Abstract
The Saccharomyces cerevisiae CRY1 gene encodes ribosomal protein rp59, a component of the 40S ribosomal subunit. Mutations in CRY1 can confer resistance to the alkaloid cryptopleurine, an inhibitor of the elongation step of translation. The nucleotide sequence of the cloned CRY1 gene was determined. The predicted amino acid sequence shows that CRY1 encodes a 14,561-dalton polypeptide that has 88% amino acid sequence homology to the hamster or human S14 ribosomal protein responsible for emetine resistance and 45% homology to Escherichia coli ribosomal protein S11. Analysis of the DNA sequences upstream from CRY1 revealed the presence of three sequences, HOMOL1 (consensus, A/TACATCC/TG/ATA/GCA), RPG (consensus, ACCCA/GTACATT/CT/A), and a thymine-rich sequence, found upstream of more than 20 other cloned yeast genes encoding components of the translational apparatus. We exploited the ability to assay the expression of CRY1 in vivo by using the cryptopleurine resistance phenotype to demonstrate that these three consensus sequences are necessary for the transcription of CRY1. We previously showed that the upstream promoter element of the yeast RP39A gene consists of these identical sequence motifs. Therefore, we suggest that these three sequences define a consensus promoter element for the genes encoding the yeast translational apparatus. CRY1 is one of several hundred yeast genes, including ribosomal protein genes, whose expression is transiently decreased 10-fold upon heat shock. We found that the HOMOL1 and RPG consensus sequences are not necessary for the heat shock response of CRY1.
Full text
PDF


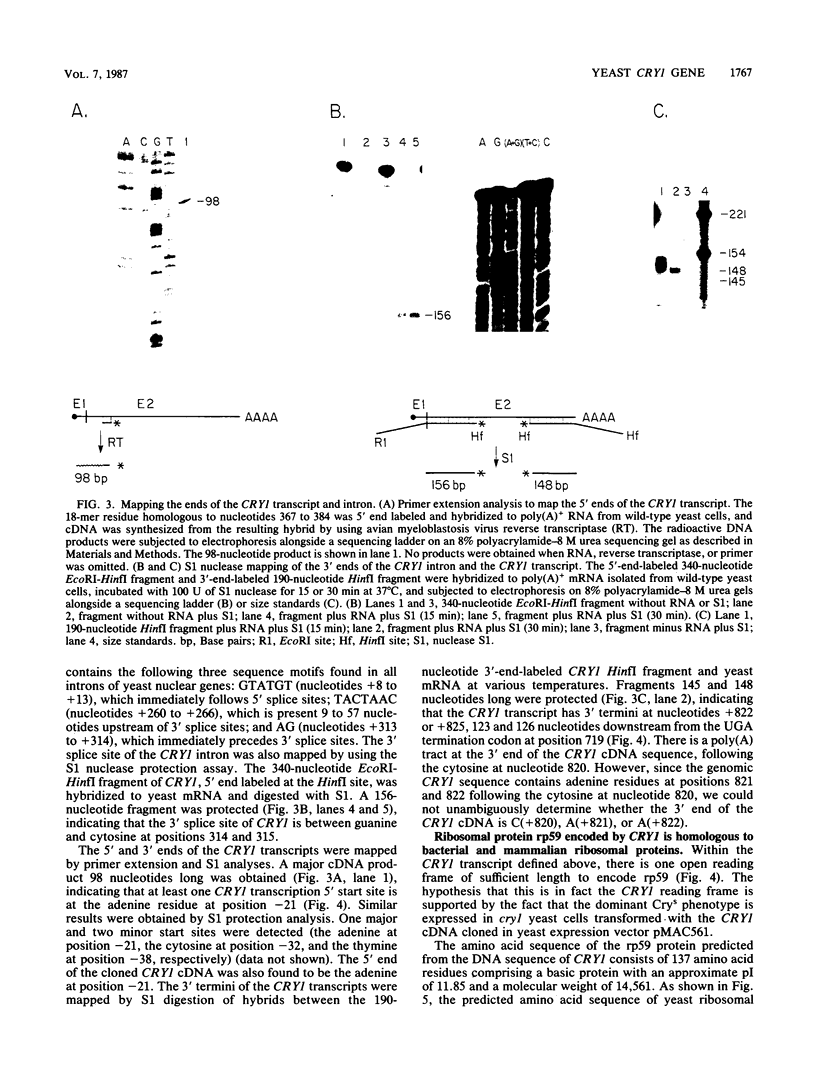








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abovich N., Rosbash M. Two genes for ribosomal protein 51 of Saccharomyces cerevisiae complement and contribute to the ribosomes. Mol Cell Biol. 1984 Sep;4(9):1871–1879. doi: 10.1128/mcb.4.9.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbacid M., Fresno M., Vazquez D. Inhibitors of polypeptide elongation on yeast polysomes. J Antibiot (Tokyo) 1975 Jun;28(6):453–462. doi: 10.7164/antibiotics.28.453. [DOI] [PubMed] [Google Scholar]
- Bingham P. M., Levis R., Rubin G. M. Cloning of DNA sequences from the white locus of D. melanogaster by a novel and general method. Cell. 1981 Sep;25(3):693–704. doi: 10.1016/0092-8674(81)90176-8. [DOI] [PubMed] [Google Scholar]
- Bollen G. H., Cohen L. H., Mager W. H., Klaassen A. W., Planta R. J. Isolation of cloned ribosomal protein genes from the yeast Saccharomyces carlsbergensis. Gene. 1981 Sep;14(4):279–287. doi: 10.1016/0378-1119(81)90160-8. [DOI] [PubMed] [Google Scholar]
- Bucher K., Skogerson L. Cryptopleurine--an inhibitor of translocation. Biochemistry. 1976 Nov 2;15(22):4755–4759. doi: 10.1021/bi00667a001. [DOI] [PubMed] [Google Scholar]
- Chen I. T., Dixit A., Rhoads D. D., Roufa D. J. Homologous ribosomal proteins in bacteria, yeast, and humans. Proc Natl Acad Sci U S A. 1986 Sep;83(18):6907–6911. doi: 10.1073/pnas.83.18.6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies M. S., Henney A., Ward W. H., Craig R. K. Characterisation of an mRNA encoding a human ribosomal protein homologous to the yeast L44 ribosomal protein. Gene. 1986;45(2):183–191. doi: 10.1016/0378-1119(86)90253-2. [DOI] [PubMed] [Google Scholar]
- Donovan D. M., Pearson N. J. Transcriptional regulation of ribosomal proteins during a nutritional upshift in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jul;6(7):2429–2435. doi: 10.1128/mcb.6.7.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dölz H., Vázquez D., Jiménez A. Quantitation of the specific interaction of [14a-3H]cryptopleurine with 80S and 40S ribosomal species from the yeast Saccharomyces cerevisiae. Biochemistry. 1982 Jun 22;21(13):3181–3187. doi: 10.1021/bi00256a023. [DOI] [PubMed] [Google Scholar]
- Fried H. M., Nam H. G., Loechel S., Teem J. Characterization of yeast strains with conditionally expressed variants of ribosomal protein genes tcm1 and cyh2. Mol Cell Biol. 1985 Jan;5(1):99–108. doi: 10.1128/mcb.5.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried H. M., Pearson N. J., Kim C. H., Warner J. R. The genes for fifteen ribosomal proteins of Saccharomyces cerevisiae. J Biol Chem. 1981 Oct 10;256(19):10176–10183. [PubMed] [Google Scholar]
- Fried H. M., Warner J. R. Cloning of yeast gene for trichodermin resistance and ribosomal protein L3. Proc Natl Acad Sci U S A. 1981 Jan;78(1):238–242. doi: 10.1073/pnas.78.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried H. M., Warner J. R. Molecular cloning and analysis of yeast gene for cycloheximide resistance and ribosomal protein L29. Nucleic Acids Res. 1982 May 25;10(10):3133–3148. doi: 10.1093/nar/10.10.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergen J. P., Stern R. H., Wensink P. C. Filter replicas and permanent collections of recombinant DNA plasmids. Nucleic Acids Res. 1979 Dec 20;7(8):2115–2136. doi: 10.1093/nar/7.8.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorenstein C., Warner J. R. Coordinate regulation of the synthesis of eukaryotic ribosomal proteins. Proc Natl Acad Sci U S A. 1976 May;73(5):1547–1551. doi: 10.1073/pnas.73.5.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorenstein C., Warner J. R. Synthesis and turnover of ribosomal proteins in the absence of 60S subunit assembly in Saccharomyces cerevisiae. Mol Gen Genet. 1977 Dec 9;157(3):327–332. doi: 10.1007/BF00268670. [DOI] [PubMed] [Google Scholar]
- Grant P., Sánchez L., Jiménez A. Cryptopleurine resistance: genetic locus for a 40S ribosomal component in Saccharomyces cerevisiae. J Bacteriol. 1974 Dec;120(3):1308–1314. doi: 10.1128/jb.120.3.1308-1314.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hereford L. M., Rosbash M. Number and distribution of polyadenylated RNA sequences in yeast. Cell. 1977 Mar;10(3):453–462. doi: 10.1016/0092-8674(77)90032-0. [DOI] [PubMed] [Google Scholar]
- Himmelfarb H. J., Vassarotti A., Friesen J. D. Molecular cloning and biosynthetic regulation of cry1 gene of Saccharomyces cerevisiae. Mol Gen Genet. 1984;195(3):500–506. doi: 10.1007/BF00341453. [DOI] [PubMed] [Google Scholar]
- Huet J., Cottrelle P., Cool M., Vignais M. L., Thiele D., Marck C., Buhler J. M., Sentenac A., Fromageot P. A general upstream binding factor for genes of the yeast translational apparatus. EMBO J. 1985 Dec 16;4(13A):3539–3547. doi: 10.1002/j.1460-2075.1985.tb04114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Otaka E., Matsui K. A. Primary structures of ribosomal protein YS25 from Saccharomyces cerevisiae and its counterparts from Schizosaccharomyces pombe and rat liver. Biochemistry. 1985 Dec 3;24(25):7418–7423. doi: 10.1021/bi00346a058. [DOI] [PubMed] [Google Scholar]
- Jones E. W., Lam K. B. Mutations affecting levels of tetrahydrofolate interconversion enzymes in Saccharomyces cerevisiae. II. Map positions on chromosome VII of ade3-41 and ADE15. Mol Gen Genet. 1973 Jul 2;123(3):209–218. doi: 10.1007/BF00271239. [DOI] [PubMed] [Google Scholar]
- KIRBY K. S. ISOLATION AND CHARACTERIZATION OF RIBOSOMAL RIBONUCLEIC ACID. Biochem J. 1965 Jul;96:266–269. doi: 10.1042/bj0960266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamp R., Wittmann-Liebold B. Primary structure of protein S11 from Escherichia coli ribosomes. FEBS Lett. 1980 Nov 17;121(1):117–122. doi: 10.1016/0014-5793(80)81278-6. [DOI] [PubMed] [Google Scholar]
- Kief D. R., Warner J. R. Coordinate control of syntheses of ribosomal ribonucleic acid and ribosomal proteins during nutritional shift-up in Saccharomyces cerevisiae. Mol Cell Biol. 1981 Nov;1(11):1007–1015. doi: 10.1128/mcb.1.11.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. H., Warner J. R. Messenger RNA for ribosomal proteins in yeast. J Mol Biol. 1983 Mar 25;165(1):79–89. doi: 10.1016/s0022-2836(83)80243-5. [DOI] [PubMed] [Google Scholar]
- Kim C. H., Warner J. R. Mild temperature shock alters the transcription of a discrete class of Saccharomyces cerevisiae genes. Mol Cell Biol. 1983 Mar;3(3):457–465. doi: 10.1128/mcb.3.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraig E., Haber J. E., Rosbash M. Sporulation and rna2 lower ribosomal protein mRNA levels by different mechanisms in Saccharomyces cerevisiae. Mol Cell Biol. 1982 Oct;2(10):1199–1204. doi: 10.1128/mcb.2.10.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer A. R., Maniatis T., Ruskin B., Green M. R. Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell. 1984 Apr;36(4):993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- Käufer N. F., Fried H. M., Schwindinger W. F., Jasin M., Warner J. R. Cycloheximide resistance in yeast: the gene and its protein. Nucleic Acids Res. 1983 May 25;11(10):3123–3135. doi: 10.1093/nar/11.10.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin J. C., Woolford J. L., Jr Molecular cloning and analysis of the CRY1 gene: a yeast ribosomal protein gene. Nucleic Acids Res. 1983 Jan 25;11(2):403–420. doi: 10.1093/nar/11.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last R. L., Stavenhagen J. B., Woolford J. L., Jr Isolation and characterization of the RNA2, RNA3, and RNA11 genes of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Nov;4(11):2396–2405. doi: 10.1128/mcb.4.11.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leer R. J., Van Raamsdonk-Duin M. M., Mager W. H., Planta R. J. Conserved sequences upstream of yeast ribosomal protein genes. Curr Genet. 1985;9(4):273–277. doi: 10.1007/BF00419955. [DOI] [PubMed] [Google Scholar]
- Leer R. J., van Raamsdonk-Duin M. M., Kraakman P., Mager W. H., Planta R. J. The genes for yeast ribosomal proteins S24 and L46 are adjacent and divergently transcribed. Nucleic Acids Res. 1985 Feb 11;13(3):701–709. doi: 10.1093/nar/13.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leer R. J., van Raamsdonk-Duin M. M., Molenaar C. M., Cohen L. H., Mager W. H., Planta R. J. The structure of the gene coding for the phosphorylated ribosomal protein S10 in yeast. Nucleic Acids Res. 1982 Oct 11;10(19):5869–5878. doi: 10.1093/nar/10.19.5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A., McNally J., Wool I. G. The primary structure of rat liver ribosomal protein L37. Homology with yeast and bacterial ribosomal proteins. J Biol Chem. 1983 Sep 10;258(17):10664–10671. [PubMed] [Google Scholar]
- Lindquist S. Regulation of protein synthesis during heat shock. Nature. 1981 Sep 24;293(5830):311–314. doi: 10.1038/293311a0. [DOI] [PubMed] [Google Scholar]
- Madjar J. J., Frahm M., McGill S., Roufa D. J. Ribosomal protein S14 is altered by two-step emetine resistance mutations in Chinese hamster cells. Mol Cell Biol. 1983 Feb;3(2):190–197. doi: 10.1128/mcb.3.2.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight G. L., McConaughy B. L. Selection of functional cDNAs by complementation in yeast. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4412–4416. doi: 10.1073/pnas.80.14.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil J. B., Smith M. Transcription initiation of the Saccharomyces cerevisiae iso-1-cytochrome c gene. Multiple, independent T-A-T-A sequences. J Mol Biol. 1986 Feb 5;187(3):363–378. doi: 10.1016/0022-2836(86)90439-0. [DOI] [PubMed] [Google Scholar]
- Miller M. J., Xuong N. H., Geiduschek E. P. Quantitative analysis of the heat shock response of Saccharomyces cerevisiae. J Bacteriol. 1982 Jul;151(1):311–327. doi: 10.1128/jb.151.1.311-327.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra G., Warner J. R. A yeast ribosomal protein gene whose intron is in the 5' leader. J Biol Chem. 1984 Jul 25;259(14):9218–9224. [PubMed] [Google Scholar]
- Nakanishi O., Oyanagi M., Kuwano Y., Tanaka T., Nakayama T., Mitsui H., Nabeshima Y., Ogata K. Molecular cloning and nucleotide sequences of cDNAs specific for rat liver ribosomal proteins S17 and L30. Gene. 1985;35(3):289–296. doi: 10.1016/0378-1119(85)90007-1. [DOI] [PubMed] [Google Scholar]
- Otaka E., Higo K., Itoh T. Yeast ribosomal proteins. VIII. Isolation of two proteins and sequence characterization of twenty-four proteins from cytoplasmic ribosomes. Mol Gen Genet. 1984;195(3):544–546. doi: 10.1007/BF00341461. [DOI] [PubMed] [Google Scholar]
- Otaka E., Higo K., Osawa S. Isolation of seventeen proteins and amino-terminal amino acid sequences of eight proteins from cytoplasmic ribosomes of yeast. Biochemistry. 1982 Sep 14;21(19):4545–4550. doi: 10.1021/bi00262a005. [DOI] [PubMed] [Google Scholar]
- Pearson N. J., Fried H. M., Warner J. R. Yeast use translational control to compensate for extra copies of a ribosomal protein gene. Cell. 1982 Jun;29(2):347–355. doi: 10.1016/0092-8674(82)90151-9. [DOI] [PubMed] [Google Scholar]
- Rhoads D. D., Dixit A., Roufa D. J. Primary structure of human ribosomal protein S14 and the gene that encodes it. Mol Cell Biol. 1986 Aug;6(8):2774–2783. doi: 10.1128/mcb.6.8.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads D. D., Roufa D. J. Emetine resistance of Chinese hamster cells: structures of wild-type and mutant ribosomal protein S14 mRNAs. Mol Cell Biol. 1985 Jul;5(7):1655–1659. doi: 10.1128/mcb.5.7.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rosbash M., Harris P. K., Woolford J. L., Jr, Teem J. L. The effect of temperature-sensitive RNA mutants on the transcription products from cloned ribosomal protein genes of yeast. Cell. 1981 Jun;24(3):679–686. doi: 10.1016/0092-8674(81)90094-5. [DOI] [PubMed] [Google Scholar]
- Rotenberg M. O., Woolford J. L., Jr Tripartite upstream promoter element essential for expression of Saccharomyces cerevisiae ribosomal protein genes. Mol Cell Biol. 1986 Feb;6(2):674–687. doi: 10.1128/mcb.6.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz L. D., Friesen J. D. Nucleotide sequence of the tcml gene (ribosomal protein L3) of Saccharomyces cerevisiae. J Bacteriol. 1983 Jul;155(1):8–14. doi: 10.1128/jb.155.1.8-14.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skogerson L., McLaughlin C., Wakatama E. Modification of ribosomes in cryptopleurine-resistant mutants of yeast. J Bacteriol. 1973 Nov;116(2):818–822. doi: 10.1128/jb.116.2.818-822.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8419–8423. doi: 10.1073/pnas.82.24.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez L., Vásquez D., Jiménez A. Genetics and biochemistry of cryptopleurine resistance in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1977 Nov 18;156(3):319–326. doi: 10.1007/BF00267188. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Ishikawa K., Ogata K. On the sequence homology of the ribosomal proteins, Escherichia coli S11, yeast rp59 and Chinese hamster S14. FEBS Lett. 1986 Jul 7;202(2):295–297. doi: 10.1016/0014-5793(86)80704-9. [DOI] [PubMed] [Google Scholar]
- Teem J. L., Abovich N., Kaufer N. F., Schwindinger W. F., Warner J. R., Levy A., Woolford J., Leer R. J., van Raamsdonk-Duin M. M., Mager W. H. A comparison of yeast ribosomal protein gene DNA sequences. Nucleic Acids Res. 1984 Nov 26;12(22):8295–8312. doi: 10.1093/nar/12.22.8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teem J. L., Rosbash M. Expression of a beta-galactosidase gene containing the ribosomal protein 51 intron is sensitive to the rna2 mutation of yeast. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4403–4407. doi: 10.1073/pnas.80.14.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R., Orkin S. H., Maniatis T. Specific transcription and RNA splicing defects in five cloned beta-thalassaemia genes. Nature. 1983 Apr 14;302(5909):591–596. doi: 10.1038/302591a0. [DOI] [PubMed] [Google Scholar]
- Udem S. A., Warner J. R. Ribosomal RNA synthesis in Saccharomyces cerevisiae. J Mol Biol. 1972 Mar 28;65(2):227–242. doi: 10.1016/0022-2836(72)90279-3. [DOI] [PubMed] [Google Scholar]
- Warner J. R., Gorenstein C. The synthesis of eucaryotic ribosomal proteins in vitro. Cell. 1977 May;11(1):201–212. doi: 10.1016/0092-8674(77)90331-2. [DOI] [PubMed] [Google Scholar]
- Warner J. R., Mitra G., Schwindinger W. F., Studeny M., Fried H. M. Saccharomyces cerevisiae coordinates accumulation of yeast ribosomal proteins by modulating mRNA splicing, translational initiation, and protein turnover. Mol Cell Biol. 1985 Jun;5(6):1512–1521. doi: 10.1128/mcb.5.6.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford J. L., Jr, Hereford L. M., Rosbash M. Isolation of cloned DNA sequences containing ribosomal protein genes from Saccharomyces cerevisiae. Cell. 1979 Dec;18(4):1247–1259. doi: 10.1016/0092-8674(79)90236-8. [DOI] [PubMed] [Google Scholar]
- Woolford J. L., Jr, Rosbash M. Ribosomal protein genes rp 39(10 - 78), rp 39(11 - 40), rp 51, and rp 52 are not contiguous to other ribosomal protein genes in the Saccharomyces cerevisiae genome. Nucleic Acids Res. 1981 Oct 10;9(19):5021–5036. doi: 10.1093/nar/9.19.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woudt L. P., Smit A. B., Mager W. H., Planta R. J. Conserved sequence elements upstream of the gene encoding yeast ribosomal protein L25 are involved in transcription activation. EMBO J. 1986 May;5(5):1037–1040. doi: 10.1002/j.1460-2075.1986.tb04319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]






