Abstract
In recent years, the highly conserved Lin28 RNA-binding proteins have emerged as factors that define stemness in several tissue lineages. Lin28 proteins repress let-7 microRNAs and influence mRNA translation, thereby regulating the self-renewal of mammalian embryonic stem cells. Subsequent discoveries revealed that Lin28a and Lin28b are also important in organismal growth and metabolism, tissue development, somatic reprogramming and cancer. In this Review, we discuss the Lin28 pathway and its regulation, outline its roles in stem cells, tissue development, and pathogenesis, and examine the ramifications for re-engineering mammalian physiology.
Introduction
A central question in stem cell biology is whether common factors exist to define “stemness” in multiple tissue lineages. Arguably, one such candidate is the RNA-binding protein Lin28, which was first identified in the nematode C. elegans through screens for lineage-modifying genes that alter developmental timing, or heterochrony (lin-28; Ambros and Horvitz 1984). Two other prominent heterochronic genes, lin-4 and let-7, were the first microRNAs to be discovered, and both directly repress lin-28 to suppress heterochronic reiterations of cell lineages. Heterochronic “reiteration” of nematode stem cells, as C. elegans geneticists first observed, was strongly reminiscent of mammalian stem cell self-renewal (Chalfie et al. 1981; Ambros and Horvitz 1984). This connection was reinforced by the discovery that mouse embryonic stem cells (ESCs) express high levels of mammalian Lin28, which decrease upon differentiation (Moss and Tang 2003). Successful reprogramming of human fibroblasts into induced pluripotent stem cells (iPSCs) using Lin28, along with Oct4, Sox2, and Nanog, further corroborated its role in pluripotent stem cells (Yu et al. 2007), but the mechanism of action for Lin28 remained unclear. A subsequent flurry of studies showing that Lin28 directly inhibits let-7 maturation in ESCs rapidly validated Lin28’s function in ESC self-renewal (Viswanathan et al. 2008; Rybak et al. 2008; Heo et al. 2008 Newman et al. 2008). With the discovery that Lin28 is also important in cancer, the germ lineage, and cellular metabolism (Viswanathan et al. 2009; West et al. 2009; Zhu et al. 2011), understanding the role of Lin28 in stem cells during development and disease pathogenesis has emerged as a new field of research. In this Review, we will discuss the Lin28 pathway and its complex molecular mechanisms, outline its known roles in stem cells, tissue development, and pathogenesis, and examine its ramifications for re-engineering mammalian physiology.
Lin28/let-7: A Conserved Bistable Switch
Current insights into Lin28 rest heavily on precedents in C. elegans genetics. Lin-28 was first discovered through mutagenesis screens for heterochronic genes (Horvitz and Sulston 1980; Sulston and Horvitz 1981; Ambros and Horvitz 1984). Loss-of-function in lin-28 accelerates differentiation of the hypodermal and vulval stem cells (called seam cells and VPCs respectively in nematodes). In contrast, gain-of-function in lin-28 promotes self-renewal and delays differentiation of the hypodermal and vulval stem cells, leading to proliferation of hypodermal stem cells and a cell-cycle delay in vulval stem cells (Moss et al. 1997). Lin-28 is highly expressed during embryogenesis and during early larval development in the hypodermal, neural and muscle cells, but gradually diminishes and disappears by adulthood.
Two heterochronic microRNAs (miRNAs) repress lin-28 post-transcriptionally via direct binding sites in its 3’ UTR: lin-4 and let-7 (Reinhart et al. 2000; Pasquinelli et al. 2000; Roush et al. 2008). Although the canonical let-7 is only expressed late in larval development to drive the transition to adulthood, three let-7 homologs (mir-48, mir-84 and mir-241) display overlapping expression with lin-28. Indeed, loss-of-function in these let-7 homologs phenocopied lin-28 gain-of-function in the hypodermal stem cells, and lin-28 was epistatic to the three let-7 homologs (Abbott et al. 2005). Mutation of the let-7 binding site in the lin-28 3’ UTR also led to an increase in lin-28 3’ UTR-lacZ reporter expression (Morita and Han 2006), suggesting that let-7 binding contributes to lin-28 repression, and underlies their opposing roles in regulating differentiation.
The role of lin-28 in mammalian stem cells was less clear until quite recently. The first glimpse of a connection came from the discovery that the mammalian lin-28 ortholog is highly expressed in mouse ESCs and human embryonal carcinoma cells (Moss and Tang 2003). The connection was further validated when human Lin28 was used with Oct4, Sox2 and Nanog to reprogram human somatic fibroblasts into pluripotent stem cells (Yu et al. 2007). Around the same time, a post-transcriptional mechanism was proposed to be responsible for the dramatic disparity between high levels of pri-let-7 transcript and the deficiency of mature let-7 microRNA in early mouse embryos and ESCs (Thomson et al. 2006; Wulczyn et al. 2007). These two lines of inquiry rapidly converged through a flurry of studies that showed that Lin28 (now routinely termed Lin28a) and its paralog Lin28b directly inhibit the posttranscriptional maturation of let-7 in ESCs (Viswanathan et al. 2008; Rybak et al. 2008; Heo et al. 2008; Newman et al. 2008). A generally similar mechanism was later verified to be conserved in C. elegans (Lehrbach et al. 2009; Van Wynsberghe et al. 2011). Since Lin28a/b inhibit the biogenesis of let-7 microRNAs, which in turn repress Lin28a/b expression, it became clear that this bistable switch represents a central mechanism that governs stem cell self-renewal from worms to mammals.
Molecular Mechanisms of Lin28 Function
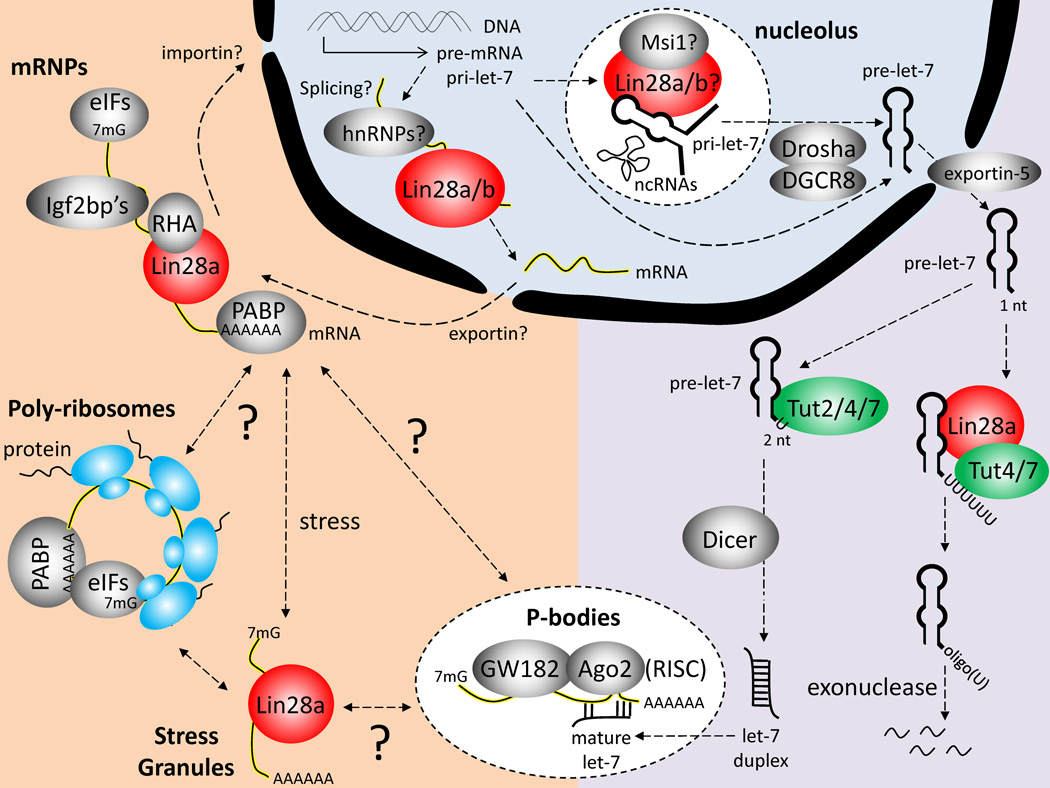
Following the discovery that Lin28a/b represses let-7 biogenesis, several groups set about to determine the detailed biochemical mechanisms underlying let-7 repression as a model for understanding miRNA biogenesis. Similar to the biogenesis of other miRNAs, let-7 is first transcribed as part of long pri-let-7 transcripts in the nucleus (Roush et al. 2008). Within the pri-let-7 transcripts is a hairpin structure that is the precursor miRNA (pre-let-7). Drosha, in complex with its RNA-binding cofactor DGCR8, cleaves and releases the ~70 nt hairpin structure to produce pre-let-7. Like other pre-miRNAs, pre-let-7 is then thought to be exported from the nucleus into the cytoplasm by exportin-5, although the majority of prelet-7 species lack the 3’ two-nucleotide overhang that exportin-5 presumably needs to export pre-miRNAs (Heo et al. 2012; Yi et al. 2003), suggesting that another mechanism might serve this function. In the cytoplasm, pre-let-7 is further processed by Dicer to produce a 22-nt double-stranded RNA duplex. Mature single-stranded let-7 is then incorporated from the duplex into the RNA-induced silencing complex (RISC) to target mRNAs for translation inhibition and/or degradation in P-bodies (Figure 1).
Figure 1. Overview of Molecular Mechanisms Underlying Lin28 Function.
Both Lin28a and Lin28b have been observed to shuttle between the nucleus and cytoplasm, binding both mRNAs and pri-/prelet-7. In the nucleus, Lin28a/b could potentially work in tandem with the heterogeneous nuclear ribonucleoproteins (hnRNPs) to regulate splicing, or with Musashi-1 (Msi1) to block pri-let-7 processing. In the cytoplasm, Lin28a recruits Tut4/7 to oligouridylate pre-let-7, and block Dicer processing to mature let-7 miRNA (right, violet). Lin28a also recruits RNA helicase A (RHA) to regulate mRNA processing in messenger ribonucleoprotein (mRNP) complexes, in tandem with the Igf2bp’s, poly(A)-binding protein (PABP), and the eukaryotic translation initiation factors (eIFs). In response to unknown signals and stimuli, the mRNAs are either shuttled into poly-ribosomes for translation, stress granules for temporary sequestering, or P-bodies for degradation, in part via miRNAs and the Ago2 endonuclease (left, orange).
In a multi-pronged fashion, Lin28a/b binds to both pri-let-7 and pre-let-7, effectively sabotaging the post-transcriptional processing of let-7 (Viswanathan et al. 2008; Rybak et al. 2008; Heo et al. 2008; Newman et al. 2008). X-ray crystallography studies further revealed that Lin28a binds pre-let-7 at the terminal loop and at the bulge GGAG motif where Dicer cleaves (Nam et al. 2011). Lin28a also recruits Tut4 (Zcchc11), a cytoplasmic terminal uridylyl transferase, to oligo-uridylate pre-let-7 and prevent its processing by Dicer (Heo et al. 2009; Hagan et al. 2009). Recent studies have further elaborated on this mechanism, suggesting that Tut7 (Zcchc6) is a redundant homolog of Tut4 that can also oligo-uridylate pre-let-7 in the presence of Lin28a (Thornton et al. 2012; Heo et al. 2012). In contrast, when Lin28a is absent, Tut4/7 or Tut2 (Papd4/Gld2) mono-uridylates pre-let-7’s at their 3’ one-nucleotide overhang to generate a two-nucleotide overhang, thereby enabling their processing by Dicer (Heo et al. 2012). Thus, one would expect oligo-uridylated pre-let-7 to accumulate when pri-let-7, Lin28a and Tut4/7 are present. But this is not observed, suggesting that an unknown nuclease must exist to degrade oligo-uridylated pre-let-7’s (Heo et al. 2008, 2009), and/or that Lin28 can sequester pri-let-7’s to prevent further processing (Viswanathan et al. 2008; Newman et al. 2008). Indeed, a study suggests that Lin28b is predominantly localized in the nucleolus where it can sequester pri-let-7 away from Drosha/DGCR8 processing, whereas Lin28a is predominantly localized in the cytoplasm where it can recruit Tut4 to oligo-uridylate pre-let-7 and prevent Dicer processing (Piskounova et al. 2011). However, mammalian Lin28a/b and C. elegans lin-28 can all enter the nucleus as well as the cytoplasm (Moss et al. 1997; Guo et al. 2006; Balzer and Moss 2007; Heo et al. 2008; Piskounova et al. 2011; Van Wynsberghe et al. 2011; Vogt et al. 2012; Hafner et al. 2013). Moreover, all three proteins possess a putative nucleolar localization signal, and all three proteins can bind to both pri- and pre-let-7 (Viswanathan et al. 2008; Rybak et al. 2008; Heo et al. 2008; Newman et al. 2008; Lehrbach et al. 2009; Van Wynsberghe et al. 2011). Thus, the mode of regulation of this division of labor between Lin28a and Lin28b remains unclear (Figure 1).
Under different conditions of metabolic stress in embryonal carcinoma cells and myoblasts, Lin28a localizes specifically to cytoplasmic stress granules where mRNAs are sequestered and mRNA translation is temporarily stalled (Balzer and Moss 2007; Polesskaya et al. 2007). Under normal growth conditions, cytoplasmic Lin28a can directly or indirectly associate with translation initiation factors eIF3B and eIF4E, elongation factors EF1α and EF1α2, ribosomal proteins, poly(A)-binding protein (PABP), Igf2bp1/2/3, Musashi1 (Msi1) and RNA helicase A (RHA) in messenger ribonucleoprotein (mRNP) complexes to regulate mRNA translation (Balzer et al. 2007; Polesskaya et al. 2007; Jin et al. 2011). When point mutations are introduced into the RNA-binding motifs, Lin28a localizes to the nucleus (Balzer and Moss 2007). These findings suggest a model in which Lin28a regulates the post-transcriptional processing of its mRNA targets, perhaps by first binding them in the nucleus and subsequently shuttling them between ribosomes, P-bodies or stress granules for translational regulation, depending on the environmental conditions. It would be interesting to know which factors sense the environmental conditions to regulate this shuttling of Lin28a, and whether these conditions alter the RNAs bound by Lin28a (Figure 1). One study, for example, suggests that retinoic acid-induced differentiation of ESCs triggers Msi1 expression, which recruits Lin28a to the nucleus to sequester and inhibit pri-miR-98 but not pri-let-7b (Kawahara et al. 2011). But what else could Lin28a/b be doing in the nucleus? Nuclear Lin28 could also regulate the alternative splicing of pre-mRNAs, or processing of small nucleolar RNAs (snoRNAs) and long non-coding RNAs (lincRNAs) to generate greater RNA diversity. The likelihood for this additional role is supported by the RNA-dependent association between Lin28a protein and nuclear splicing factors like hnRNP F and hnRNP H1 (Polesskaya et al. 2007), and its direct regulation of splicing factors and snoRNAs (Wilbert et al. 2012; Hafner et al. 2013). How these various RNA-processing mechanisms relate to stem cell self-renewal and plasticity in response to environmental changes remains an important avenue for future research.
Regulatory Signals Upstream of Lin28
Throughout their lifespan stem cells must decide whether to self-renew, proliferate, differentiate or die. The regulation of stem cell homeostasis is a complex process that involves integrating intrinsic and extrinsic signals, so that stem cells can correctly adapt to the environment. The central role of the Lin28/let-7 bistable switch in governing stem cell self-renewal raises the provocative question: what signaling pathways converge upstream to regulate the switch?
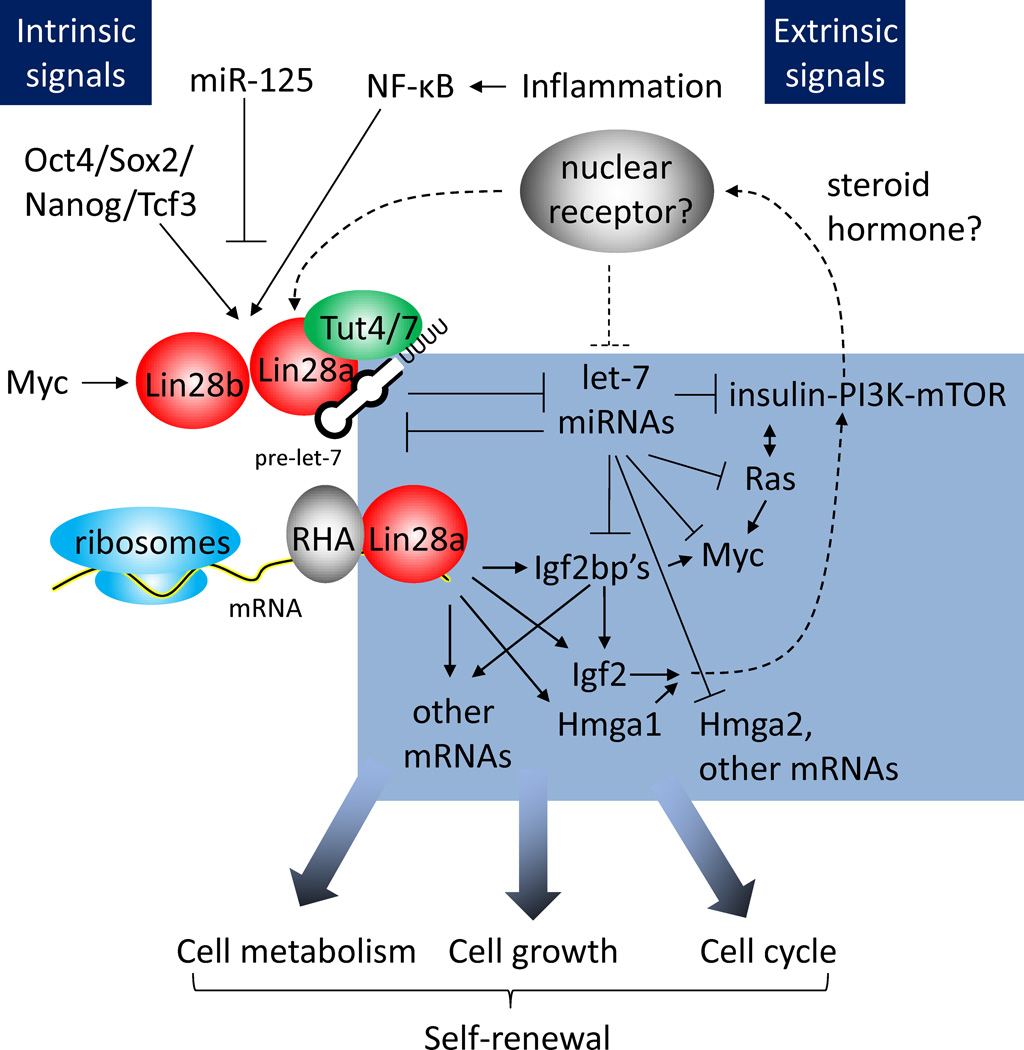
In C. elegans, an important intrinsic signal upstream of lin-28 that regulates hypodermal stem cell self-renewal is the microRNA lin-4. In vertebrates, lin-4 is conserved as miR-125a and miR-125b (Lagos-Quintana et al. 2002). Vertebrate miR-125/lin-4 has been shown to be critical for regulating processes as disparate as neurogenesis, somitogenesis, hematopoiesis, myogenesis and epidermal stem cell self-renewal (Rybak et al. 2008; Le et al. 2009a; Le et al. 2009b; Klusmann et al. 2010; Bousquet et al. 2010; O’Connell et al. 2010; Ooi et al. 2010; Ge et al. 2011; Zhang et al. 2011; Guo et al. 2010; Chaudhuri et al. 2012). During these developmental processes, miR-125/lin-4 appears to regulate stem cell self-renewal and progenitor differentiation by repressing a variety of different targets, including Lin28a/b, as well as the p53 network (Le et al. 2011). In mammalian ESCs, the pluripotency factors Oct4, Sox2, Nanog and Tcf3 have also been shown to regulate the transcription of Lin28a (Marson et al. 2008). Amongst these pluripotency factors, Sox2 appears to be the most critical for regulating Lin28a expression, based on Bayesian probabilistic network modeling of single-cell gene expression data during iPS reprogramming (Buganim et al. 2012). Interestingly, Sox2 directly binds to Lin28a in a nuclear protein-protein complex, suggesting a close relationship between Sox2 and Lin28a in pluripotency (Cox et al. 2010). Repression of the Dot1L H3K79 histone methyltransferase upregulates Lin28a during reprogramming, although the mechanism is indirect via mesenchymal regulators downstream of TGFβ signaling (Onder et al. 2012). Finally, c-Myc and NF-κB transactivate Lin28b in transformed cancer cells (Chang et al. 2009; Iliopoulos et al. 2009), suggesting that Lin28a and Lin28b possess distinct cis-regulatory elements to drive their transcription (Figure 2). Beyond these studies, relatively little is known about the transcription factors that regulate Lin28a and Lin28b expression during mammalian development, and further investigation is warranted.
Figure 2. Signals Upstream and Targets Downstream of Lin28 in the Lin28 Pathway.
The lin-4 homolog miR-125a/b represses both Lin28a and Lin28b during stem cell differentiation. The core pluripotency transcription factors Oct4, Sox2, Nanog and Tcf3 can activate Lin28a transcription in ESCs and iPSCs, whereas the growth regulator Myc and the inflammation-/stress-responsive NF-κB can transactivate Lin28b. A putative steroid hormone-activated nuclear receptor, conserved from C. elegans daf-12, might also regulate both Lin28a/b and let-7 expression. Downstream of Lin28a/b, the let-7 family represses a network of proto-oncogenes, including the insulin-PI3K-mTOR pathway, Ras, Myc, Hmga2, and the Igf2bp’s. At the same time, Lin28a can also directly bind to and regulate translation of mRNAs, including Igf2bp’s, Igf2, Hmga1, and mRNAs encoding metabolic enzymes, ribosomal peptides, and cell-cycle regulators. Together, this broad network of targets allows Lin28 to program both metabolism and growth to regulate self-renewal.
The C. elegans nuclear receptor daf-12 feeds extrinsic signals from steroid hormones to lin-28 and let-7, to regulate diapause or dauer arrest (Antebi et al. 1998, 2000; Gerisch et al. 2007; Bethke et al. 2009; Hammell et al. 2009). Although this mechanism is well-characterized in C. elegans development, and a similar ecdysone-let-7 mechanism operates in Drosophila metamorphosis (Sokol et al. 2008; Chawla et al. 2012), it is unclear if a similar mechanism exists to hormonally regulate the Lin28/let-7 switch in mammals (Figure 2). Studies have shown that the homologous retinoic acid receptors and estrogen receptor α regulate let-7 expression in vitro (Thomson et al. 2006; Wulczyn et al. 2007; Gehrke et al. 2010; Schulz et al. 2011; Bhat-Nakshatri et al. 2009), but both the directness and the physiological relevance of these mechanisms remain to be shown. It would also be interesting to test if any of the nuclear hormone receptors implicated in pluripotency and especially the naive state (Feng et al. 2009; Heng et al. 2010; Guo and Smith 2010; Wang et al. 2011; Martello et al. 2012), a state associated with rodent diapause (Nichols et al. 2001, 2009), also regulates the Lin28/let-7 switch (Figure 2).
Coordinate Regulation of Metabolism and Cell Cycle by Downstream Targets of Lin28
Given our wealth of understanding of the upstream regulators of lin-28 in worms, it is surprising that relatively little was discerned about the downstream RNA targets in the C. elegans literature. It was only after the discovery that lin-28 directly binds to and represses pre-let-7, that the more well-known let-7 targets could be placed downstream of lin-28. Only some of these let-7 targets are known to be conserved in mammals, including lin-41 (Trim71), let-60 (Ras), and Lin28 itself. The regulation of NRAS and KRAS by let-7 in human cancer cells has led to the proposal that let-7 functions as a tumor suppressor in humans (Johnson et al. 2005). Indeed, a network of let-7 targets involving numerous other proto-oncogenes has been uncovered in mammalian cells, including Myc, Hmga2, Igf2bp’s, cyclins (Sampson et al. 2007; Mayr et al. 2007; Lee et al. 2007; Johnson et al. 2007; Yu et al. 2007; Boyerinas et al. 2008; Iliopoulos et al. 2009; Legesse-Miller et al. 2009; Chang et al. 2012), and components of the insulin-PI3K-mTOR pathway like Igf1r, Insr, Irs2, Akt2 and Rictor (Zhu et al. 2011; Frost and Olson 2011). These studies fit with the suggestion that Lin28a/b function as oncogenes in multiple cancers by repressing the let-7 network (Viswanathan et al. 2009). A majority of these studies, however, were conducted in vitro and many of these claims still await validation in vivo through Lin28a/b or let-7 mouse models.
Adding a second layer of complexity downstream of Lin28, some studies have shown that Lin28a directly binds to many mRNAs, including Igf2 in myoblasts and neural progenitors, and cyclin A/B in ESCs, to directly enhance their translation independently of let-7 (Polesskaya et al. 2007; Xu et al. 2009; Balzer et al. 2010). Genome-wide RNA-immunoprecipitation studies further reveal thousands of mRNA targets bound directly by Lin28. In human ESCs and cancer cells, LIN28A directly binds and promotes the mRNA translation of numerous metabolic enzymes, ribosomal peptides, cyclins as well as splicing factors (Peng et al. 2011; Li et al. 2012; Wilbert et al. 2012; Hafner et al. 2013). Curiously, in mouse ESCs, Lin28a was also recently found to bind and subtly repress the ribosomal occupancy of numerous membrane protein mRNAs (Cho et al. 2012). Given the plethora of mRNA targets that are emerging for Lin28a, including nearly 50% of the human transcriptome in one study (Cho et al. 2012), an important task lying ahead is to determine whether all these targets or only a subset contribute to the Lin28 phenotypes observed in vivo. Such an undertaking might require a return to the powerful genetics of C. elegans to search for conserved mRNA targets of lin-28. This idea is supported by the finding that let-7-independent mechanisms must account for the lin-28 phenotype in C. elegans as well (Vadla et al. 2012).
Several key insights into Lin28 function are emerging from the small set of well-validated targets, most notably that Lin28 coordinates both proliferative growth and metabolism. Lin28a/b can upregulate a large number of cell-cycle regulators through let-7 repression, including Myc, Ras, cyclin D1/2, Cdk6, Cdc25a, Cdc34, Trim71 (which represses p21Cip1), Hmga2 (which represses p16Ink4a and p19Arf) and PI3K/Akt signaling (Johnson et al. 2005; Sampson et al. 2007; Johnson et al. 2007; Chang et al. 2012; Mayr et al. 2007; Nishino et al. 2008; Zhu et al. 2011). A recent study even suggested that let-7 can directly bind to and silence Rb1/E2F target genes via heterochromatin during senescence (Benhamed et al. 2012). Lin28a also directly binds and promotes the translation of mRNAs encoding cyclin A/B/D, Cdk1/2/4, Cdc2 and Cdc20, thereby coordinating the cell-cycle at multiple checkpoints (Xu et al. 2009; Li et al. 2012; Hafner et al. 2013). Besides the cell-cycle, Lin28a/b might also control cellular growth by regulating ribosomal synthesis of proteins. Lin28a directly binds to the mRNAs of numerous ribosomal peptides in human ESCs (Peng et al. 2011). In addition, Lin28a/b increases mTOR signaling via let-7 (Zhu et al. 2011; Frost and Olson 2011), which can activate ribosomal biogenesis and translation in many contexts. In parallel with its extensive control of cell-cycle and cell growth regulators, Lin28a/b appears to also coordinate cellular metabolism, both via let-7 and by directly stimulating mRNA translation. Through let-7, Lin28a/b upregulates the insulin/PI3K, Ras and Myc pathways – all of which are oncogenic regulators of metabolism (Vander Heiden et al. 2009; Dang 2011). By directly binding mRNAs and influencing translation of the Igf2bp’s, Igf2, glycolysis enzymes, and mitochondrial enzymes, Lin28a can directly potentiate cellular metabolism (Zhu et al. 2011; Peng et al. 2011; Polesskaya et al. 2007; Janiszewska et al. 2012; Hafner et al. 2013). Hmga1, another mRNA target of Lin28a, can also upregulate insulin/PI3K signaling (Liau et al. 2006; Chiefari et al. 2011; Peng et al. 2011). Given how growth signaling pathways are intertwined with cellular metabolism, it is perhaps not surprising that Lin28 would have to program both arms of genes to regulate self-renewal. Thus a model is emerging, albeit an inchoate one, whereby Lin28 programs both metabolism and proliferative growth to regulate stem/progenitor cell self-renewal (Figure 2).
Lin28 in Embryonic Stem Cell Metabolism
The functional role of Lin28 in cellular metabolism is evidenced by recent studies in ESCs. Recent work has shown that aerobic glycolysis, akin to the Warburg effect in cancer, is critical to ESCs and iPSCs (Zhu et al. 2010b; Folmes et al. 2011). This is perhaps not surprising, given the high proliferative capacity of ESCs, and the importance of glycolysis in providing carbon intermediates for anabolic growth (Vander Heiden et al. 2009). What is surprising is that some studies have shown that mitochondrial oxidative metabolism is also critical to ESCs (Wang et al. 2009; Alexander et al. 2011; Zhang et al. 2011b), despite the immature morphology of ESC mitochondria. Interestingly, Lin28a binds to a large number of mRNAs encoding mitochondrial enzymes in human ESCs (Peng et al. 2011). One possibility is that ESC mitochondrial oxidation could be operating to recycle mitochondrial NAD+ and keep the Krebs cycle running in order to generate fatty acids and various amino acids for ESCs (Shyh-Chang et al. 2011). Curiously, mouse ESCs uniquely rely upon mitochondrial oxidation of threonine (Thr) into glycine (Gly), via threonine dehydrogenase (Tdh), to generate one carbon/folate intermediates to fuel rapid nucleotide synthesis (Wang et al. 2009). This seminal early work led to findings that the 5-methyl-THF generated by mitochondrial Thr oxidation also fuels the synthesis of S-adenosyl-methionine (SAM) to regulate histone H3K4 methylation and the pluripotency of ESCs (Shyh-Chang et al. 2013a). Surprisingly, Lin28a overexpression in ESCs leads to a dramatic accumulation of many metabolites in the Thr-Gly-SAM pathway, whereas overexpression of let-7 reduces the abundance of these metabolites, suggesting that the Thr-Gly-SAM pathway is at least indirectly regulated by the Lin28/let-7 switch to maintain ESC self-renewal. These findings might also have relevance to cancer, since lung cancer stem cells have been found to express and depend upon high levels of both Lin28b and glycine decarboxylase (Gldc) in the Thr-Gly pathway to initiate tumorigenesis (Zhang et al. 2012). In fact several enzymes in Gly metabolism have recently been implicated in human tumorigenesis (Locasale et al. 2011; Possemato et al. 2011; Jain et al. 2012). Lin28 could thus potentially regulate glucose and amino acid metabolism in a variety of stem and progenitor cells, both normal and malignant (Shyh-Chang et al. 2013b).
Lin28 in Early Embryogenesis, Pluripotent Stem Cells and Reprogramming
The earliest phases of embryogenesis feature high levels of Lin28a due to protein inheritance through the maternal oocyte. From the mouse zygote to the pre-implantation blastocyst, Lin28a is exclusively localized in the nucleolus where it is thought to regulate nucleolar maturation (Vogt et al. 2012). Morpholino knockdown of Lin28a in the zygote produces defects in nucleolar morphology and developmental arrest at the 2-cell and 4-cell stages, suggesting that Lin28a is required for proper nucleolar genesis and function and early embryogenesis. Curiously, Lin28a is localized in the nucleolus of mouse ESCs as well, but not primate ESCs (Vogt et al. 2012). Given that Lin28a is an RNA-binding protein, these observations suggest that maternal Lin28a might also be involved in ribosomal RNA processing in the nucleolus to regulate zygotic genome activation during the maternal-zygotic transition, although this remains speculative.
After zygotic genome activation, mammalian blastocysts show high levels of Lin28a and Lin28b ranscription in the pluripotent cells of the inner cell mass (ICM) and epiblast and their in vitro correlates – the indefinitely self-renewing ESCs. Studies suggest that Lin28a/b acts as a repressor of let-7 icroRNAs to prevent premature differentiation in the pluripotent ICM and epiblast (Suh et al. 2010; Melton et al. 2010). When the pluripotent ICM is cultured in vitro for ESC derivation, Lin28a is further upregulated, in parallel with acquisition of indefinite self-renewal capacity in vitro (Tang et al. 2010). Furthermore, overexpression of Lin28a with a cocktail of the core pluripotency-associated transcription factors Oct4, Sox2 and Nanog, helps promote reprogramming of human somatic fibroblasts into indefinitely self-renewing iPSCs (Yu et al. 2007; Hanna et al. 2009). These data suggest that Lin28a is critical to pluripotent stem cell self-renewal.
But is Lin28 also required for pluripotency? Lin28 knockout mouse models suggest the answer is no. Lin28a knockout mice progress through the blastocyst stage without obvious developmental defects in utero, although they weigh 20% less at birth (Zhu et al. 2010). Lin28b knockout mice are viable and fertile. Thus Lin28a/b do not appear to be essential for pluripotency per se, in vivo.
Do pluripotent stem cells then require Lin28a/b for indefinite self-renewal? The answer depends on context, since overexpression of mature let-7 does not inhibit mouse ESC self-renewal unless DGCR8 is knocked out and miRNA biogenesis is prevented (Melton et al. 2010). Another class of miRNAs called the miR-290 family can respond to and compensate for the effects of let-7 overexpression. Although the breadth of genes targeted by the miR-290 family, and the connections with Lin28a/b (if any) remain unclear, it is thought that let-7 promotes ESC differentiation in the absence of DGCR8 by directly repressing Sall4, Nmyc, and Lin28a. Conversely, Lin28a or Lin28b knockout in the presence of DGCR8 should not lead to defects in self-renewal via let-7 upregulation alone, even if we ignore the compensatory redundancy observed between Lin28a and Lin28b (Wilbert et al. 2012). RNAi against Lin28, however, does lead to proliferative defects in both mouse and human ESC (Xu et al. 2009; Peng et al. 2011), suggesting that Lin28a/b might synergistically promote ESC self-renewal through a combination of let-7 repression and let-7-independent mechanisms such as direct binding of mRNAs involved in metabolism and growth. Thus it will also be interesting to see if Lin28a/b double knockout leads to defects in ESC self-renewal. On the other hand, let-7 knockdown in fibroblasts promotes iPS reprogramming (Melton et al. 2010), suggesting that Lin28a might promote self-renewal via repression of let-7 during iPS reprogramming, without compensatory effects from ESC-specific miR-290. Indeed, studies have shown that Lin28a can accelerate the early stochastic phase of iPS reprogramming by accelerating the cell-cycle, and that Lin28a is one of the earliest markers of the deterministic phase of iPS reprogramming after endogenous Sox2 expression is induced (Hanna et al. 2009; Buganim et al. 2012; Golipour et al. 2012). It remains to be verified, however, whether let-7 is the relevant target of Lin28a during reprogramming, and what downstream targets of let-7 drive iPS reprogramming. It is also unknown whether Lin28/let-7 is implicated in the much more rapid and deterministic process of reprogramming by somatic cell nuclear transfer (SCNT).
Lin28 in Normal and Transformed Tissue Progenitors
Contrary to popular belief, Lin28a/b’s expression and influence on development is far from unique to pluripotent cells in the blastocyst ICM, but rather extends to a variety of tissues. For instance, the trophoblast and the resultant placental tissues show high levels of Lin28a/b. Studies suggest that the high Lin28a levels in trophoblast stem cells decrease to permit differentiation into trophoblast giant cells (Fromme et al. 2009; Winger et al. 2010), and Lin28a levels increase again during the invasive phase of placenta development to regulate cell migration (Seabrook et al., 2011). Moreover, the LIN28B locus shows imprinting and paternal monoallelic expression in the human placenta (Barbaux et al. 2012). However the placental function of Lin28 remains unclear in vivo.
Germline stem cells also retain high levels of Lin28 expression during mammalian development. Lin28a promotes primordial germ cell (PGC) specification via let-7 regulation of the master regulator Blimp1 (West et al. 2009), and remains high specifically in the spermatogonial stem cells of adult male testes (Zheng et al. 2009). Both Lin28a knockout and let-7 overexpression led to a reduction in PGCs during embryogenesis, and a reduction in proliferating spermatogonia and germ cells before adulthood (Shinoda et al., 2013). Interestingly, aberrant overexpression of Lin28a/b is associated with the malignancy of human germ cell tumors, such as choriocarcinomas, embryonal carcinomas, seminomas, yolk-sac tumors, and mixed germ cell tumors (West et al. 2009; Cao et al. 2011a; Cao et al. 2011b; Gillis et al. 2011; Xue et al. 2011). Overexpression of Lin28a produces higher grade teratomas whereas Lin28a knockdown leads to smaller teratomas, suggesting that Lin28a acts as an oncogene in germ cell tumors by enhancing the self-renewal of PGCs and spermatogonial stem cells (West et al. 2009).
Although Lin28a/b declines rapidly upon implantation (Tang et al. 2010), high levels of Lin28 persist in the neural tube and neural crest (Yang and Moss 2003; Balzer et al. 2010). miR-125 is thought to promote neural differentiation, in part by downregulating Lin28a in neural stem cells (Wulczyn et al. 2007; Rybak et al. 2008). Lin28a/b overexpression, in turn, regulates the balance of neurogenesis and gliogenesis in vitro (Balzer et al. 2010), and leads to an abundance of primitive neural tissue in teratomas formed by ESCs (West et al. 2009). By increasing Nmyc, conditional overexpression of Lin28b in neural crest progenitors in mice could inhibit neuronal differentiation and lead to neuroblastoma (Molenaar et al. 2012). Interestingly, a genome-wide association study (GWAS) also found that a Lin28b variant with higher expression is associated with higher neuroblastoma risk in humans (Diskin et al. 2012). This suggests that dysregulation of Lin28b in neural crest progenitors, which normally show only limited self-renewal, can provoke transformation into neuroblastoma. Lin28a is also highly expressed in aggressive primitive neuroectodermal brain tumors and medulloblastoma, although its oncogenic role in these tumors remains less clear (Picard et al. 2012; Rodini et al. 2012).
Within the developing mesodermal tissues, fetal hematopoietic stem and progenitor cells (HSPCs) express high levels of Lin28b, whereas adult HSPCs do not (Yuan et al. 2012). Overexpression of Lin28a alone in adult Lin- bone marrow cells can reprogram some of them into fetal-like lymphoid progenitors (Yuan et al. 2012), which may be relevant to Lin28’s oncogenic role in T-cell lymphoma and leukemia (Beachy et al. 2012; Rao et al. 2012). In acute myeloid leukemia (AML) however, Lin28a appears to act as an oncogene in MLL-driven AML on one end of the spectrum, and a tumor suppressor in miR-125-driven AML on the other (Jiang et al. 2012; Chaudhuri et al. 2012). It is especially interesting that miR-125 overexpression alone can promote self-renewal of long-term adult HSCs, to cause a variety of myeloid and lymphoid malignancies in both mice and humans (Bousquet et al. 2008, 2010; Guo et al. 2010; O’Connell et al. 2010; Ooi et al. 2010; Chaudhuri et al. 2012), even though the miR-125 homolog lin-4 promotes differentiation in nematode stem cells. The unresolved questions surrounding miR-125 and Lin28’s roles in hematopoiesis indicate that our understanding of how the miR-125-Lin28-let-7 pathway regulates hematopoiesis remains incomplete.
In another mesoderm-derived tissue, muscle stem cells or satellite cells have not been observed to express Lin28a/b, but proliferative myoblasts do upregulate Lin28a during muscle regeneration (Polesskaya et al. 2007). Loss of Lin28a by siRNA knockdown inhibits myogenesis, whereas Lin28a overexpression promotes myogenesis, at least in vitro. This process depends on direct stimulation of Igf2 translation (Polesskaya et al. 2007), and probably other Lin28a targets in cellular growth and metabolism (Zhu et al. 2011). Although muscle development does not seem overtly affected in Lin28a transgenic or knockout mice (Zhu et al. 2010; Zhu et al. 2011), it remains to be tested whether Lin28a/b are functionally important in muscle regeneration upon injury, or in rhabdomyosarcoma growth in vivo.
Despite tremendous progress in our knowledge of Lin28 function in tissues of germline, ectodermal and mesodermal origin, little is known about Lin28 function in endodermal tissues. Lin28 expression has been detected in the fetal liver, kidney, intestines and lung by immunohistochemistry (Yang and Moss 2003). A variety of cancers involving these tissues express Lin28b, including hepatocellular carcinoma, Wilm’s tumors, colorectal cancer and lung cancer – suggesting that Lin28a/b might play a role in both the normal development and malignancy of endodermal tissues (Guo et al. 2006; Viswanathan et al. 2009; King et al. 2011; Zhang et al. 2012). Given the limitations of immunohistochemistry in small cellular compartments, and the expectation that Lin28 may only be active in stem or progenitor cells, careful analysis using tissue specific Cre- or Cre-ER-driven mouse models are needed to rigorously address the role of Lin28a/b in tissue development by lineage-tracing.
Lin28 in Re-engineering of Mammalian Physiology
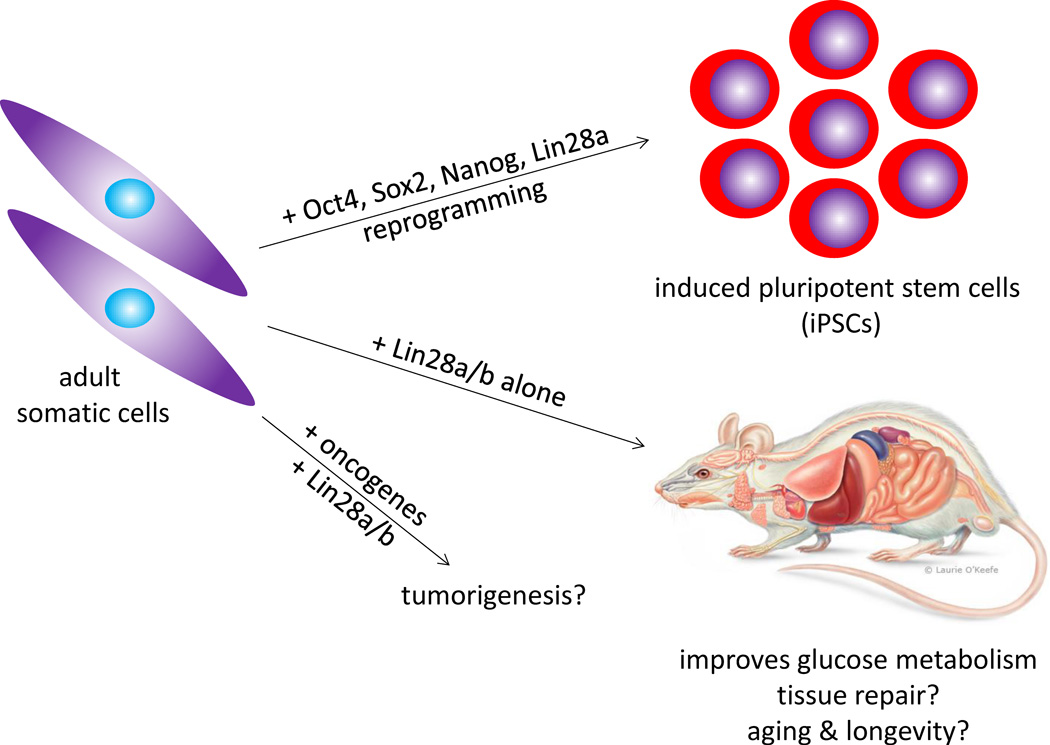
It has been proposed that Lin28 is an oncofetal gene with little physiological relevance in normal adult tissues (Boyerinas et al. 2008; Peter 2009). Since Lin28a/b are primarily expressed during embryogenesis and largely silent in most adult tissues, one could argue that these proteins bear little relevance to normal adult human physiology except when reactivated in the setting of malignancy. Evolutionarily, this could be due to Lin28’s potency in promoting stem cell self-renewal, and hence tumorigenesis, if dysregulated. Another gene endowed with similar properties is the catalytic component of telomerase, Tert, which is likewise predominantly expressed during early embryogenesis, in small compartments of adult stem cells, and in cancers (Kolquist et al. 1998; Schaetzlein et al. 2004). Despite its apparent irrelevance to adult physiology initially – telomerase knockout mice are healthy and viable for the first few generations (Blasco et al. 1997; Yuan et al. 1999) – Tert has gained preeminence as an agent for re-engineering mammalian adult cells, by immortalizing cells and extending lifespan in vivo, along with being a potential target in cancer therapy (Sahin et al. 2010). Could Lin28 show similar potential in reengineering adult human cells (Figure 3)?
Figure 3. Potential of Lin28 in Re-Engineering Adult Mammalian Physiology.
Lin28a, in conjunction with the pluripotency factors Oct4, Sox2 and Nanog, can reprogram somatic cells into iPSCs. Alone, Lin28a/b can reprogram adult HSPCs into a fetal-like state, and enhance insulin sensitivity in the skeletal muscles to improve glucose homeostasis, resist obesity and prevent diabetes. Emergent clues suggest that optimal doses of Lin28a/b might have the potential to re-engineer adult mammalian tissue repair capacities and extend longevity, although Lin28a/b could also cooperate with oncogenes to initiate tumorigenesis. Future work might elucidate these mysteries.
The answer appears to be in the affirmative. Taken together, the demonstrations that Lin28a overexpression with Oct4, Sox2 and Nanog can help reprogram adult human fibroblasts into ESC-like iPSCs (Yu et al. 2007), that Lin28a overexpression can reprogram adult HSPCs into a fetal-like HSPCs (Yuan et al. 2012), and that Lin28b overexpression can expand neural crest progenitors (Molenaar et al. 2012), suggest that Lin28a/b overexpression might be useful for promoting stem cell or progenitor self-renewal in vitro. This is conceptually distinct from Tert-based immortalization of any somatic cell, since Lin28a/b appears to counteract cellular differentiation whereas Tert counteracts replicative senescence. If Lin28a/b can enhance self-renewal, what effects would it exert on adult tissue repair in vivo? Although Lin28’s effects on mammalian tissue repair remains unexplored, a study has shown that zebrafish Lin28 can promote retinal regeneration by repressing let-7 (Ramachandran et al. 2010). This finding hints at the possibility that Lin28 might also extend the limits of mammalian tissue repair upon injury – a hypothesis that awaits further testing (Figure 3).
In the same vein, the Lin28/let-7 pathway could impact longevity, although it remains unclear whether Lin28 overexpression would promote or delay aging. Given that Lin28 upregulates and let-7 downregulates insulin-PI3K signaling in mammals (Zhu et al. 2011), and insulin-PI3K signaling also regulates mammalian aging in an evolutionarily conserved fashion, one might expect Lin28 to promote and let-7 to delay aging in mammals. This would concur with several studies showing that several long-lived mouse strains like the Ames, Snell, GHRKO and Igfr1+/− mice are all dwarfed (Bartke 2012), similar to mice overexpressing let-7 (Zhu et al. 2011; Frost and Olson 2011). Yet improved insulin sensitivity, a prominent phenotype of mice overexpressing Lin28a/b, is associated with longevity (Barbieri et al. 2003), indicative of how longevity is regulated by a multifaceted network of factors. Adding to this complexity, deficiency in tissue repair can accelerate aging as shown in telomerase-deficient and p53-overexpressing mice (Rudolph et al. 1999; Tyner et al. 2002), but hyperactive tissue regeneration can also lead to stem cell exhaustion and shorter lifespans, as shown in models of Pten−/− myelodysplasias and muscular dystrophy (Yilmaz et al. 2006; Sacco et al. 2010). It could well be that only the precise dosage of Lin28/let-7 that strikes an optimal equilibrium between insulin signaling and tissue repair would enhance mammalian longevity.
As is the case with telomerase, a wide variety of cancers reactivate Lin28 to re-engineer their cellular states (Viswanathan et al. 2009). Although the oncogenic role of Lin28a/b has only been demonstrated in vivo for a small subset of cancers including neuroblastoma, T-cell acute lymphoblastic leukemia and peripheral T-cell lymphoma (Molenaar et al. 2012; Beachy et al. 2012; Rao et al. 2012), the cancer stem cell model seems to apply in all these cases, with Lin28 as a stem cell factor promoting self-renewal. The cancer stem cell model posits that cancers are maintained by a small population of tumor-sustaining cancer stem cells with self-renewal capacity (Rosen et al. 2009). Although the cancer stem cell model has engendered debate and does not apply universally to all cancers, as shown most convincingly in advanced melanoma and lymphoma (Quintana et al. 2008, 2010; Boiko et al. 2010), leukemias, germ cell tumors and some other solid cancers appear to follow the cancer stem cell model (Ishizawa et al. 2010). Irving Weissman and colleagues have proposed that in the earliest stages of melanoma, rare cancer stem cells differentiate into nonmalignant progeny to form the bulk of the tumor, whereas in the advanced stages cancer stem cell clones dominate and constitute the bulk of the tumor (Boiko et al. 2010). These findings suggest that targeting cancer cell heterogeneity may be a relevant approach for eradicating tumors only during a cancer’s early stages. If Lin28’s role in promoting stem cell and progenitor self-renewal is also its essential mechanism for promoting tumorigenesis, then Lin28 would represent a promising universal factor for therapeutic targeting in a wide variety of cancer stem cells. This exciting prospect warrants attempts to drug the Lin28 pathway.
Conclusion
Seminal observations made nearly three decades ago by Victor Ambros and Robert Horvitz on the role of Lin28 in C. elegans heterochronic reiterations have spawned great leaps in our understanding of how post-transcriptional RNA processing can regulate stem cells. Recent advances are painting a detailed picture of how Lin28 regulates let-7 microRNA biogenesis and mRNA translation, to coordinate both cellular metabolism and proliferative growth pathways for the purpose of stem cell self-renewal. Although much effort has been expended to elucidate the let-7 microRNA regulatory mechanism, it is imperative that we also understand how Lin28a/b regulates mRNA processing and trafficking between the nucleus, ribosomes, P-bodies and stress granules – a mystery that remains unresolved. And what conditions or pathways regulate the mechanism of Lin28a or Lin28b within the nucleolus, the cytoplasm, and P-bodies? How do these mechanisms affect Lin28a/b function in tissue stem cells and progenitors, especially those derived from the endoderm given its preponderance in colon, kidney, liver and lung cancers? And can we harness Lin28 to re-engineer and improve mammalian tissue repair and longevity? Or target it for cancer therapy? We hope to answer these questions, and more, in the coming decade.
Acknowledgements
We apologize to those authors whose papers could not be cited due to space constraints. We thank H. Zhu, K. Tsanov, J.T. Powers, and three anonymous reviewers for constructive feedback on the manuscript. N.S.C. is supported by the NSS Scholarship from the Agency for Science, Technology and Research, Singapore. G.Q.D. is an investigator of the HHMI. Support was provided by the Ellison Medical Foundation and the Progenitor Cell Biology Consortium of the National Heart Lung Blood Institute (UO1- HL100001).
References
- Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V. The let-7 microRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev. Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander PB, Wang J, McKnight SL. Targeted killing of a mammalian cell based upon its specialized metabolic state. Proc Natl Acad Sci USA. 2011;108:15828–15833. doi: 10.1073/pnas.1111312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- Antebi A, Culotti JG, Hedgecock EM. Daf-12 regulates developmental age and the dauer alternative in Caenorhabditis elegans. Development. 1998;125:1191–1205. doi: 10.1242/dev.125.7.1191. [DOI] [PubMed] [Google Scholar]
- Antebi A, Yeh WH, Tait D, Hedgecock EM, Riddle DL. Daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev. 2000;14:1512–1527. [PMC free article] [PubMed] [Google Scholar]
- Balzer E, Moss EG. Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biol. 2007;4:16–25. doi: 10.4161/rna.4.1.4364. [DOI] [PubMed] [Google Scholar]
- Balzer E, Heine C, Jiang Q, Lee VM, Moss EG. LIN28 alters cell fate succession and acts independently of the let-7 microRNA during neurogliogenesis in vitro. Stem Cells. 2010;900:891–900. doi: 10.1242/dev.042895. [DOI] [PubMed] [Google Scholar]
- Barbaux S, Gascoin-Lachambre G, Buffat C, Monnier P, Mondon F, Tonanny MB, Pinard A, Auer J, Bessieres B, Barlier A, et al. A genome-wide approach reveals novel imprinted genes expressed in the human placenta. Epigenetics. 2012;7:1079–1090. doi: 10.4161/epi.21495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri M, Rizzo MR, Manzella D, Grella R, Ragno E, Carbonella M, Abbatecola AM, Paolisso G. Glucose regulation and oxidative stress in healthy centenarians. Exp. Gerontol. 2003;38:137–143. doi: 10.1016/s0531-5565(02)00153-5. [DOI] [PubMed] [Google Scholar]
- Bartke A. Healthy aging: is smaller better? - a mini-review. Gerontology. 2012;58:337–343. doi: 10.1159/000335166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beachy SH, Onozawa M, Chung YJ, Slape C, Bilke S, Francis P, Pineda M, Walker RL, Meltzer P, Aplan PD. Enforced expression of Lin28b leads to impaired T-cell development, release of inflammatory cytokines, and peripheral T-cell lymphoma. Blood. 2012;120:1048–1059. doi: 10.1182/blood-2012-01-401760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamed M, Herbig U, Ye T, Dejean A, Bischof O. Senescence is an endogenous trigger for microRNA-directed transcriptional gene silencing in human cells. Nat. Cell Biol. 2012;14:266–275. doi: 10.1038/ncb2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke A, Fielenbach N, Wang Z, Mangelsdorf DJ, Antebi A. Nuclear hormone receptor regulation of microRNAs controls developmental progression. Science. 2009;324:95–98. doi: 10.1126/science.1164899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat-Nakshatri P, Wang G, Collins NR, Thomson MJ, Geistlinger TR, Carroll JS, Brown M, Hammond S, Srour EF, Liu Y, et al. Estradiol-regulated microRNAs control estradiol response in breast cancer cells. Nucleic Acids Res. 2009;37:4850–4861. doi: 10.1093/nar/gkp500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- Boiko AD, Razorenova OV, van de Rijn M, Swetter SM, Johnson DL, Ly DP, Butler PD, Yang GP, Joshua B, Kaplan MJ, et al. Human melanoma-initiating cells express neural crest nerve growth factor receptor CD271. Nature. 2010;466:133–137. doi: 10.1038/nature09161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet M, Quelen C, Rosati R, Mansat-De Mas V, La Starza R, Bastard C, Lippert E, Talmant P, Lafage-Pochitaloff M, Leroux D, et al. Myeloid cell differentiation arrest by miR-125b-1 in myelodysplastic syndrome and acute myeloid leukemia with the t(2;11)(p21;q23) translocation. J Exp Med. 2008;205:2499–2506. doi: 10.1084/jem.20080285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousquet M, Harris MH, Zhou B, Lodish HF. MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci USA. 2010;107:21558–21563. doi: 10.1073/pnas.1016611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyerinas B, Park SM, Shomron N, Hedegaard MM, Vinther J, Andersen JS, Feig C, Xu J, Burge CB, Peter ME. Identification of let-7-regulated oncofetal genes. Cancer Res. 2008;68:2587–2591. doi: 10.1158/0008-5472.CAN-08-0264. [DOI] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Liu A, Wang F, Allan RW, Mei K, Peng Y, Du J, Guo S, Abel TW, Lane Z, et al. RNA-binding protein LIN28 is a marker for primary extragonadal germ cell tumors: an immunohistochemical study of 131 cases. Mod. Pathol. 2011a;24:288–296. doi: 10.1038/modpathol.2010.195. [DOI] [PubMed] [Google Scholar]
- Cao D, Allan RW, Cheng L, Peng Y, Guo CC, Dahiya N, Akhi S, Li J. RNA-binding protein LIN28 is a marker for testicular germ cell tumors. Hum. Pathol. 2011b;42:710–718. doi: 10.1016/j.humpath.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Horvitz HR, Sulston JE. Mutations that lead to reiterations in the cell lineages of C. elegans. Cell. 1981;24:59–69. doi: 10.1016/0092-8674(81)90501-8. [DOI] [PubMed] [Google Scholar]
- Chang HM, Martinez NJ, Thornton JE, Hagan JP, Nguyen KD, Gregory RI. Trim71 cooperates with microRNAs to repress Cdkn1a expression and promote embryonic stem cell proliferation. Nat. Commun. 2012;3:923. doi: 10.1038/ncomms1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Zeitels LR, Hwang HW, Chivukula RR, Wentzel EA, Dews M, Jung J, Gao P, Dang CV, Beer MA, et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci USA. 2009;106:3384–3389. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri AA, So AY, Mehta A, Minisandram A, Sinha N, Jonsson VD, Rao DS, O’Connell RM, Baltimore D. Oncomir miR-125b regulates hematopoiesis by targeting the gene Lin28A. Proc Natl Acad Sci USA. 2012;109:4233–4238. doi: 10.1073/pnas.1200677109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla G, Sokol NS. Hormonal activation of let-7-C microRNAs via EcR is required for adult Drosophila melanogaster morphology and function. Development. 2012;139:1788–1797. doi: 10.1242/dev.077743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiefari E, Tanyolac S, Paonessa F, Pullinger CR, Capula C, Iiritano S, Mazza T, Forlin M, Fusco A, Durlach V, et al. Functional variants of the HMGA1 gene and type 2 diabetes mellitus. JAMA. 2011;305:903–912. doi: 10.1001/jama.2011.207. [DOI] [PubMed] [Google Scholar]
- Cho J, Chang H, Kwon SC, Kim B, Kim Y, Choe J, Ha M, Kim YK, Kim VN. LIN28A Is a Suppressor of ERAssociated Translation in Embryonic Stem Cells. Cell. 2012;151:765–777. doi: 10.1016/j.cell.2012.10.019. [DOI] [PubMed] [Google Scholar]
- Cox JL, Mallanna SK, Luo X, Rizzino A. Sox2 uses multiple domains to associate with proteins present in Sox2-protein complexes. PloS One. 2010;5:e15486. doi: 10.1371/journal.pone.0015486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. Therapeutic targeting of Myc-reprogrammed cancer cell metabolism. Cold Spring Harb Symp Quant Biol. 2011;76:369–374. doi: 10.1101/sqb.2011.76.011296. [DOI] [PubMed] [Google Scholar]
- Diskin SJ, Capasso M, Schnepp RW, Cole KA, Attiyeh EF, Hou C, Diamond M, Carpenter EL, Winter C, Lee H, et al. Common variation at 6q16 within HACE1 and LIN28B influences susceptibility to neuroblastoma. Nat. Genet. 2012;44:1126–1130. doi: 10.1038/ng.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B, Jiang J, Kraus P, Ng JH, Heng JC, Chan YS, Yaw LP, Zhang W, Loh YH, Han J, et al. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat. Cell Biol. 2009;11:197–203. doi: 10.1038/ncb1827. [DOI] [PubMed] [Google Scholar]
- Folmes CDL, Nelson TJ, Martinez-Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, Ikeda Y, Perez-Terzic C, Terzic A. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011;14:264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci USA. 2011;108:21075–21080. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Sun Y, Chen J. IGF-II is regulated by microRNA-125b in skeletal myogenesis. J. Cell Biol. 2011;192:69–81. doi: 10.1083/jcb.201007165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke S, Imai Y, Sokol N, Lu B. Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature. 2010;466:637–641. doi: 10.1038/nature09191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerisch B, Rottiers V, Li D, Motola DL, Cummins CL, Lehrach H, Mangelsdorf DJ, Antebi A. A bile acid-like steroid modulates Caenorhabditis elegans lifespan through nuclear receptor signaling. Proc Natl Acad Sci USA. 2007;104:5014–5019. doi: 10.1073/pnas.0700847104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis AJ, Stoop H, Biermann K, van Gurp RJ, Swartzman E, Cribbes S, Ferlinz A, Shannon M, Oosterhuis JW, Looijenga LH. Expression and interdependencies of pluripotency factors LIN28, OCT3/4, NANOG and SOX2 in human testicular germ cells and tumours of the testis. Int. J. Androl. 2011;34:e160–e174. doi: 10.1111/j.1365-2605.2011.01148.x. [DOI] [PubMed] [Google Scholar]
- Golipour A, David L, Liu Y, Jayakumaran G, Hirsch CL, Trcka D, Wrana JL. A late transition in somatic cell reprogramming requires regulators distinct from the pluripotency network. Cell Stem Cell. 2012;11:769–782. doi: 10.1016/j.stem.2012.11.008. [DOI] [PubMed] [Google Scholar]
- Guo G, Smith A. A genome-wide screen in EpiSCs identifies Nr5a nuclear receptors as potent inducers of ground state pluripotency. Development. 2010;137:3185–3192. doi: 10.1242/dev.052753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Lu J, Schlanger R, Zhang H, Wang JY, Fox MC, Purton LE, Fleming HH, Cobb B, Merkenschlager M, et al. MicroRNA miR-125a controls hematopoietic stem cell number. Proc Natl Acad Sci USA. 2010;107:14229–14234. doi: 10.1073/pnas.0913574107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Chen Y, Ito H, Watanabe A, Ge X, Kodama T, Aburatani H. Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene. 2006;384:51–61. doi: 10.1016/j.gene.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell CM, Karp X, Ambros V. A feedback circuit involving let-7-family miRNAs and DAF-12 integrates environmental signals and developmental timing in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2009;106:18668–18673. doi: 10.1073/pnas.0908131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Max KE, Bandaru P, Morozov P, Gerstberger S, Brown M, Molina H, Tuschl T. Identification of mRNAs bound and regulated by human LIN28 proteins and molecular requirements for RNA recognition. RNA. 2013 doi: 10.1261/rna.036491.112. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng JC, Feng B, Han J, Jiang J, Kraus P, Ng JH, Orlov YL, Huss M, Yang L, Lufkin T, et al. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell. 2010;6:167–174. doi: 10.1016/j.stem.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han Jinju, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol. Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim Y-K, Ha M, Yoon M-J, Cho J, Yeom K-H, Han Jinju, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Heo I, Ha M, Lim J, Yoon MJ, Park JE, Kwon SC, Chang H, Kim VN. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell. 2012;151:521–532. doi: 10.1016/j.cell.2012.09.022. [DOI] [PubMed] [Google Scholar]
- Horvitz HR, Sulston JE. Isolation and genetic characterization of cell-lineage mutants of the nematode Caenorhabditis elegans. Genetics. 1980;96:435–454. doi: 10.1093/genetics/96.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D, Hirsch Ha, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 microRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizawa K, Rasheed ZA, Karisch R, Wang Q, Kowalski J, Susky E, Pereira K, Karamboulas C, Moghal N, Rajeshkumar NV, et al. Tumor-initiating cells are rare in many human tumors. Cell Stem Cell. 2010;7:279–282. doi: 10.1016/j.stem.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janiszewska M, Suva ML, Riggi N, Houtkooper RH, Radovanovic I, Rheinbay E, Auwerx J, Cle V. Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes Dev. 2012;26:1926–1944. doi: 10.1101/gad.188292.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Huang H, Li Z, Li Y, Wang X, Gurbuxani S, Chen P, He C, You D, Zhang S, et al. Blockade of miR-150 maturation by MLL-fusion/MYC/LIN-28 is required for MLL-associated leukemia. Cancer Cell. 2012;22:524–535. doi: 10.1016/j.ccr.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Jing W, Lei XX, Feng C, Peng S, Boris-Lawrie K, Huang Y. Evidence that Lin28 stimulates translation by recruiting RNA helicase A to polysomes. Nucleic Acids Res. 2011;39:3724–3734. doi: 10.1093/nar/gkq1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Lin SY, Slack FJ. The time of appearance of the C. elegans let-7 microRNA is transcriptionally controlled utilizing a temporal regulatory element in its promoter. Dev. Biol. 2003;259:364–379. doi: 10.1016/s0012-1606(03)00202-1. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Kawahara H, Okada Y, Imai T, Iwanami A, Mischel PS, Okano H. Musashi1 cooperates in abnormal cell lineage protein 28 (Lin28)-mediated let-7 family microRNA biogenesis in early neural differentiation. J. Biol. Chem. 2011;286:16121–16130. doi: 10.1074/jbc.M110.199166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C, Cuatrecasas M, Castells A, Sepulveda A, Lee JS, Rustgi AK. Lin28b promotes colon cancer progression and metastasis. Cancer Res. 2011;71:4260–4268. doi: 10.1158/0008-5472.CAN-10-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klusmann JH, Li Z, Bohmer K, Maroz A, Koch ML, Emmrich S, Godinho FJ, Orkin SH, Reinhardt D. miR-125b-2 is a potential oncomiR on human chromosome 21 in megakaryoblastic leukemia. Genes Dev. 2010;24:478–490. doi: 10.1101/gad.1856210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolquist KA, Ellisen LW, Counter CM, Meyerson M, Tan LK, Weinberg RA, Haber DA, Gerald WL. Expression of TERT in early premalignant lesions and a subset of cells in normal tissues. Nat. Genet. 1998;19:182–186. doi: 10.1038/554. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Le MT, Xie H, Zhou B, Chia PH, Rizk P, Um M, Udolph G, Yang H, Lim B, Lodish HF. MicroRNA-125b promotes neuronal differentiation in human cells by repressing multiple targets. Mol. Cell. Biol. 2009a;29:5290–5305. doi: 10.1128/MCB.01694-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V, Lodish HF, Lim B. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009b;23:862–876. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le MT, Shyh-Chang N, Khaw SL, Chin L, Teh C, Tay J, O'Day E, Korzh V, Yang H, Lal A, et al. Conserved regulation of p53 network dosage by microRNA-125b occurs through evolving miRNA-target gene pairs. PLoS Genet. 2011;7:e1002242. doi: 10.1371/journal.pgen.1002242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legesse-Miller A, Elemento O, Pfau SJ, Forman JJ, Tavazoie S, Coller HA. Let-7 overexpression leads to an increased fraction of cells in G2/M, direct down-regulation of Cdc34, and stabilization of Wee1 kinase in primary fibroblasts. J. Biol. Chem. 2009;284:6605–6609. doi: 10.1074/jbc.C900002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrbach NJ, Armisen J, Lightfoot HL, Murfitt KJ, Bugaut A, Balasubramanian S, Miska EA. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2009;16:1016–1020. doi: 10.1038/nsmb.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang J, Gao L, McClellan S, Finan MA, Butler TW, Owen LB, Piazza GA, Xi Y. MiR-181 mediates cell differentiation by interrupting the Lin28 and let-7 feedback circuit. Cell Death Differ. 2011;19:378–386. doi: 10.1038/cdd.2011.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhong X, Lin X, Guo J, Zou L, Tanyi JL, Shao Z, Liang S, Wang LP, Hwang WT, et al. Lin-28 homologue A (LIN28A) promotes cell cycle progression via regulation of cyclin-dependent kinase 2 (CDK2), cyclin D1 (CCND1), and cell division cycle 25 homolog A (CDC25A) expression in cancer. J. Biol. Chem. 2012;287:17386–17397. doi: 10.1074/jbc.M111.321158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liau SS, Jazag A, Whang EE. HMGA1 is a determinant of cellular invasiveness and in vivo metastatic potential in pancreatic adenocarcinoma. Cancer Res. 2006;66:11613–11622. doi: 10.1158/0008-5472.CAN-06-1460. [DOI] [PubMed] [Google Scholar]
- Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW, Cantley LC. Metabolic flux and the regulation of mammalian cell growth. Cell Metab. 2011;14:443–451. doi: 10.1016/j.cmet.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G, Sugimoto T, Diamanti E, Joshi A, Hannah R, Ohtsuka S, Gottgens B, Niwa H, Smith A. Esrrb is a pivotal target of the Gsk3/Tcf3 axis regulating embryonic stem cell self-renewal. Cell Stem Cell. 2012;11:491–504. doi: 10.1016/j.stem.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar JJ, Domingo-Fernandez R, Ebus ME, Lindner S, Koster J, Drabek K, Mestdagh P, van Sluis P, Valentijn LJ, van Nes J, et al. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat Genet. 2012;44:1199–1206. doi: 10.1038/ng.2436. [DOI] [PubMed] [Google Scholar]
- Morita K, Han M. Multiple mechanisms are involved in regulating the expression of the developmental timing regulator lin-28 in Caenorhabditis elegans. EMBO J. 2006;25:5794–5804. doi: 10.1038/sj.emboj.7601451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- Moss EG, Tang L. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev. Biol. 2003;258:432–442. doi: 10.1016/s0012-1606(03)00126-x. [DOI] [PubMed] [Google Scholar]
- Nam Y, Chen C, Gregory RI, Chou JJ, Sliz P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell. 2011;147:1080–1091. doi: 10.1016/j.cell.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Chambers I, Taga T, Smith A. Physiological rationale for responsiveness of mouse embryonic stem cells to gp130 cytokines. Development. 2001;128:2333–2339. doi: 10.1242/dev.128.12.2333. [DOI] [PubMed] [Google Scholar]
- Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Nishino J, Kim I, Chada K, Morrison SJ. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell. 2008;135:227–239. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Marcarci BO, Unternaehrer J, Gupta PB, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi AG, Sahoo D, Adorno M, Wang Y, Weissman IL, Park CY. MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. Proc Natl Acad Sci USA. 2010;107:21505–21510. doi: 10.1073/pnas.1016218107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RM, Chaudhuri AA, Rao DS, Gibson WSJ, Balazs AB, Baltimore D. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc Natl Acad Sci USA. 2010;107:14235–14240. doi: 10.1073/pnas.1009798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Peng S, Chen LL, Lei XX, Yang L, Lin H, Carmichael GG, Huang Y. Genome-wide studies reveal that Lin28 enhances the translation of genes important for growth and survival of human embryonic stem cells. Stem Cells. 2011;29:496–504. doi: 10.1002/stem.591. [DOI] [PubMed] [Google Scholar]
- Peter ME. Let-7 and miR-200 microRNAs: Guardians against pluripotency and cancer progression. Cell Cycle. 2009;8:843–852. doi: 10.4161/cc.8.6.7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D, Miller S, Hawkins CE, Bouffet E, Rogers HA, Chan TS, Kim SK, Ra YS, Fangusaro J, Korshunov A, et al. Markers of survival and metastatic potential in childhood CNS primitive neuro-ectodermal brain tumours: an integrative genomic analysis. Lancet Oncol. 2012;13:838–848. doi: 10.1016/S1470-2045(12)70257-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E, Polytarchou C, Thornton JE, Lapierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D, Gregory RI. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147:1066–1079. doi: 10.1016/j.cell.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesskaya A, Cuvellier S, Naguibneva I, Duquet A, Moss EG, Harel-Bellan A. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 2007;21:1125–1138. doi: 10.1101/gad.415007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana E, Shackleton M, Foster HR, Fullen DR, Sabel MS, Johnson TM, Morrison SJ. Phenotypic heterogeneity among tumorigenic melanoma cells from patients that is reversible and not hierarchically organized. Cancer Cell. 2010;18:510–523. doi: 10.1016/j.ccr.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Fausett BV, Goldman D. Ascl1a regulates Muller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nat. Cell Biol. 2010;12:1101–1107. doi: 10.1038/ncb2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Lee SY, Gutierrez A, Perrigoue J, Thapa RJ, Tu Z, Jeffers JR, Rhodes M, Anderson S, Oravecz T, et al. Inactivation of ribosomal protein L22 promotes transformation by induction of the stemness factor, Lin28B. Blood. 2012;120:3764–3773. doi: 10.1182/blood-2012-03-415349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rodini CO, Suzuki DE, Saba-Silva N, Cappellano A, de Souza JES, Cavalheiro S, Toledo SRC, Okamoto OK. Expression analysis of stem cell-related genes reveal OCT4 as a predictor of poor clinical outcome in medulloblastoma. J. Neurooncol. 2012;106:71–79. doi: 10.1007/s11060-011-0647-9. [DOI] [PubMed] [Google Scholar]
- Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324:1670–1673. doi: 10.1126/science.1171837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Rudolph KL, Chang S, Lee HW, Blasco M, Gottlieb GJ, Greider C, DePinho RA. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell. 1999;96:701–712. doi: 10.1016/s0092-8674(00)80580-2. [DOI] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nature Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- Sacco A, Mourkioti F, Tran R, Choi J, Llewellyn M, Kraft P, Shkreli M, Delp S, Pomerantz JH, Artandi SE, et al. Short telomeres and stem cell exhaustion model Duchenne muscular dystrophy in mdx/mTR mice. Cell. 2010;143:1059–1071. doi: 10.1016/j.cell.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin E, Depinho RA. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature. 2010;464:520–528. doi: 10.1038/nature08982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- Schaetzlein S, Lucas-Hahn A, Lemme E, Kues WA, Dorsch M, Manns MP, Niemann H, Rudolph KL. Telomere length is reset during early mammalian embryogenesis. Proc Natl Acad Sci USA. 2004;101:8034–8038. doi: 10.1073/pnas.0402400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz C, Paus M, Frey K, Schmid R, Kohl Z, Mennerich D, Winkler J, Gillardon F. Leucine-rich repeat kinase 2 modulates retinoic acid-induced neuronal differentiation of murine embryonic stem cells. PloS One. 2011;6:e20820. doi: 10.1371/journal.pone.0020820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda G, de Soysa TY, Seligson MT, Yabuuchi A, Fujiwara Y, Yi Huang P, Hagan JP, Gregory RI, Moss EG, Daley GQ. Lin28a Regulates Germ Cell Pool Size and Fertility. Stem Cells. 2013 doi: 10.1002/stem.1343. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Zheng Y, Locasale JW, Cantley LC. Human pluripotent stem cells uncouple respiration from energy production. EMBO J. 2011;30:4851–4852. doi: 10.1038/emboj.2011.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, Zhang J, Onder T, Unternaehrer JJ, Zhu H, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013a;339:222–226. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Daley GQ, Cantley LC. Stem cell metabolism during tissue development and aging. Development. 2013b doi: 10.1242/dev.091777. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol NS, Xu P, Jan YN, Ambros Victor. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 2008;22:1591–1596. doi: 10.1101/gad.1671708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh N, Baehner L, Moltzahn F, Melton C, Shenoy A, Chen J, Blelloch R. MicroRNA function is globally suppressed in mouse oocytes and early embryos. Curr. Biol. 2010;20:271–277. doi: 10.1016/j.cub.2009.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Abnormal cell lineages in mutants of the nematode Caenorhabditis elegans. Dev. Biol. 1981;55:41–55. doi: 10.1016/0012-1606(81)90427-9. [DOI] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Bao S, Lee C, Nordman E, Wang Xiaohui, Lao K, Surani MA. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell. 2010;6:468–478. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive posttranscriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JE, Chang HM, Piskounova E, Gregory RI. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7) RNA. 2012;18:1875–1885. doi: 10.1261/rna.034538.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, Lu X, Soron G, Cooper B, Brayton C, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wynsberghe PM, Kai ZS, Massirer KB, Burton VH, Yeo GW, Pasquinelli AE. LIN-28 cotranscriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans. Nat. Struct. Mol. Bio. 2011;18:302–308. doi: 10.1038/nsmb.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley George Q, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O'Sullivan M, Lu J, Phillips LA, Lockhart VL, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet. 2009;41:843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt EJ, Meglicki M, Hartung KI, Borsuk E, Behr R. Importance of the pluripotency factor LIN28 in the mammalian nucleolus during early embryonic development. Development. 2012;139:4514–4523. doi: 10.1242/dev.083279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Alexander P, Wu L, Hammer R, Cleaver O, McKnight SL. Dependence of mouse embryonic stem cells on threonine catabolism. Science. 2009;325:435–439. doi: 10.1126/science.1173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yang J, Liu H, Lu D, Chen X, Zenonos Z, Campos LS, Rad R, Guo G, Zhang S, et al. Rapid and efficient reprogramming of somatic cells to induced pluripotent stem cells by retinoic acid receptor gamma and liver receptor homolog 1. Proc Natl Acad Sci USA. 2011;108:18283–18288. doi: 10.1073/pnas.1100893108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JA, Viswanathan SR, Yabuuchi A, Cunniff K, Takeuchi A, Park IH, Sero JE, Zhu H, Perez-Atayde A, Frazier AL, et al. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature. 2009;460:909–913. doi: 10.1038/nature08210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbert ML, Huelga SC, Kapeli K, Stark TJ, Liang TY, Chen SX, Yan BY, Nathanson JL, Hutt KR, Lovci MT, et al. LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol. Cell. 2012;48:195–206. doi: 10.1016/j.molcel.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O, Strehle M, Seiler A, Schumacher S, Nitsch R. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J. 2007;21:415–426. doi: 10.1096/fj.06-6130com. [DOI] [PubMed] [Google Scholar]
- Vadla B, Kemper K, Alaimo J, Heine C, Moss EG. lin-28 controls the succession of cell fate choices via two distinct activities. PLoS Genet. 2012;8:e1002588. doi: 10.1371/journal.pgen.1002588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Zhang Kexiong, Huang Y. Lin28 modulates cell growth and associates with a subset of cell cycle regulator mRNAs in mouse embryonic stem cells. RNA. 2009;15:357–361. doi: 10.1261/rna.1368009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue D, Peng Y, Wang F, Allan RW, Cao D. RNA-binding protein LIN28 is a sensitive marker of ovarian primitive germ cell tumours. Histopathology. 2011;59:452–459. doi: 10.1111/j.1365-2559.2011.03949.x. [DOI] [PubMed] [Google Scholar]
- Yanes O, Clark J, Wong DM, Patti GJ, Sanchez-Ruiz A, Benton HP, Trauger SA, Desponts C, Ding S, Siuzdak G. Metabolic oxidation regulates embryonic stem cell differentiation. Nat Chem. Biol. 2010;6:411–417. doi: 10.1038/nchembio.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Moss EG. Temporally regulated expression of Lin-28 in diverse tissues of the developing mouse. Gene Expr. Patterns. 2003;3:719–726. doi: 10.1016/s1567-133x(03)00140-6. [DOI] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, Morrison SJ. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, et al. Let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Yuan J, Nguyen CK, Liu X, Kanellopoulou C, Muljo SA. Lin28b reprograms adult bone marrow hematopoietic progenitors to mediate fetal lymphopoiesis. Science. 2012;335:1195–1200. doi: 10.1126/science.1216557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Ishibashi S, Hatakeyama S, Saito M, Nakayama J, Nikaido R, Haruyama T, Watanabe Y, Iwata H, Iida M, et al. Presence of telomeric G-strand tails in the telomerase catalytic subunit TERT knockout mice. Genes Cells. 1999;4:563–572. doi: 10.1046/j.1365-2443.1999.00284.x. [DOI] [PubMed] [Google Scholar]
- Zhang L, Stokes N, Polak L, Fuchs E. Specific microRNAs are preferentially expressed by skin stem cells to balance self-renewal and early lineage commitment. Cell Stem Cell. 2011a;8:294–308. doi: 10.1016/j.stem.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Khvorostov I, Hong JS, Oktay Y, Vergnes L, Nuebel E, Wahjudi PN, Setoguchi K, Wang G, Do A, et al. UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. EMBO J. 2011b;30:4860–4873. doi: 10.1038/emboj.2011.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WC, Shyh-Chang N, Yang H, Rai A, Umashankar S, Ma S, Soh BS, Sun LL, Tai BC, Nga ME, et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148:259–272. doi: 10.1016/j.cell.2011.11.050. [DOI] [PubMed] [Google Scholar]
- Zheng K, Wu X, Kaestner KH, Wang PJ. The pluripotency factor LIN28 marks undifferentiated spermatogonia in mouse. BMC Dev. Biol. 2009;9:38. doi: 10.1186/1471-213X-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Shah S, Shyh-Chang N, Shinoda G, Einhorn WS, Viswanathan SR, Takeuchi A, Grasemann C, Rinn JL, Lopez MF, et al. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat. Genet. 2010;42:626–630. doi: 10.1038/ng.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Shyh-Chang N, Segre AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, et al. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Li W, Zhou H, Wei W, Ambasudhan R, Lin T, Kim J, Zhang K, Ding S. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell. 2010;7:651–655. doi: 10.1016/j.stem.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]