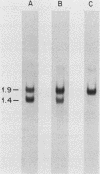
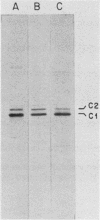
Abstract
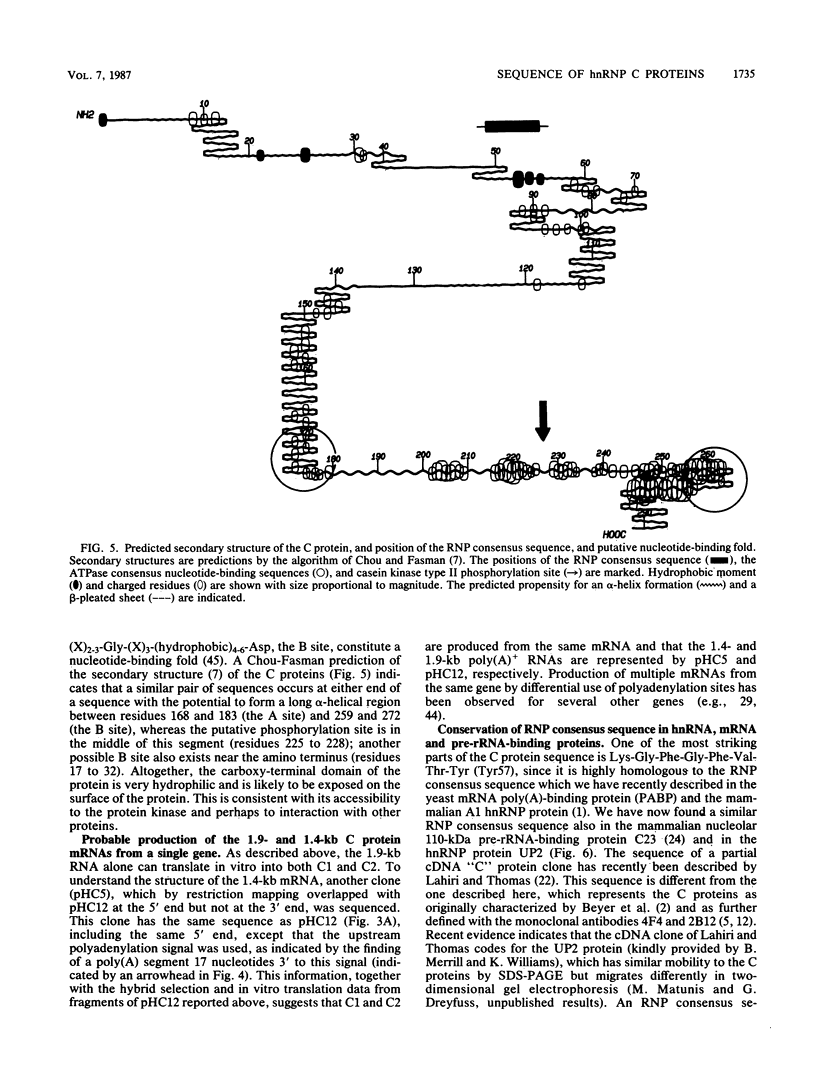
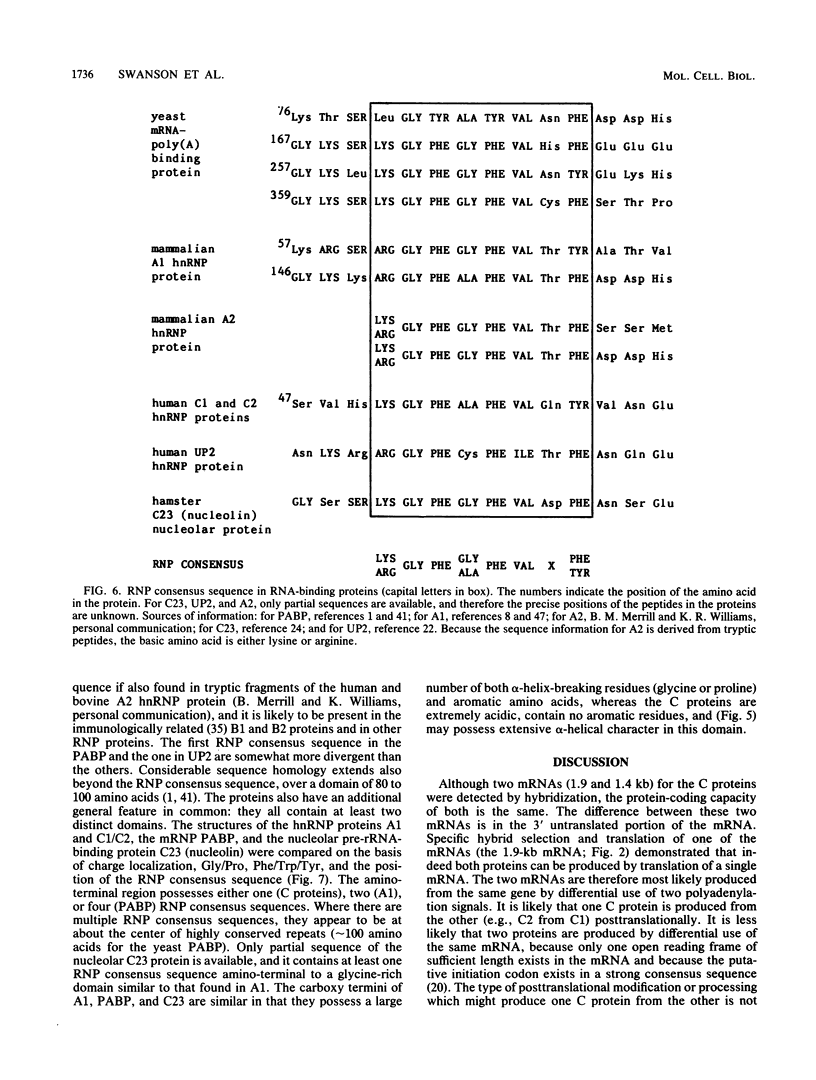
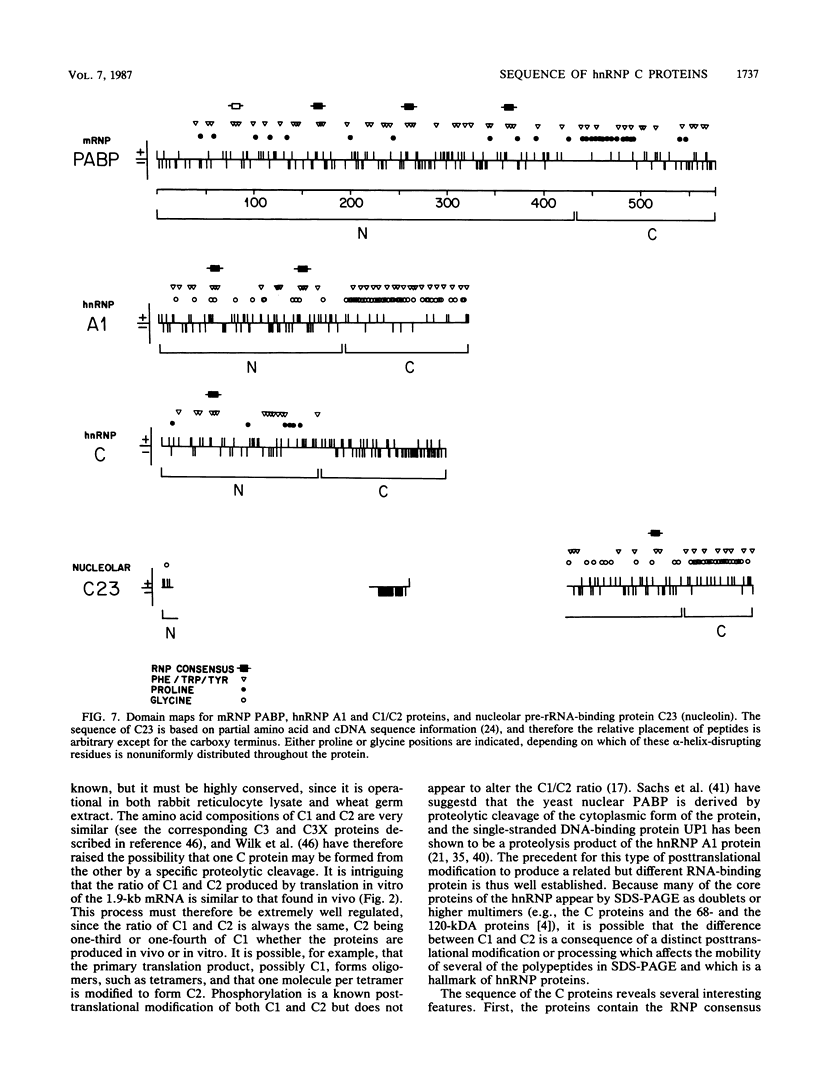
In the eucaryotic nucleus, heterogeneous nuclear RNAs exist in a complex with a specific set of proteins to form heterogeneous nuclear ribonucleoprotein particles (hnRNPs). The C proteins, C1 and C2, are major constituents of hnRNPs and appear to play a role in RNA splicing as suggested by antibody inhibition and immunodepletion experiments. With the use of a previously described partial cDNA clone as a hybridization probe, full-length cDNAs for the human C proteins were isolated. All of the cDNAs isolated hybridized to two poly(A)+ RNAs of 1.9 and 1.4 kilobases (kb). DNA sequencing of a cDNA clone for the 1.9-kb mRNA (pHC12) revealed a single open reading frame of 290 amino acids coding for a protein of 31,931 daltons and two polyadenylation signals, AAUAAA, approximately 400 base pairs apart in the 3' untranslated region of the mRNA. DNA sequencing of a clone corresponding to the 1.4-kb mRNA (pHC5) indicated that the sequence of this mRNA is identical to that of the 1.9-kb mRNA up to the first polyadenylation signal which it uses. Both mRNAs therefore have the same coding capacity and are probably transcribed from a single gene. Translation in vitro of the 1.9-kb mRNA selected by hybridization with a 3'-end subfragment of pHC12 demonstrated that it by itself can direct the synthesis of both C1 and C2. The difference between the C1 and C2 proteins which results in their electrophoretic separation is not known, but most likely one of them is generated from the other posttranslationally. Since several hnRNP proteins appeared by sodium dodecyl sulfate-polyacrylamide gel electrophoresis as multiple antigenically related polypeptides, this raises the possibility that some of these other groups of hnRNP proteins are also each produced from a single mRNA. The predicted amino acid sequence of the protein indicates that it is composed of two distinct domains: an amino terminus that contains what we have recently described as a RNP consensus sequence, which is the putative RNA-binding site, and a carboxy terminus that is very negatively charged, contains no aromatic amino acids or prolines, and contains a putative nucleoside triphosphate-binding fold, as well as a phosphorylation site for casein kinase type II. The RNP consensus sequence was also found in the yeast poly(A)-binding protein (PABP), the heterogeneous nuclear RNA-binding proteins A1 and A2, and the pre-rRNA binding protein C23. All of these proteins are also composed of at least two distinct domains: an amino terminus, which possesses one or more RNP consensus sequences, and a carboxy terminus, which is unique to each protein, being very acidic in the C proteins and rich in glycine in A1, and C23 and rich in proline in the poly(A)-binding protein. These findings suggest that the amino terminus of these proteins possesses a highly conserved RNA-binding domain, whereas the carboxy terminus contains a region essential to the unique function and interactions of each of the RNA-binding proteins.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adam S. A., Nakagawa T., Swanson M. S., Woodruff T. K., Dreyfuss G. mRNA polyadenylate-binding protein: gene isolation and sequencing and identification of a ribonucleoprotein consensus sequence. Mol Cell Biol. 1986 Aug;6(8):2932–2943. doi: 10.1128/mcb.6.8.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer A. L., Christensen M. E., Walker B. W., LeStourgeon W. M. Identification and characterization of the packaging proteins of core 40S hnRNP particles. Cell. 1977 May;11(1):127–138. doi: 10.1016/0092-8674(77)90323-3. [DOI] [PubMed] [Google Scholar]
- Billings P. B., Martin T. E. Proteins of nuclear ribonucleoprotein subcomplexes. Methods Cell Biol. 1978;17:349–376. doi: 10.1016/s0091-679x(08)61154-1. [DOI] [PubMed] [Google Scholar]
- Choi Y. D., Dreyfuss G. Isolation of the heterogeneous nuclear RNA-ribonucleoprotein complex (hnRNP): a unique supramolecular assembly. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7471–7475. doi: 10.1073/pnas.81.23.7471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. D., Dreyfuss G. Monoclonal antibody characterization of the C proteins of heterogeneous nuclear ribonucleoprotein complexes in vertebrate cells. J Cell Biol. 1984 Dec;99(6):1997–1204. doi: 10.1083/jcb.99.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. D., Grabowski P. J., Sharp P. A., Dreyfuss G. Heterogeneous nuclear ribonucleoproteins: role in RNA splicing. Science. 1986 Mar 28;231(4745):1534–1539. doi: 10.1126/science.3952495. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Cobianchi F., SenGupta D. N., Zmudzka B. Z., Wilson S. H. Structure of rodent helix-destabilizing protein revealed by cDNA cloning. J Biol Chem. 1986 Mar 15;261(8):3536–3543. [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Adam S. A., Choi Y. D. Physical change in cytoplasmic messenger ribonucleoproteins in cells treated with inhibitors of mRNA transcription. Mol Cell Biol. 1984 Mar;4(3):415–423. doi: 10.1128/mcb.4.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G., Choi Y. D., Adam S. A. Characterization of heterogeneous nuclear RNA-protein complexes in vivo with monoclonal antibodies. Mol Cell Biol. 1984 Jun;4(6):1104–1114. doi: 10.1128/mcb.4.6.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economidis I. V., Pederson T. Structure of nuclear ribonucleoprotein: heterogeneous nuclear RNA is complexed with a major sextet of proteins in vivo. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1599–1602. doi: 10.1073/pnas.80.6.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe V. E., Wilkinson L. E., Laird C. D. Comparative organization of active transcription units in Oncopeltus fasciatus. Cell. 1976 Sep;9(1):131–146. doi: 10.1016/0092-8674(76)90059-3. [DOI] [PubMed] [Google Scholar]
- GALL J. G., CALLAN H. G. H3 uridine incorporation in lampbrush chromosomes. Proc Natl Acad Sci U S A. 1962 Apr 15;48:562–570. doi: 10.1073/pnas.48.4.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hathaway G. M., Traugh J. A. Casein kinases--multipotential protein kinases. Curr Top Cell Regul. 1982;21:101–127. [PubMed] [Google Scholar]
- Holcomb E. R., Friedman D. L. Phosphorylation of the C-proteins of HeLa cell hnRNP particles. Involvement of a casein kinase II-type enzyme. J Biol Chem. 1984 Jan 10;259(1):31–40. [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Karn J., Vidali G., Boffa L. C., Allfrey V. G. Characterization of the non-histone nuclear proteins associated with rapidly labeled heterogeneous nuclear RNA. J Biol Chem. 1977 Oct 25;252(20):7307–7322. [PubMed] [Google Scholar]
- Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983 Mar;47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Williams K. R., Szer W. Purification and domain structure of core hnRNP proteins A1 and A2 and their relationship to single-stranded DNA-binding proteins. J Biol Chem. 1986 Aug 25;261(24):11266–11273. [PubMed] [Google Scholar]
- Lahiri D. K., Thomas J. O. A cDNA clone of the hnRNP C proteins and its homology with the single-stranded DNA binding protein UP2. Nucleic Acids Res. 1986 May 27;14(10):4077–4094. doi: 10.1093/nar/14.10.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb M. M., Daneholt B. Characterization of active transcription units in Balbiani rings of Chironomus tentans. Cell. 1979 Aug;17(4):835–848. doi: 10.1016/0092-8674(79)90324-6. [DOI] [PubMed] [Google Scholar]
- Lapeyre B., Amalric F., Ghaffari S. H., Rao S. V., Dumbar T. S., Olson M. O. Protein and cDNA sequence of a glycine-rich, dimethylarginine-containing region located near the carboxyl-terminal end of nucleolin (C23 and 100 kDa). J Biol Chem. 1986 Jul 15;261(20):9167–9173. [PubMed] [Google Scholar]
- Lehrach H., Diamond D., Wozney J. M., Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977 Oct 18;16(21):4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Martin T., Billings P., Levey A., Ozarslan S., Quinlan T., Swift H., Urbas L. Some properties of RNA:protein complexes from the nucleus of eukaryotic cells. Cold Spring Harb Symp Quant Biol. 1974;38:921–932. doi: 10.1101/sqb.1974.038.01.094. [DOI] [PubMed] [Google Scholar]
- Martin T., Billings P., Pullman J., Stevens B., Kinniburgh A. Substructure of nuclear ribonucleoprotein complexes. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):899–909. doi: 10.1101/sqb.1978.042.01.091. [DOI] [PubMed] [Google Scholar]
- Masters J. N., Yang J. K., Cellini A., Attardi G. A human dihydrofolate reductase pseudogene and its relationship to the multiple forms of specific messenger RNA. J Mol Biol. 1983 Jun 15;167(1):23–36. doi: 10.1016/s0022-2836(83)80032-1. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Miller O. L., Jr Ultrastructural patterns of RNA synthesis during early embryogenesis of Drosophila melanogaster. Cell. 1976 Jun;8(2):305–319. doi: 10.1016/0092-8674(76)90014-3. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Nakagawa T. Y., Swanson M. S., Wold B. J., Dreyfuss G. Molecular cloning of cDNA for the nuclear ribonucleoprotein particle C proteins: a conserved gene family. Proc Natl Acad Sci U S A. 1986 Apr;83(7):2007–2011. doi: 10.1073/pnas.83.7.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orrick L. R., Olson M. O., Busch H. Comparison of nucleolar proteins of normal rat liver and Novikoff hepatoma ascites cells by two-dimensional polyacrylamide gel electrophoresis. Proc Natl Acad Sci U S A. 1973 May;70(5):1316–1320. doi: 10.1073/pnas.70.5.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandolfo M., Valentini O., Biamonti G., Morandi C., Riva S. Single stranded DNA binding proteins derive from hnRNP proteins by proteolysis in mammalian cells. Nucleic Acids Res. 1985 Sep 25;13(18):6577–6590. doi: 10.1093/nar/13.18.6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. Proteins associated with heterogeneous nuclear RNA in eukaryotic cells. J Mol Biol. 1974 Feb 25;83(2):163–183. doi: 10.1016/0022-2836(74)90386-6. [DOI] [PubMed] [Google Scholar]
- Pelham H. R., Jackson R. J. An efficient mRNA-dependent translation system from reticulocyte lysates. Eur J Biochem. 1976 Aug 1;67(1):247–256. doi: 10.1111/j.1432-1033.1976.tb10656.x. [DOI] [PubMed] [Google Scholar]
- Ricciardi R. P., Miller J. S., Roberts B. E. Purification and mapping of specific mRNAs by hybridization-selection and cell-free translation. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4927–4931. doi: 10.1073/pnas.76.10.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Riva S., Morandi C., Tsoulfas P., Pandolfo M., Biamonti G., Merrill B., Williams K. R., Multhaup G., Beyreuther K., Werr H. Mammalian single-stranded DNA binding protein UP I is derived from the hnRNP core protein A1. EMBO J. 1986 Sep;5(9):2267–2273. doi: 10.1002/j.1460-2075.1986.tb04494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A. B., Bond M. W., Kornberg R. D. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: domain structure and expression. Cell. 1986 Jun 20;45(6):827–835. doi: 10.1016/0092-8674(86)90557-x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla R. C. Processed pseudogenes for rat cytochrome c are preferentially derived from one of three alternate mRNAs. Mol Cell Biol. 1984 Nov;4(11):2279–2288. doi: 10.1128/mcb.4.11.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen H., Etzerodt M., Reuter R., Schneider C., Lottspeich F., Argos P., Lührmann R., Philipson L. Cloning of the human cDNA for the U1 RNA-associated 70K protein. EMBO J. 1986 Dec 1;5(12):3209–3217. doi: 10.1002/j.1460-2075.1986.tb04631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk H. E., Werr H., Friedrich D., Kiltz H. H., Schäfer K. P. The core proteins of 35S hnRNP complexes. Characterization of nine different species. Eur J Biochem. 1985 Jan 2;146(1):71–81. doi: 10.1111/j.1432-1033.1985.tb08621.x. [DOI] [PubMed] [Google Scholar]
- Williams K. R., LoPresti M. B., Setoguchi M., Konigsberg W. H. Amino acid sequence of the T4 DNA helix-destabilizing protein. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4614–4617. doi: 10.1073/pnas.77.8.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K. R., Stone K. L., LoPresti M. B., Merrill B. M., Planck S. R. Amino acid sequence of the UP1 calf thymus helix-destabilizing protein and its homology to an analogous protein from mouse myeloma. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5666–5670. doi: 10.1073/pnas.82.17.5666. [DOI] [PMC free article] [PubMed] [Google Scholar]