et al. A lysosomal storage disorder in mice characterized by a dual deficiency of sphingomyelinase and glucocerebrosidase. Biochim. Biophys. Acta. 1980;619:669–679. doi: 10.1016/0005-2760(80)90116-2. [DOI] [PubMed] [Google Scholar]
- 20.Reagan JW, Jr, Hubbert ML, Shelness GS. Posttranslational regulation of acid sphingomyelinase in niemann-pick type C1 fibroblasts and free cholesterol-enriched chinese hamster ovary cells. J Biol Chem. 2000;275:38104–38110. doi: 10.1074/jbc.M005296200. [DOI] [PubMed] [Google Scholar]
- 21.Devlin C, Pipalia NH, Liao X, Schuchman EH, et al. Improvement in lipid and protein trafficking in Niemann-Pick C1 cells by correction of a secondary enzyme defect. Traffic. 2010;11:601–615. doi: 10.1111/j.1600-0854.2010.01046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dixit SS, Jadot M, Sohar I, Sleat DE, et al. Loss of niemann-pick c1 or c2 protein results in similar biochemical changes suggesting that these proteins function in a common lysosomal pathway. PLoS One. 2011;6:e23677. doi: 10.1371/journal.pone.0023677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loftus SK, Morris JA, Carstea ED, Gu JZ, et al. Murine model of Niemann-Pick C disease: mutation in a cholesterol homeostasis gene. Science. 1997;277:232–235. doi: 10.1126/science.277.5323.232. [DOI] [PubMed] [Google Scholar]
- 24.Sleat DE, Wiseman JA, El-Banna M, Price SM, et al. Genetic evidence for nonredundant functional cooperativity between NPC1 and NPC2 in lipid transport. Proc Natl Acad Sci U S A. 2004;101:5886–5891. doi: 10.1073/pnas.0308456101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sleat DE, Della Valle MC, Zheng H, Moore DF, Lobel P. The mannose 6-phosphate glycoprotein proteome. J Proteome Res. 2008;7:3010–3021. doi: 10.1021/pr800135v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beavis RC. Using the global proteome machine for protein identification. Methods Mol Biol. 2006;328:217–228. doi: 10.1385/1-59745-026-X:217. [DOI] [PubMed] [Google Scholar]
- 27.Craig R, Cortens JP, Beavis RC. Open source system for analyzing, validating, and storing protein identification data. J Proteome Res. 2004;3:1234–1242. doi: 10.1021/pr049882h. [DOI] [PubMed] [Google Scholar]
- 28.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 29.Zhang B, VerBerkmoes NC, Langston MA, Uberbacher E, et al. Detecting differential and correlated protein expression in label-free shotgun proteomics. J Proteome Res. 2006;5:2909–2918. doi: 10.1021/pr0600273. [DOI] [PubMed] [Google Scholar]
- 30.Sleat DE, Zheng H, Qian M, Lobel P. Identification of sites of mannose 6-phosphorylation on lysosomal proteins. Mol Cell Proteomics. 2006;5:686–701. doi: 10.1074/mcp.M500343-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.McCullagh P, Nelder JA. Generalized linear models. New York: Chapman and Hall; 1989. [Google Scholar]
- 32.Sleat DE, Sohar I, Lackland H, Majercak J, Lobel P. Rat brain contains high levels of mannose-6-phosphorylated glycoproteins including lysosomal enzymes and palmitoyl-protein thioesterase, an enzyme implicated in infantile neuronal lipofuscinosis. J Biol Chem. 1996;271:19191–19198. doi: 10.1074/jbc.271.32.19191. [DOI] [PubMed] [Google Scholar]
- 33.Kim KH, Pham CT, Sleat DE, Lobel P. Dipeptidyl-peptidase I does not functionally compensate for the loss of tripeptidyl-peptidase I in the neurodegenerative disease late-infantile neuronal ceroid lipofuscinosis. Biochem J. 2008;415:225–232. doi: 10.1042/BJ20080411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dando PM, Fortunato M, Smith L, Knight CG, et al. Pig kidney legumain: an asparaginyl endopeptidase with restricted specificity. Biochem J. 1999;339(Pt 3):743–749. [PMC free article] [PubMed] [Google Scholar]
- 35.Wendeler M, Sandhoff K. Hexosaminidase assays. Glycoconj J. 2009;26:945–952. doi: 10.1007/s10719-008-9137-5. [DOI] [PubMed] [Google Scholar]
- 36.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.German DC, Liang CL, Song T, Yazdani U, et al. Neurodegeneration in the Niemann-Pick C mouse: glial involvement. Neuroscience. 2002;109:437–450. doi: 10.1016/s0306-4522(01)00517-6. [DOI] [PubMed] [Google Scholar]
- 38.Lopez ME, Klein AD, Dimbil UJ, Scott MP. Anatomically defined neuron-based rescue of neurodegenerative Niemann-Pick type C disorder. J Neurosci. 2011;31:4367–4378. doi: 10.1523/JNEUROSCI.5981-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reczek D, Schwake M, Schroder J, Hughes H, et al. LIMP-2 is a receptor for lysosomal mannose-6-phosphate-independent targeting of beta-glucocerebrosidase. Cell. 2007;131:770–783. doi: 10.1016/j.cell.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 40.Sleat DE, Sohar I, Pullarkat PS, Lobel P, Pullarkat RK. Specific alterations in levels of mannose 6-phosphorylated glycoproteins in different neuronal ceroid lipofuscinoses. Biochem J. 1998;334(Pt 3):547–551. doi: 10.1042/bj3340547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schulze H, Kolter T, Sandhoff K. Principles of lysosomal membrane degradation: Cellular topology and biochemistry of lysosomal lipid degradation. Biochim Biophys Acta. 2009;1793:674–683. doi: 10.1016/j.bbamcr.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 42.Banno Y, Sasaki N, Miyawaki S, Kitagawa T, Nozawa Y. Properties of lysosomal beta-hexosaminidase accumulated in Niemann-Pick mouse liver. Biochem Med Metab Biol. 1986;36:322–332. doi: 10.1016/0885-4505(86)90143-x. [DOI] [PubMed] [Google Scholar]
- 43.Matsuda J, Yoneshige A, Suzuki K. The function of sphingolipids in the nervous system: lessons learnt from mouse models of specific sphingolipid activator protein deficiencies. J Neurochem. 2007;103(Suppl 1):32–38. doi: 10.1111/j.1471-4159.2007.04709.x. [DOI] [PubMed] [Google Scholar]
- 44.Morimoto S, Yamamoto Y, O'Brien JS, Kishimoto Y. Distribution of saposin proteins (sphingolipid activator proteins) in lysosomal storage and other diseases. Proc Natl Acad Sci U S A. 1990;87:3493–3497. doi: 10.1073/pnas.87.9.3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Repa JJ, Valasek MA, Beltroy EP, et al. Molecular, anatomical, and biochemical events associated with neurodegeneration in mice with Niemann-Pick type C disease. J Neuropathol Exp Neurol. 2005;64:323–333. doi: 10.1093/jnen/64.4.323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.