Abstract
Suppressor of cytokine signalling 3 (SOCS3) is responsible for regulating the cellular response to a variety of cytokines, including interleukin 6 and leukaemia inhibitory factor. Identification of the SOCS box domain led to the hypothesis that SOCS3 can associate with functional E3 ubiquitin ligases and thereby induce the degradation of bound signalling proteins. This model relies upon an interaction between the SOCS box, elonginBC and a cullin protein that forms the E3 ligase scaffold. We have investigated this interaction in vitro using purified components and show that SOCS3 binds to elonginBC and cullin5 with high affinity. The SOCS3–elonginBC interaction was further characterised by determining the solution structure of the SOCS box–elonginBC ternary complex and by deletion and alanine scanning mutagenesis of the SOCS box. These studies revealed that conformational flexibility is a key feature of the SOCS–elonginBC interaction. In particular, the SOCS box is disordered in isolation and only becomes structured upon elonginBC association. The interaction depends upon the first 12 residues of the SOCS box domain and particularly on a deeply buried, conserved leucine. The SOCS box, when bound to elonginBC, binds tightly to cullin5 with 100 nM affinity. Domains upstream of the SOCS box are not required for elonginBC or cullin5 association, indicating that the SOCS box acts as an independent binding domain capable of recruiting elonginBC and cullin5 to promote E3 ligase formation.
Keywords: SOCS, cytokine signalling, ubiquitin ligase, elongin, cullin
Introduction
The SOCS (suppressors of cytokine signalling) proteins were first identified based on their ability to suppress cytokine signalling through the Janus kinase/signal transduction and activator of transcription (JAK/STAT) pathway.1 Cytokines bind to their cell surface receptors and induce receptor dimerisation, which allows trans-phosphorylation of receptor-associated JAKs. This leads to tyrosine phosphorylation of the intracellular receptor subunits, which are then bound by members of the STAT family. STAT phosphorylation ensues, followed by dimerisation and translocation into the nucleus.2 The eight SOCS proteins, SOCS1–7 and cytokine-inducible SH2-containing protein (CIS), are able to inhibit this pathway. SOCS3 is known to inhibit signalling induced by a variety of cytokines including interleukin 6, granulocyte colony-stimulating factor (G-CSF), leptin and LIF.3,4 SOCS3 knockout mice die in utero as a result of placental defects.5 Conditional knockout studies have shown that a lack of SOCS3 can induce a variety of metabolic and inflammatory disorders. For example, mice with haematopoietic deletion of SOCS3 are overresponsive to G-CSF stimulation.4 Lack of SOCS3 in neural cells leads to enhanced leptin signaling and a severe loss in body weight, while SOCS3-deficient adipocytes are protected against tumor necrosis factor α-induced insulin resistance.6 Intracellular delivery of SOCS3 in mice can reduce the production of inflammatory cytokines in response to bacterial toxins and attenuate liver apoptosis and hemorrhagic necrosis.7
Each SOCS protein consists of an N-terminal domain, a central SH2 domain and a C-terminal SOCS box. The N-terminal and SH2 domains are responsible for direct or competitive inhibition of signalling proteins by interaction with the JAKs or the receptors themselves.8–13 The SOCS box mediates signalling suppression by a different mechanism; it is believed to promote the degradation of bound signalling intermediates via an interaction with cellular ubiquitination machinery.14–19 The SOCS box is a small, 40- to 60-residue domain with significant sequence and structural homology to the α domain of the von Hippel–Lindau (VHL) protein and, to a lesser extent, the F-box from Skp2.18 Mutational analysis has shown that the SOCS box contains two distinct interaction motifs. The N-terminal half of the SOCS box is responsible for elonginBC binding, whilst the C-terminal portion encodes specificity for binding a cullin protein20. Cullin proteins act as the scaffold for a large family of E3 ubiquitin ligases that catalyse the ubiquitination of protein substrates.21 They are large (~100 kDa) proteins consisting of two domains; the N-terminal domain binds via adaptor proteins to the protein that is to be ubiquitinated, while the C-terminal domain binds to a RING finger protein such as Rbx2 to create a docking site for the ubiquitin-conjugating enzyme, E2. A comparison to the more well studied SCF (Skp–cullin–F-box) family of ubiquitin ligases suggests that in ECS (elongin-cullin-SOCS) ligases, substrate specificity will be defined by interaction domains upstream of the SOCS box. As SOCS3 can interact with JAK and the gp130 receptor, it is assumed that these proteins will be polyubiquitinated following SOCS3 association and then degraded by the proteasome.
The in vivo role of the SOCS box in cytokine regulation has been investigated by studies involving SOCS3 SOCS box null mice. These mice did not develop inflammatory disease but were hyperresponsive to G-CSF, and interleukin 6, at a level intermediate between that of wild-type and conditional SOCS null animals.22 Similar studies involving knockout of the SOCS1 SOCS box show a similar phenomenon; in this case, the mice were hyperresponsive to interferon γ, again to a level intermediate between that of wild-type and SOCS1 null knockout animals.15 Together, these studies suggest that the presence of the SOCS box, at least for SOCS1 and SOCS3, is required for signalling inhibition. Although the structure of the extended SH2 domain of SOCS3 was solved recently by NMR23 and X-ray crystallography,24 to date there is no detailed information regarding the SOCS3 SOCS box structure and its interactions.
It is known that SOCS3 can bind elonginBC and cullin5 in vivo20; however, there has been no structural or biophysical analysis of this interaction. In this article, we dissect this interaction in vitro, using mutagenesis, structural studies and affinity measurements with purified components. These studies show that conformational malleability is a key feature of the interaction. Analysis of the SOCS3 SOCS box by mutagenesis indicates that the elonginBC binding epitope is confined to the 12 N-terminal residues. Elucidation of the solution structure of the SOCS box–elonginB–elonginC ternary complex shows the SOCS box epitope to be a single α-helix preceded by three residues. Interestingly, this epitope was largely tolerant of mutation apart from a strictly conserved leucine residue that inserts deeply into a hydrophobic pocket on the surface of elonginC. Our results show that the SOCS box acts as an independent domain that recruits elonginBC and cullin5.
Results
ElonginBC expressed in isolation undergoes conformational exchange and does not interact with SOCS3
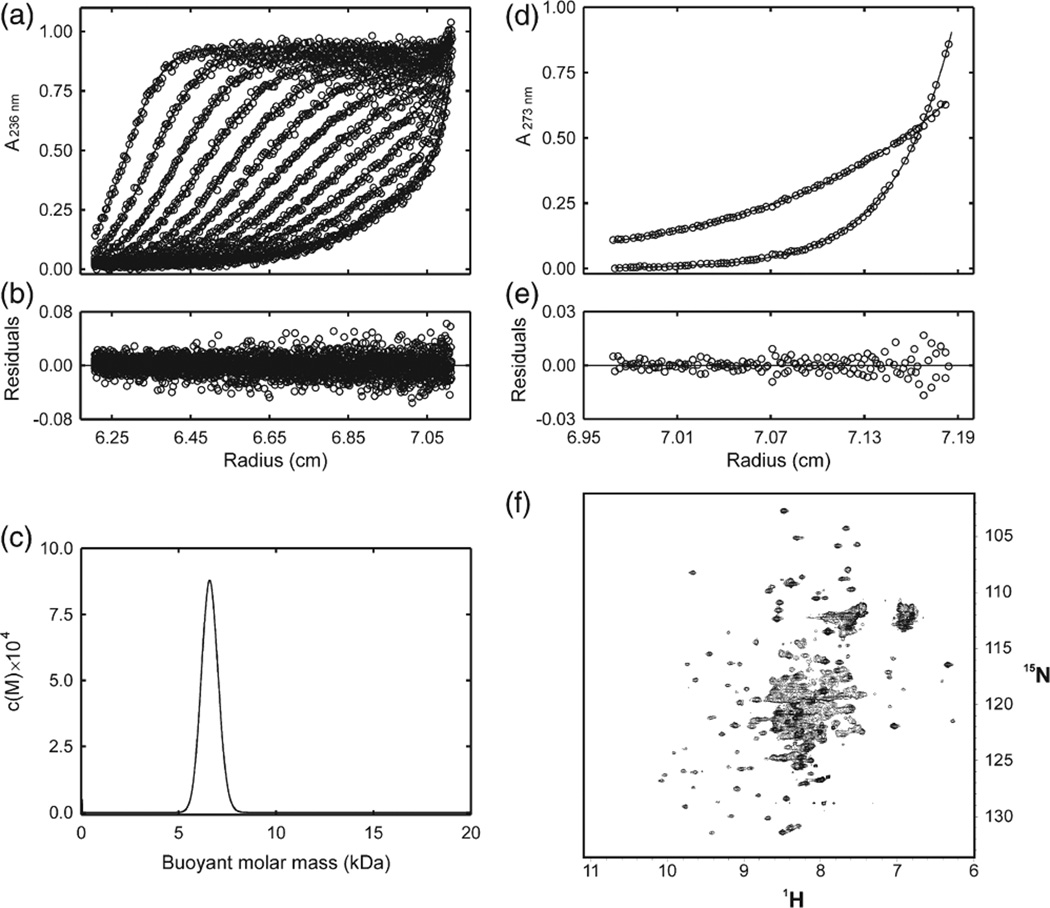
ElonginB and elonginC were co-expressed in Escherichia coli and formed a stable complex. However, this complex did not bind recombinant SOCS3 protein, as shown by isothermal titration calorimetry (ITC), glutathione S-transferase (GST) pull-down assay and NMR. The elonginBC complex behaved aberrantly by gel filtration, eluting at an apparent molecular mass of ca 45 kDa, which would be consistent with that of a 2:2 dimer (Fig. S1). Nevertheless, sedimentation velocity and equilibrium ultracentrifugation studies showed conclusively that elonginBC exists as a 1:1 heterodimer, even at concentrations of >1 mg/mL, and did not associate to higher oligomers (Fig. 1a–e). The 15N-heteronuclear single quantum coherence (HSQC) NMR spectrum of elonginBC showed that the complex was at least partially folded but contained broad linewidths and variable peak intensities, which suggests the protein was undergoing conformational exchange (Fig. 1f). This is similar to previous studies involving yeast elonginC, which had shown that trifluoroethanol was required to stabilise the tertiary structure of the protein and that in the absence of bound VHL peptide (an SOCS box homologue) there was widespread chemical/conformational exchange.25 Thus, elonginBC, when expressed in isolation, does not interact with the SOCS box and is in a state exhibiting conformational exchange by NMR and runs aberrantly on a gel-filtration column, due to either shape and/or dynamic effects.
Fig. 1.
NMR and analytical ultracentrifugation analysis of elonginBC. (a) Radial profiles of 0.92 mg/mL elonginBC monitored at 236 nm during centrifugation at 40,000 rpm (○). The sedimentation velocity data were analyzed in terms of a continuous buoyant molar mass distribution (−) using the continuous mass distribution model [c(M)] resident in the program SEDFIT. Ninety scans were used in the analysis; however, for clarity, only every seventh is presented. (b) Plot of residuals for the analysis of the experimental data in (a). (c) c(M) distribution describing the experimental data in (b). The distribution exhibits a single, well-defined maximum corresponding to a buoyant (reduced) molar mass of 6.5 kDa. This corresponds to a physical mass of 24.5 kDa under the conditions used. The predicted mass of a 1:1 complex is 25.6 kDa. (d) Radial profiles of 0.92 mg/mL elonginBC monitored at 273 nm after centrifugation for 24 h at 20,000 and at 35,000 rpm. The sedimentation equilibrium data were globally analyzed in terms of a single ideal solute (−) using the species analysis model resident in the program SEDPHAT. The buoyant molar mass recovered from the analysis was 6.7 kDa. This corresponds to a physical mass of 25.1 kDa under the conditions used. (e) Plot of residuals for the analysis of the experimental data in (d). (f) 1H–15N HSQC analysis of elonginBC. Recombinant elonginBC, 0.25 mM in 20 mM potassium phosphate, 2.5 mM DTT, pH 6.7, was recorded at 310 K on a Bruker Avance 500 spectrometer. Note the concentration of broad overlapped peaks in the centre of the spectrum indicating conformational exchange.
SOCS3ΔPEST forms a ternary complex with elonginBC when co-expressed, which binds cullin5
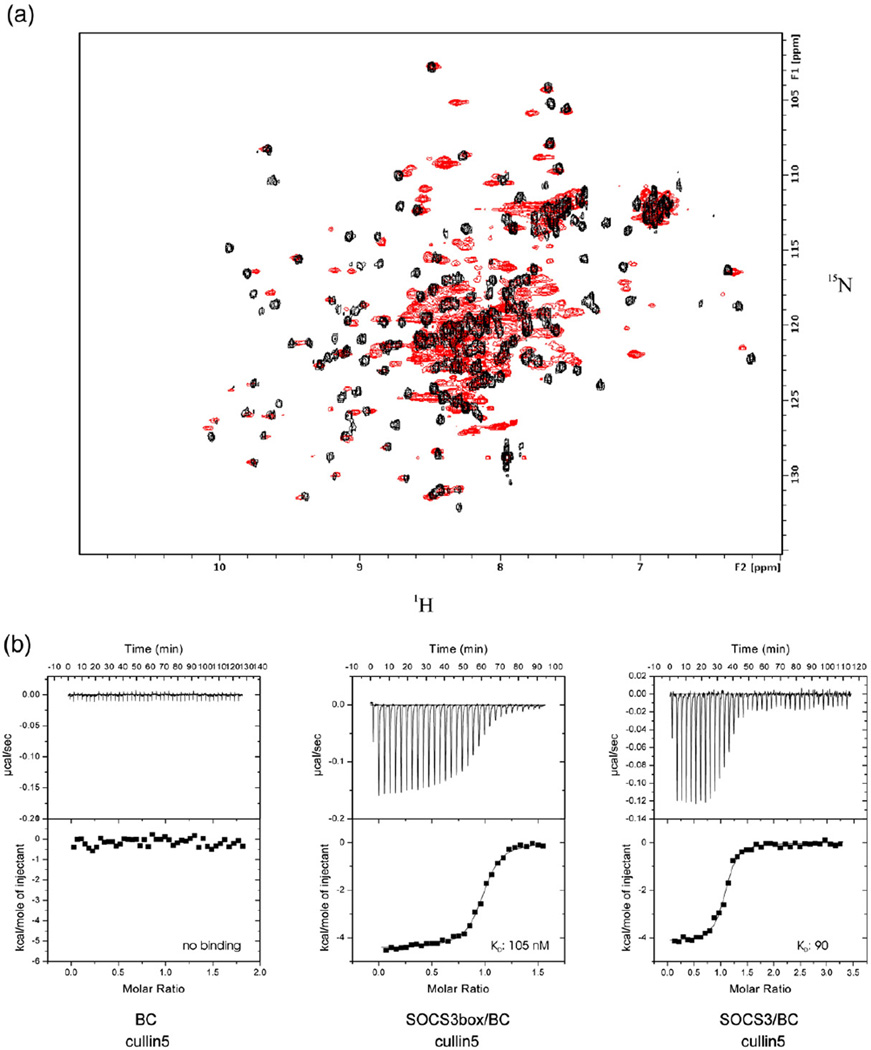
As SOCS3 and elonginBC did not interact when expressed in isolation, co-expression of the three species was tested. Although wild-type SOCS3 is poorly soluble and forms inclusion bodies, SOCS3ΔPEST, which lacks the PEST motif (residues 129–163) is significantly more soluble, and co-expression with elonginBC was found to yield a stable ternary complex. Likewise, co-expression of just the SOCS box domain with elonginBC produced a stable ternary complex. Both of these ternary complexes (termed SOCS3–elonginBC and SOCS box–elonginBC) were stable, as assessed by size-exclusion chromatography, even at concentrations of ~10 µM, suggesting that the interaction was of high affinity, and they eluted at the expected apparent molecular weight for a 1:1:1 complex. The conformational exchange seen in the elonginBC NMR spectrum was also absent in the ternary complexes (Fig. 2a). Quantification of the SOCS3–elonginBC affinity cannot be achieved using titration methods, as isolated elonginBC does not bind SOCS3 as detailed in the previous section. However, such methods can be used to probe the expected biological activity of such a complex, which is to bind cullin5, the E3 ligase scaffold. Therefore, using purified recombinant cullin5 and ITC, we examined whether our material was active in this regard. The N-terminal domain of cullin5, which contains the expected binding site for SOCS3–elonginBC, was expressed in E. coli as a GST fusion protein and used for this purpose. Both full-length SOCS3–elonginBC and SOCS3 box–elonginBC ternary complexes bound cullin5 with high affinity (90±10 and 105±10 nM, respectively; Fig. 2b). In addition, both these ternary complexes bound cullin5 with the same enthalpy (−4.1±0.2 and −3.7± 0.5 kcal/mol, respectively; duplicate experiments in each case). The identical affinities and enthalpies of the isolated domain and the full-length protein show that the SOCS box, when bound to elonginBC, acts as an independent cullin5 binding domain that does not require the upstream SH2 and N-terminal domains in order to be active.
Fig. 2.
SOCS3 and elonginBC forms a stable ternary complex that binds tightly to cullin5. Overlay of the 15N HSQC spectrum of elonginBC alone shown in red with that of elonginBC (15N labelled) in complex with the SOCS3 box (unlabelled) shown in black. The broad peaks in the centre of the elonginBC-alone spectrum are shifted and well resolved in the ternary complex, indicating little, if any, conformational exchange once the SOCS box is bound. (b) ITC analysis of the SOCS3 SOCS box–elonginBC–cullin5 interaction. GST–cullin5 (80 µM, NTD) was titrated into 10 µM elonginBC, SOCS3box/elonginBC or SOCS3/elonginBC in 30×10-µL injections using a VP-ITC unit (Microcal). Both protein and peptide were prepared in 20 mM potassium phosphate and 100 mM NaCl supplemented with 2 mM 2-mercaptoethanol. Only the SOCS3box–elonginBC and SOCS3–elonginBC ternary complexes bound to cullin5 with measurable affinity. These titration curves fitted well to a single-site model with a Kd of 105±10 nM, ΔH −3700±500 cal/mol, N=0.96 and Kd of 90±10 nM, ΔH −4100±200 cal/mol, N=1.05, respectively (average of duplicate experiments).
Defining the minimal elonginBC binding epitope on the SOCS box
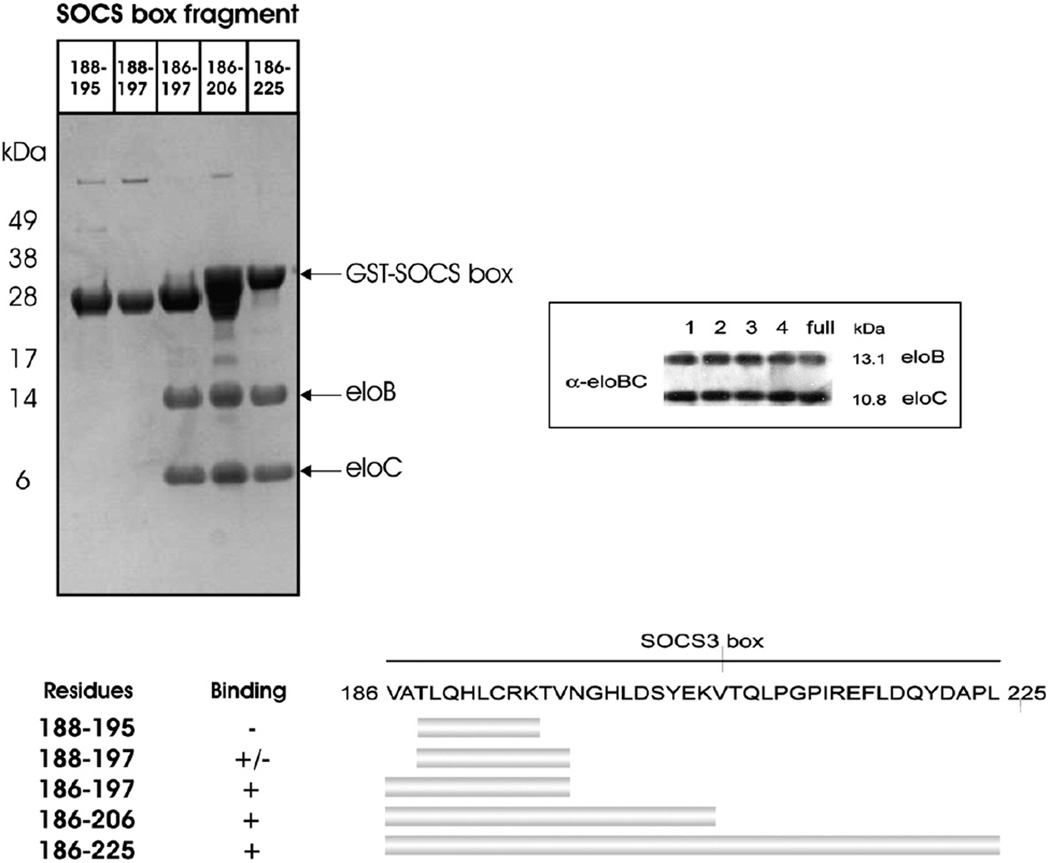
In order to determine the minimal elonginBC binding epitope on the SOCS box, deletion analyses were performed. Constructs of full-length and shorter fragments of the SOCS box were produced as GST fusion proteins and tested for association with elonginBC by coexpression and GST pull-down methodology. As shown in Fig. 3, the first 12 residues of the SOCS3 SOCS box were necessary and sufficient for elonginBC binding. Therefore, the minimal high-affinity binding epitope for elonginBC on SOCS3 is VATLQHLCRKTV 197, corresponding to residues 1–12 of the SOCS box domain.
Fig. 3.
Determination of the minimal elonginBC binding epitope on the SOCS3 SOCS box. The SOCS box fragments were produced as GST fusions to aid stability during expression and co-expressed with elonginBC. Glutathione Sepharose was used to pull down GST-labelled proteins present in the cell lysate (upper left panel). As shown, a fragment comprising residues 1–12 was the smallest segment of the SOCS box capable of co-purifying with elonginBC with similar affinity to the entire SOCS box (1–40). The successful co-expression of elonginBC in all cultures was shown by Western blot (upper right panel) and a schematic view of the mutants used and the results are also shown (bottom panel).
The SOCS box is unstructured in isolation but becomes structured upon elonginBC association
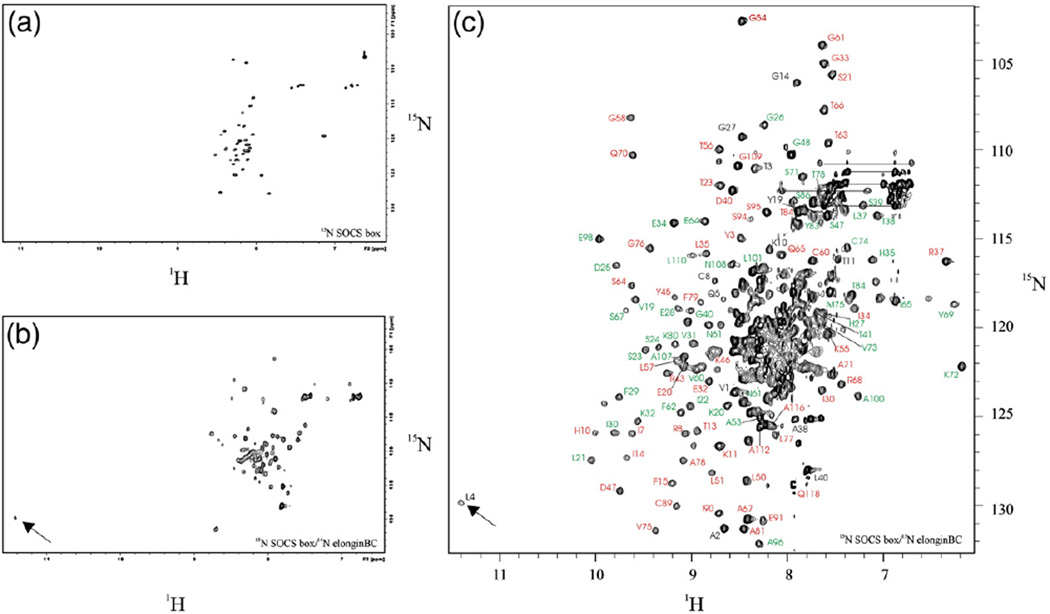
Having established that the SOCS box in isolation binds elonginBC and cullin5, we undertook structural studies on the isolated domain. A 42-residue 15N-labelled recombinant SOCS box peptide was produced; the 1H–15N HSQC spectrum of this isolated peptide is characteristic of that of an unstructured protein (Fig. 4a). Extensive analysis of 1H–1H total correlated spectroscopy (TOCSY) and nuclear Overhauser enhancement spectroscopy (NOESY) experiments of unlabelled SOCS box domain also suggested that the peptide was completely unstructured in solution (data not shown). The labelled peptide was used to displace the SOCS box domain in an unlabelled SOCS box–elonginBC ternary complex in order to form a partially labelled complex. Subsequent HSQC analysis of this (15N-SOCS box–14N-elonginBC) complex demonstrated extensive SOCS box peak dispersion after elonginBC association (Fig. 4b). In particular, there was a highly downfield shifted amide resonance (11.3 ppm) arising from Leu4 of the SOCS box. The number of peaks was higher than expected and there was some variability in intensity and linewidth, suggesting that part of the SOCS domain was still undergoing conformational exchange in solution. An analysis of uniformly 15N or 15N/13C labelled SOCS box–elonginBC supported this hypothesis. The spectrum was well resolved and the resonances of elonginB and elonginC (88% and 90% of backbone, respectively) were readily assigned, but only the first 15 residues of the SOCS box could be assigned contiguously, including the highly downfield shifted Leu4 (Fig. 4c). The C-terminal half of the SOCS box could not be assigned completely because of overlap and peak broadening. This suggests that it may be sampling a range of conformations in solution.
Fig. 4.
The SOCS boxis unstructured inisolation and becomes partially structured upon elonginBC association. 1H–15N HSQC spectra of the SOCS box, in isolation and bound to elonginBC. (a) Recombinant 15N-labelled SOCS3 SOCS box was prepared by thrombin digestion of a GST–SOCS box fusion protein. 15N HSQC spectrum of 0.1 mM protein in 20 mM potassium phosphate and 2.5 mM DTT, pH 6.7, was recorded at 310 K and 500 MHz. (b) The sample from (a) was mixed with a threefold excess of unlabelled SOCS box–elonginBC ternary complex and HSQC analysis was performed as before. The 15N-labelled SOCS box was able to compete with the unlabelled domain in the ternary complex and thus bind elonginBC. The downfield amide peak at 11.3 ppm arises from Leu4 of the SOCS box (arrow). The range of peak intensities seen is a further indication of conformational exchange. (c) Backbone amide assignment of the SOCS box–elonginBC ternary complex. HSQC analysis of a fully 15N labelled ternary complex was performed using conditions identical to that of (a) and (b). SOCS box assignments are shown in black, elonginC assignments in green and elonginB assignments in red. Some assignments are omitted for clarity.
To determine whether this conformational exchange was a consequence of the fact that the isolated SOCS3 box rather than full-length SOCS3 was used in these studies, HSQC spectra of full-length SOCS3 in complex with elonginBC were examined. The spectrum of SOCS3/elonginBC was an almost perfect sum of the HSQC spectrum of the SOCS box–elonginBC complex and that of SOCS322–185 (which lacks the SOCS box; Fig. S2). Although resonances of the SOCS3–elonginBC complex were not assigned because of its size (45 kDa), resonances from the SOCS box that were well resolved in the spectrum (Thr3, Leu4, Gln5, Cys8, Gly14, Gly27 and Leu40) overlaid perfectly between complexes of full-length SOCS3 and the SOCS box alone (Fig. S2). The only exceptions were Val1 and Ala2, which were shifted in the full-length protein as the sequence immediately preceding them had changed. Two residues from the distal half of the SOCS3 box, Gly27 and Leu40, which could be assigned but were unstructured in the SOCS box–elonginBC complex (based on the absence of NOEs to non-adjacent residues), had identical chemical shifts in the full-length SOCS3–elonginBC complex, indicating that they were similarly unstructured in the complex with full-length protein. These data imply that the C-terminal half of the SOCS box undergoes significant conformational exchange in the absence of bound cullin5 whether as an isolated domain or as part of full-length SOCS3.
Tertiary structure of the SOCS3 SOCS box bound to elonginBC
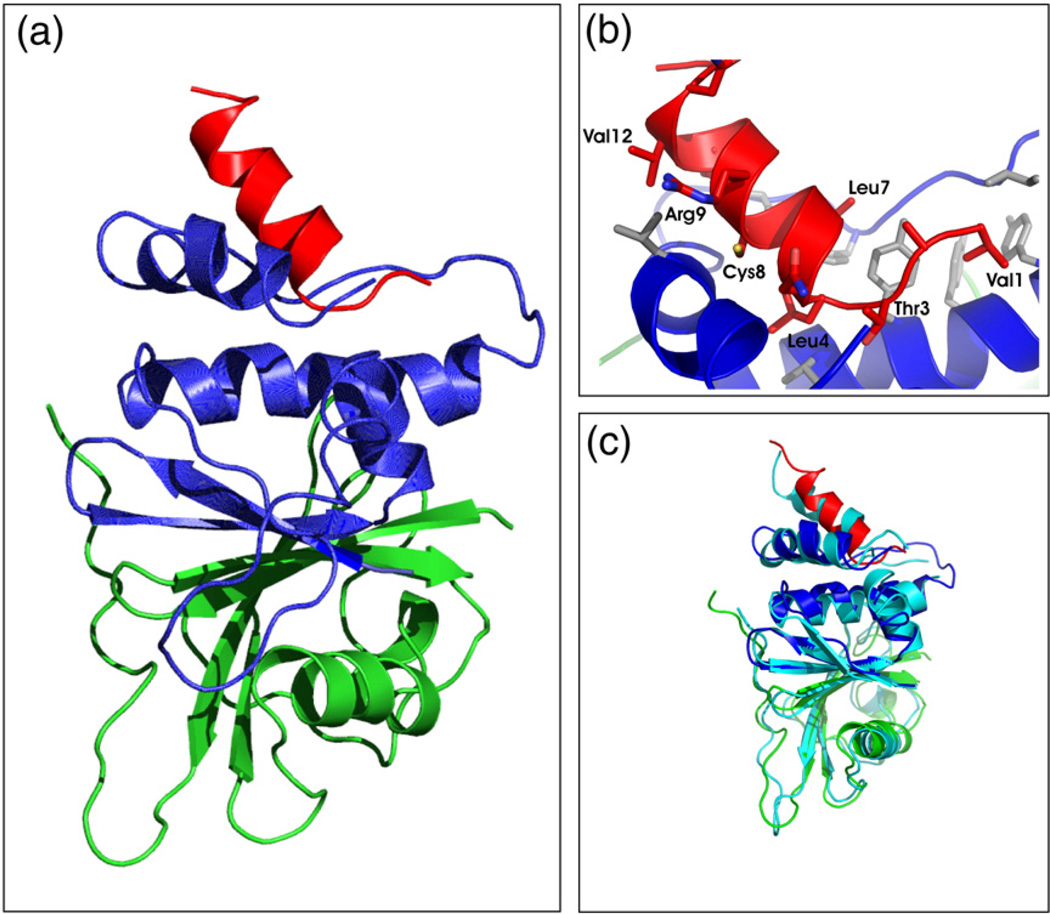
Using NMR, we determined the structure of the SOCS box–elonginBC ternary complex (Fig. 5 and Fig. S3). The C-terminal half of the SOCS box domain was excluded from the structure calculations due to incomplete resonance assignments (see above). Resonances that could be assigned in this region showed α-helical chemical shifts but were significantly broadened and displayed NOEs only to adjacent residues. In contrast, the N-terminal half of the SOCS box, which contains the elonginBC-binding motif (the BC box, residues 1–12) was completely assigned and well structured. As only the first 12 residues of the SOCS box (referred to as the BC box) are required for elonginBC binding (Fig. 5), the assignment of residues 1–12 was sufficient to analyse the SOCS–elonginBC interaction.
Fig. 5.
Tertiary structure of an SOCS3 SOCS box–elonginBC complex. (a) Ribbon diagram of the SOCS3 SOCS box (red) in complex with elonginC (blue) and elonginB (green). The interaction of the SOCS box occurs exclusively with elonginC and is mediated mostly by hydrophobic interactions. ElonginBC forms a tightly associated complex with the core of the association being a continuous β-sheet formed by residues from both proteins. A number of side-chain hydrophobic interactions further stabilise the complex. (b) Close view of the SOCS box elonginC interface with hydrophobic side chains from the SOCS box (red) labelled. The hydrophobic residues from elonginC (blue) that form the interface are shown in grey stick representation. (c) Ribbon diagram of the SOCS3 SOCS box in complex with elonginBC [same color scheme as in (a)] shown overlaid on the SOCS2/elonginBC structure (cyan).
This region, the SOCS3 elonginBC binding motif (termed the BC box), which is completely unstructured as an isolated peptide and only adopts structure upon binding, exists as a nine-residue α-helix with a three-residue N-terminal extension. This 12-residue motif presents an almost completely hydrophobic face to elonginC, with Val1, Leu4, Leu7, Cys8 and Val12 being the most deeply buried at the interface. In particular, Leu4 occupies a very deep hydrophobic pocket on the surface of elonginC (Fig. 5b). The backbone amide 1H resonance of Leu4 is shifted significantly down-field, to 11.3 ppm, only when bound to elonginBC, as it is then located very close to Tyr76 of elonginC, resulting in a ring-current-induced downfield shift. It therefore acts as a highly distinctive marker of binding.
The structure of the elonginBC subunits of the complex is very similar to previously described structures of elonginBC in complex with VHL,26 SOCS2 and SOCS427; the Cα RMSD was 1.5 Å over 158 residues compared to the SOCS2–elonginBC structure, with major differences seen only in loops and poorly structured regions. In elonginB, the 23 most C-terminal residues were unstructured and there is a 12-residue unstructured loop in the elonginC subunit (Leu46-Thr57). This is consistent with the same regions in the SOCS2–elonginBC structure,27 where lack of electron density or higher than average B-factors were observed. A stretch of residues in elonginC that could not be completely assigned because of peak broadening (Ser87-Phe93) had significantly higher than average B-factors in the SOCS2–elonginBC crystal structure. This suggests that the loop is mobile and that in full-length SOCS3, where the SH2 domain immediately precedes Val1 of the SOCS box, it could move to accommodate the presence of the rest of SOCS3.
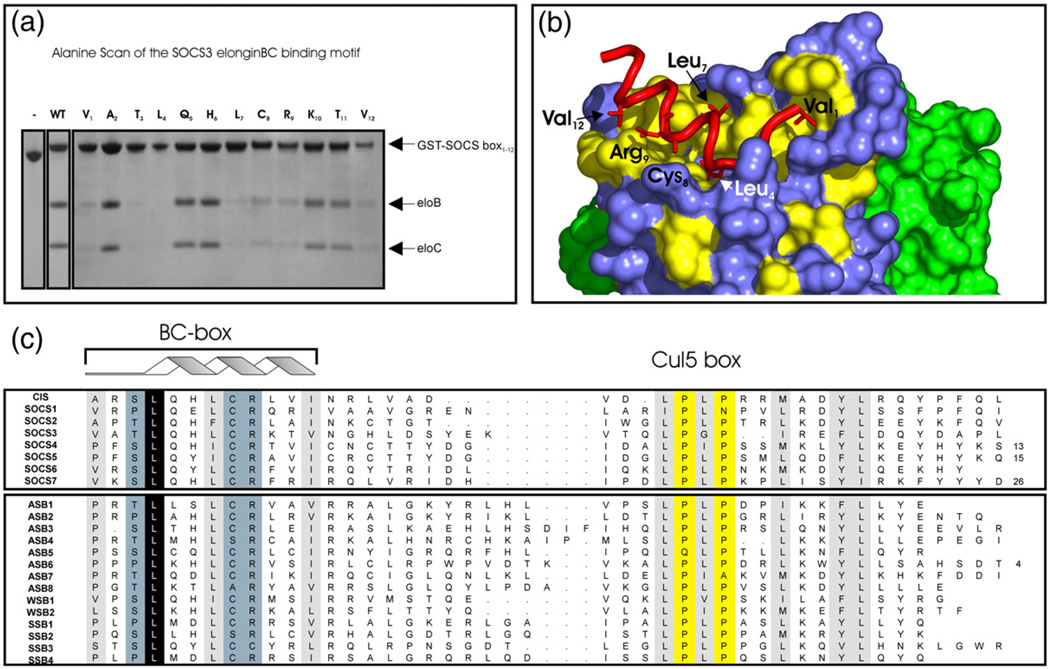
Alanine scanning mutagenesis of the elonginBC binding epitope of SOCS3
Having determined the structure of the SOCS3 BC box in complex with elonginBC, we wished to define the residues that were critical for binding. Therefore, the BC box was further investigated by alanine scanning mutagenesis. As shown in Fig. 6, there were several residues whose mutation to alanine clearly interfered with binding: Val1, Thr3, Leu4, Leu7, Cys8, Arg9 and Val12. Interestingly, only mutation of Leu4 to alanine completely disrupted the interaction; other mutations led to reduced affinity but did not entirely abrogate the association. The alanine scanning results are consistent with our structure of the complex and also with sequence conservation across the family. Of the seven key residues identified, five (Val1, Leu4, Leu7, Cys8, and Val12) contain side chains that interact with elonginC, most lying on one face of the α-helix. The only key residues that did not have side chains in direct contact with elonginC were Thr3 and Arg9. Thr3 is almost always Ser or Thr across the SOCS-box-containing family and analysis of the available structures shows that the side-chain oxygen makes a hydrogen bond with the backbone amide of His6, reminiscent of a helical N-cap. The reason for the importance of Arg9 is unclear.
Fig. 6.
Key residues required for elonginBC binding. An Ala scan of the SOCS3 SOCS box to determine residues required for elonginBC binding was performed. Co-expression of 12 SOCS box domain constructs, each containing a single Ala mutation, with elonginBC was performed in E. coli, and glutathione Sepharose was used to pull down GST-labelled proteins present in the cell lysate. (a) The following mutations completely interfered with elonginBC binding: Val1, Thr3, Leu4, Leu7, Cys8, Arg9 and Val12. Of these, only the L4A mutation completely abolished binding. (c) Residues identified by Ala scan are highlighted on a surface representation of elonginBC where hydrophobic residues on the surface of elonginC are shown in yellow. The BC box of SOCS3 (red) is shown in cartoon representation with important side chains displayed in “stick” representation. ElonginC is shown in blue and elonginB in green.
ElonginBC binding to full-length SOCS3 does not affect the SH2–pY interaction
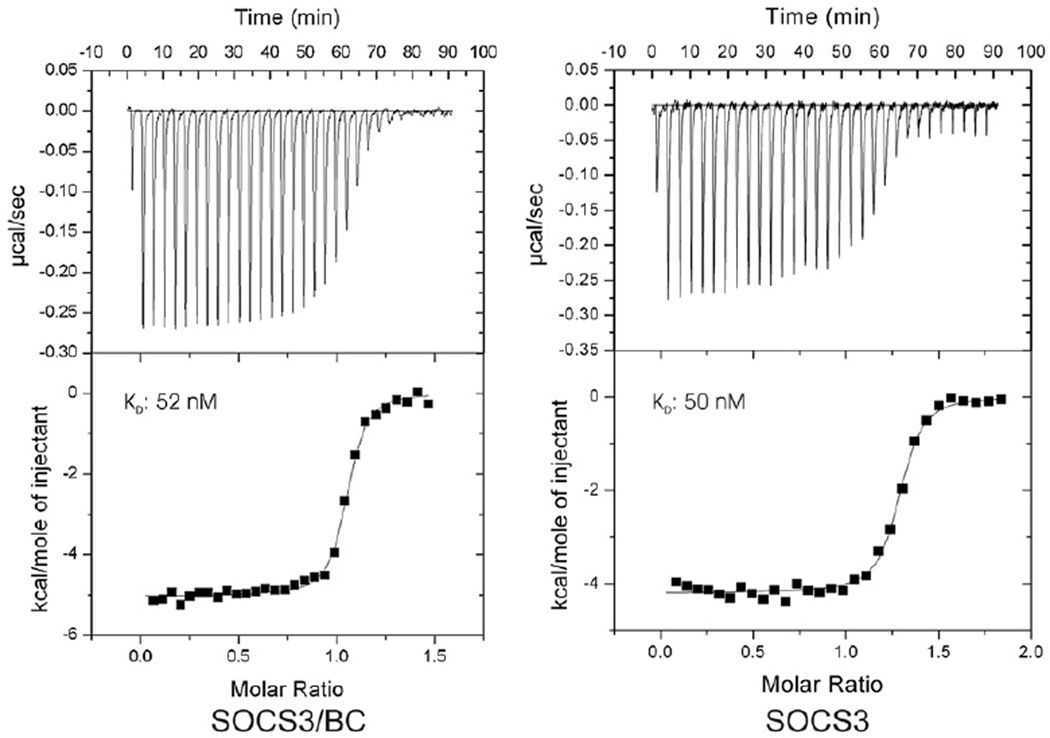
In the structures of both SOCS2/BC and SOCS4/BC, the SOCS box interacted with the SH2 domain. This led to the hypothesis that there may be “crosstalk” between the two domains, suggesting a general mechanism of SOCS activity in which the SH2 and SOCS box domains engage in cross-talk such that elonginBC binding can disrupt (or enhance) phos-photyrosine binding or vice versa. This may be especially relevant for CIS, where the presence of the SOCS box, and more specifically the C-terminal tyrosines, is necessary for receptor binding.28 In order to determine whether this is the case for SOCS3, we performed ITC experiments to measure the affinity of SOCS3 alone and SOCS3–elonginBC for a tyrosine-phosphorylated fragment of the gp130 receptor. SOCS3–elonginBC bound the phosphopeptide with a Kd of 50 nM (Fig. 7a), identical within experimental error to the affinity of SOCS3 alone (Fig. 7b). Gel filtration showed that phosphotyrosine binding did not result in dissociation of elonginBC. Thus, phosphotyrosine peptide binding by the SH2 domain of SOCS3 does not seem to be affected by the presence of elonginBC and vice versa.
Fig. 7.
Isothermal titration analyses of the SOCS3–gp130 interaction in the presence and absence of elonginBC. ITC analyses show that a ternary SOCS–elonginBC complex is able to bind a tyrosine-phosphorylated peptide from the gp130 receptor with similar affinity to SOCS3 alone. Phosphopeptide (160 µM) was titrated into 20 µM SOCS3/elonginBC (left) or 20 µM SOCS3 (right) in 30×10-µL injections using a VP-ITC unit (Microcal). Both protein and peptide were prepared in Tris-buffered saline supplemented with 2 mM 2-mercaptoethanol. The titration curves fitted well to a single-site model with a Kd of 50±4 nM, ΔH −5030±30 kcal/mol, N 1.03 (SOCS3/BC) and Kd of 52±7 nM, ΔH −4200±30 kcal/mol, N 1.26. SOCS3, both as an isolated protein and in complex with elonginBC, lacked the PEST motif.
Discussion
In-depth structural analysis of yeast elonginC had previously shown that in the absence of ligand, there was widespread conformational exchange.25,29 Our data show that this is also true for mammalian elongin BC. ElonginC is a member of the BTB-domain-containing family of proteins. BTB domains are exclusively protein–protein interaction domains that promote self- or hetero-association. ElonginC is distinct from the rest of this family in not containing an additional C-terminal α-helix. In other BTB-containing proteins, such as Skp and the Shaker channel, this α-helix occupies a position similar to that of the BC-box from the SOCS proteins with a similarly placed leucine. Thus, it is really the BC box–elonginC complex that adopts a BTB structure, whereas elonginC alone is missing a secondary structural element present in all other family members. The lack of this helix exposes a large hydrophobic surface on elonginC and may explain its inability to form a stable structure in the absence of bound ligand. Conformational exchange is also a feature of the SOCS box domain. Although the SOCS2/BC and SOCS4/BC27,30 crystal structures show that the SOCS box is well ordered when bound to elonginBC, we note there has been no structure of an SOCS box determined in the absence of elonginBC. Our NMR data show clearly that the isolated SOCS3 SOCS box peptide is disordered in solution and probably has similar disorder when part of the full-length protein (Fig. S4), but nevertheless binds tightly to elonginBC and becomes partially structured. Thus, we conclude that structural plasticity is a hallmark of the SOCS–elonginBC interaction. Advantages that have been proposed for unstructured or partially structured binding domains include that they create larger intermolecular interfaces,31 which can increase the speed of interaction with potential binding partners even in the absence of tight binding, and provide flexibility in binding diverse ligands32,33; which of these is most important for the disordered SOCS box remains to be determined. The structural disorder seen in the SOCS3 SOCS box may also enable it to be more easily phosphorylated, a required event for Nck and Crk-L association.34
A sequence comparison of the SOCS box, elonginA and the VHL α-domain had localised the elonginBC binding epitope to the N-terminal half of the SOCS box, while mutagenesis experiments had shown that the specificity for cullin5 was driven by the C-terminal region, in particular the LPXP motif.20 However, prior to this study, there had been no systematic analyses of the elonginBC binding motif by mutagenesis. Our structural and mutational analyses define the elonginBC binding epitope of the SOCS box as the first 12 residues of this domain. The first two residues had not previously been implicated as necessary for elonginBC binding20 but our results show that Val1 in particular could not be deleted or mutated to alanine without some loss of affinity for elonginBC. The interaction surface on elonginC for Val1 is identical to that seen for Ala1 and Pro1 in the SOCS2 and SOCS4 elonginBC complex structures, occupying one end of a continuous binding groove on elonginC that is markedly hydrophobic. As SOCS2 and CIS contain alanine at this position, we predict that they would bind elonginBC with lower affinity than do the rest of the family, which have a bulkier hydrophobic residue at this position. This remains to be determined as there are no quantitative affinity data in the literature regarding elonginBC binding. The most interesting feature of the alanine scanning mutagenesis results was the tolerance for mutation shown by the elonginBC binding motif. Only leucine 4 was absolutely required to maintain the SOCS3–elonginBC interaction. This leucine is the only conserved residue across all human SOCS proteins; indeed it is the only residue absolutely conserved across the wider SOCS-box-containing protein family.18 The elonginBC binding motif in SOCS3 is a simple one, a short amphipathic helix with an N-terminal extension, and the tolerance for mutation within this motif suggests there may be wider range of elonginBC binding proteins in the proteome than currently thought. Recent work by Mahrour et al.35 found that the spacing between the elonginBC and the cullin binding motifs can be highly variable and together with our results this suggests that there may be a number of hitherto unknown proteins that will interact with the elonginBC–cullin E3 ubiquitin ligases. Identification of leucine 4 as the key residue in the elonginBC binding motif should help in bioinformatics approaches to identify such proteins.
The cellular response to cytokine stimulation is temporally regulated with exquisite precision. The SOCS proteins play a key role in this regulation via mechanisms involving two protein domains, the SH2 domain/kinase inhibitory region and the SOCS box. The SH2 domain/kinase inhibitory region has the ability to inhibit the signalling cascade independently by either blocking STAT docking or directly inhibiting JAK kinase activity. It also confers substrate specificity. The SOCS box, acting as a general degradation signal, can take advantage of this specificity to target individual signalling intermediates for ubiquitination, achieved by interacting with subunits of an E3 ubiquitin ligase. Kamura et al. showed that the SOCS box of SOCS3 directs interaction specifically to elonginBC and cullin5.20 We wished to understand structurally how the proteins associate, determine the affinity of the interaction and show that it is direct and not mediated by other proteins—data that can only be obtained using in vitro studies with purified components. Taken together our data imply that the SOCS box domain acts as an independent binding unit as the isolated domain binds elonginBC and cullin5 with high affinity, binds cullin5 with identical affinity and enthalpy as when it is part of the full-length protein, and does not affect (and is not affected by) the presence of bound ligand on the upstream SH2 domain. We therefore conclude that it acts independently of the upstream SH2 and N-terminal domains.
The data presented here provide a basis for the development of SOCS box mimetics that could function to inhibit the activity of SOCS proteins in vivo. Inhibiting SOCS3 might be used clinically, for example, to prolong the action of G-CSF in the treatment of chemotherapy-induced neutropenia and the mobilisation of haematopoietic stem cells into the peripheral blood.36,37 Moreover, SOCS3 also acts as a critical inhibitor of insulin signalling. Thus, SOCS3 inhibitors might potentiate insulin signalling and could be used to treat insulin resistance and type 2 diabetes. An elonginC blocking compound would interfere with the ability of all SOCS proteins to form functional E3 ligases, and engineering specificity to block a single SOCS protein based on the SOCS box may therefore not be feasible. Nonetheless, an elonginBC binding compound would have wider potential than in cytokine signalling alone. For example, elonginC is known to be overexpressed in prostate cancer.38 In addition, it has been shown that the HIV protein Vif hijacks the elonginBC–cullin5 system in infected cells to effect the degradation of the antiviral cytidine deaminase APOBEC3G.39–42 Thus, an elonginC binding compound may be relevant in terms of anti-HIV therapy. Our NMR assignment of elonginBC, in complex with the SOCS box, now enables the use of “SAR by NMR”43 to screen for small-molecule inhibitors of elonginC binding and the better understanding of the structural aspects of this interaction will aid design of therapeutic agents to inhibit this association.
Materials and Methods
Cloning and expression
DNA encoding full-length murine SOCS3 or the SOCS3 SOCS box were cloned as GST fusion proteins in pGEX-4T (ElonginB) modified to contain an internal ribosome entry site preceding the elonginB. Both proteins cloned in this vector contain five vector-encoded residues after the thrombin cleavage site, SMARQ. These were co-transformed with pBB75(mouseElonginC17–112) into BL21(DE3) cells to produce the ternary SOCS–elonginB–elonginC complex. Cells were grown in the presence of ampicillin and kanamycin. Cells were harvested 8 h after IPTG induction by centrifugation at 6200g at 4 °C for 30 min. For 15N and 13C labelling, cells were grown to an OD600 of 0.6 in Neidhardt’s medium44 containing 1 g/L NH4Cl and/or 2 g/L 13C glucose as the sole nitrogen and carbon sources, respectively. SOCS3 lacking the PEST motif45 in complex with elonginBC was produced in the same manner using the same vector. ElonginBC alone was produced using pGEX-4T(elonginB) with elonginC (full-length or 17–112) cloned into site 1. The N-terminal domain of murine cullin5 (Met1-Leu385), was expressed as a GST fusion in pGEX-4T. The protein was expressed at room temperature with two point mutations introduced to allow for soluble expression: V341R and L345D.46 Cells were resuspended in phosphate-buffered saline and lysed by french press. The lysates were centrifuged at 20,000g and purified using glutathione–Sepharose chromatography. Thrombin cleavage was used to remove the GST followed by size-exclusion chromatography on a Superdex 75 or Superdex 200 16/60 column (GE Healthcare).
Analytical ultracentrifugation
Analytical ultracentrifugation experiments were carried out in an Optima XL-A analytical ultracentrifuge (Beckman-Coulter) equipped with an An-Ti60 rotor and doublesector 12-mm path-length cells containing quartz windows and charcoal-filled Epon centerpieces. Elongin BC mixtures were prepared at a concentration of 0.92 mg/mL in 20 mM Tris–HCl, pH 8.0,150 mM NaCl and 1 mM tris (2-carboxyethyl)phosphine. For sedimentation velocity measurements, samples of 400 µL were centrifuged at 20 °C for 10 h at 40,000 rpm. Radial absorbance scans were acquired at 236 nm at 6-min intervals over the range 5.3–7.3 cm using a radial resolution of 0.005 cm. Data were analysed using the continuous mass distribution model resident in the program SEDFIT. The analysis was performed using maximum entropy regularization. A confidence level (F ratio) of 0.95 was used with 200 molar mass increments. Time-independent and radial-independent noise were fitted and the position of the sample meniscus was fixed. For sedimentation equilibrium experiments, samples of 100 µL were centrifuged at 20 °C for 24 h at 20,000 rpm. Once the sample had reached equilibrium, radial absorbance scans were acquired at 273 nm over the range 6.8–7.3 cm using a radial resolution of 0.001 cm and 10 averages. The rotor speed was then increased to 35,000 rpm. Dual-speed sedimentation equilibrium data were globally analysed using the species analysis model resident in the program SEDPHAT.47 Data were analysed in terms of a single ideal species. For both sedimentation velocity and equilibrium experiments, data were analysed in terms of the buoyant rather than the physical molar mass, the reason being that the former allows the size and hence composition of the complex to be deduced without a priori knowledge of the stoichiometry. The requisite parameters for the conversion of buoyant molar mass to physical molar mass were computed using the program SEDNTERP.48
Alanine scanning, deletion mutagenesis and GST pull-down experiments
Alanine and deletion mutants of the SOCS3 SOCS box were constructed by ligation of overlapping oligonucleotides and cloned into pGEX-4TelonginB by standard ligation procedures. Following co-expression with pBB75elonginC, the cells were lysed and GST–Sepharose was used to pull down any proteins in the lysate associated with the GST–SOCS box fragment. Binding of ternary SOCS–elonginBC complexes to cullin5 was assayed by passing 10 mL of 0.1 mg/mL complex over 0.1 mg GST–cullin bound to 0.1 mL glutathione–Sepharose beads. The beads were washed with 50 column volumes of phosphate-buffered saline and then dissolved in SDS containing protein gel loading buffer and the results visualised by SDS-PAGE followed by Coomassie staining.
Isothermal titration calorimetry
Isothermal calorimetric titrations were performed with a Microcal omega VP-ITC (MicroCal Inc., Northampton, MA). SOCS3ΔPEST was dialysed against buffer (Tris-buffered saline, 5 mM 2-mercaptoethanol, pH 8.0) and the dialysis buffer was used to dissolve the tyrosine-phosphorylated gp130 peptide. Experiments were performed at 298 K. Solutions of 10 to 20 µM SOCS3 in the cell were titrated by injection of a total of 290 µL of 80–160 µM of gp130 peptide. The heat of dilution of the phosphopeptide into buffer was subtracted from the raw data of the binding experiment. The data were analysed using the evaluation software, Microcal Origin version 5.0. The binding curve fitted a single-site binding mode and Kd values were determined from experiments repeated twice.
NMR spectroscopy
Spectra were recorded at 310 K on Bruker Avance 500 (with cryoprobe), DRX-600 and Avance 800 spectrometers. Spectra were processed using XWINNMR or TOPSPIN (Bruker AG, Karlsruhe, Germany) and analysed using XEASY (version 1.3.13). Spectra were referenced to the water signal at 4.657 ppm (310 K). Sequence-specific resonance assignments for the backbone were accomplished using HNCA, HN(CO) CA, CBCA(CO)NH, HN(CA)CO and HNCO experiments, and side-chain assignments using NOESY–HSQC and HCCH–TOCSY. For structure calculations, distance restraints were obtained from 1H–15N and 1H–13C edited NOESY–HSQC experiments. Cα, Cβ, Hα, C′ and N chemical shifts were used in the program TALOS49 to obtain backbone psi/phi angles. The assignments have been deposited with the Biological Magnetic Resonance Bank (accession no. 15606).
Structure calculations
The solution structure of the mouse SOCS3 SOCS box in complex with elonginBC was calculated using a combined torsion angle and Cartesian coordinate dynamics protocol executed in X-PLOR-NIH. Structures were calculated from random starting coordinates using 1542 NOE distance restraints, including 242 medium-range (residue i to residue i+j, where 1 <j<4) and 307 long-range (residue i to residue i+j, where j>4) connectivities, 112 intermolecular restraints and 227 dihedral angle restraints, obtained using TALOS, composed of 113 φ and 114 ψ angles. The structures were displayed and analysed using PyMol,50 MolMol51 and PROCHECK-NMR.52 The final family comprised the 20 structures of lowest total energy (Table 1).
Table 1.
Summary of experimental restraints and structural statistics for murine SOCS3 SOCS box in complex with elonginBC
| No. of distance restraints | 1542 | |
| Intraresidue (i=j) | 436 | |
| Sequential (|i−j|=1) | 445 | |
| Medium range (1<|i−j|<4) | 242 | |
| Long range (|i−j|>4) | 307 | |
| Intermolecular | 112 | |
| Total dihedral angle restraints | 227 | |
| ϕ angles | 113 | |
| ψ angles | 114 | |
| Violations | ||
| NOE violations >0.4 Å | 2 | |
| Dihedral violations >5° | 2 | |
| Deviations from ideal geometrya | ||
| Bonds (Å) | 0.002549±0.00003 | |
| Angles (deg) | 0.583±0.003 | |
| Impropers (deg) | 0.409±0.003 | |
| Average RMSD to mean atomic coordinates (Å) | Ordered regionsb | Total |
| Backbone (N, Cα, C′) | 1.13 | |
| All heavy atoms | 1.68 | |
| Ramachandran plotc | ||
| Most favored (%) | 80 | 73.9 |
| Additionally allowed (%) | 13.5 | 17.7 |
| Generously allowed (%) | 4.7 | 6.1 |
| Disallowed (%) | 1.7 | 2.2 |
The values for the bonds, angles, and impropers show the deviations from ideal values based on perfect stereochemistry.
Ordered regions were considered as 1–15 (SOCS box), 1–82 (eloB), 17–44 and 59–112 (eloC).
As determined by the program PROCHECK-NMR for all residues except Gly and Pro.
Protein Data Bank accession code
The coordinates have been deposited with the RCSB Protein Data Bank (accession code 2JZ3).
Supplementary Material
Acknowledgements
We thank Tracy Willson for the gift of plasmids. This work was supported in part by the National Health and Medical Research Council (NHMRC), Australia (program grant 257500 and project grant 461260) and the National Institutes of Health (Bethesda, MD) grant CA22556. R.S.N. and N.A.N acknowledge fellowship support from the NHMRC.
Glossary
- ITC
isothermal titration calorimetry
- JAK
Janus kinase
- SOCS
suppressor of cytokine signalling
- STAT
signal transduction and activator of transcription
- CIS
cytokine-inducible SH2-containing protein
- VHL
von Hippel–Lindau
- G-CSF
granulocyte colony-stimulating factor
- GST
glutathione S-transferase
- HSQC
heteronuclear single quantum coherence
- TOCSY
total correlated spectroscopy
- NOESY
nuclear Overhauser enhancement spectroscopy
Footnotes
Supplementary Data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2008.06.038
References
- 1.Starr R, Willson TA, Viney EM, Murray LJ, Rayner JR, Jenkins BJ, et al. A family of cytokine-inducible inhibitors of signalling. Nature. 1997;387:917–921. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- 2.Leonard WJ, O’Shea JJ. Jaks and STATs: biological implications. Annu. Rev. Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 3.Kimura A, Kinjyo I, Matsumura Y, Mori H, Mashima R, Harada M, et al. SOCS3 is a physiological negative regulator for granulopoiesis and granulocyte colony-stimulating factor receptor signaling. J. Biol. Chem. 2004;279:6905–6910. doi: 10.1074/jbc.C300496200. [DOI] [PubMed] [Google Scholar]
- 4.Croker BA, Metcalf D, Robb L, Wei W, Mifsud S, DiRago L, et al. SOCS3 is a critical physiological negative regulator of G-CSF signaling and emergency granulopoiesis. Immunity. 2004;20:153–165. doi: 10.1016/s1074-7613(04)00022-6. [DOI] [PubMed] [Google Scholar]
- 5.Roberts AW, Robb L, Rakar S, Hartley L, Cluse L, Nicola NA, et al. Placental defects and embryonic lethality in mice lacking suppressor of cytokine signaling 3. Proc. Natl Acad. Sci. USA. 2001;98:9324–9329. doi: 10.1073/pnas.161271798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mori H, Hanada R, Hanada T, Aki D, Mashima R, Nishinakamura H, et al. Socs3 deficiency in the brain elevates leptin sensitivity and confers resistance to diet-induced obesity. Nat. Med. 2004;10:739–743. doi: 10.1038/nm1071. [DOI] [PubMed] [Google Scholar]
- 7.Jo D, Liu DY, Yao S, Collins RD, Hawiger J. Intracellular protein therapy with SOCS3 inhibits inflammation and apoptosis. Nat. Med. 2005;11:892–898. doi: 10.1038/nm1269. [DOI] [PubMed] [Google Scholar]
- 8.Sasaki A, Yasukawa H, Shouda T, Kitamura T, Dikic I, Yoshimura A. CIS3/SOCS-3 suppresses erythropoietin (EPO) signaling by binding the EPO receptor and JAK2. J. Biol. Chem. 2000;275:29338–29347. doi: 10.1074/jbc.M003456200. [DOI] [PubMed] [Google Scholar]
- 9.Nicholson SE, Willson TA, Farley A, Starr R, Zhang JG, Baca M, et al. Mutational analyses of the SOCS proteins suggest a dual domain requirement but distinct mechanisms for inhibition of LIF and IL-6 signal transduction. EMBO J. 1999;18:375–385. doi: 10.1093/emboj/18.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lehmann U, Schmitz J, Weissenbach M, Sobota RM, Hortner M, Friederichs K, et al. SHP2 and SOCS3 contribute to Tyr-759-dependent attenuation of interleukin-6 signaling through gp130. J. Biol. Chem. 2003;278:661–671. doi: 10.1074/jbc.M210552200. [DOI] [PubMed] [Google Scholar]
- 11.Sasaki A, Yasukawa H, Suzuki A, Kamizono S, Syoda T, Kinjyo I, et al. Cytokine-inducible SH2 protein-3 (CIS3/SOCS3) inhibits Janus tyrosine kinase by binding through the N-terminal kinase inhibitory region as well as SH2 domain. Genes Cells. 1999;4:339–351. doi: 10.1046/j.1365-2443.1999.00263.x. [DOI] [PubMed] [Google Scholar]
- 12.Hilton DJ. Negative regulators of cytokine signal transduction. Cell Mol. Life Sci. 1999;55:1568–1577. doi: 10.1007/s000180050396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasukawa H, Misawa H, Sakamoto H, Masuhara M, Sasaki A, Wakioka T, et al. The JAK-binding protein JAB inhibits Janus tyrosine kinase activity through binding in the activation loop. EMBO J. 1999;18:1309–1320. doi: 10.1093/emboj/18.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang JG, Farley A, Nicholson SE, Willson TA, Zugaro LM, Simpson RJ, et al. The conserved SOCS box motif in suppressors of cytokine signaling binds to elongins B and C and may couple bound proteins to proteasomal degradation. Proc. Natl Acad. Sci. USA. 1999;96:2071–2076. doi: 10.1073/pnas.96.5.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang JG, Metcalf D, Rakar S, Asimakis M, Greenhalgh CJ, Willson TA, et al. The SOCS box of suppressor of cytokine signaling-1 is important for inhibition of cytokine action in vivo. Proc. Natl Acad. Sci. USA. 2001;98:13261–13265. doi: 10.1073/pnas.231486498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van de Geijn GJ, Gits J, Touw IP. Distinct activities of suppressor of cytokine signaling (SOCS) proteins and involvement of the SOCS box in controlling G-CSF signaling. J. Leukoc. Biol. 2004;76:237–244. doi: 10.1189/jlb.0104041. [DOI] [PubMed] [Google Scholar]
- 17.Rui L, Yuan M, Frantz D, Shoelson S, White MF. SOCS-1 and SOCS-3 block insulin signaling by ubiquitin-mediated degradation of IRS1 and IRS2. J. Biol. Chem. 2002;277:42394–42398. doi: 10.1074/jbc.C200444200. [DOI] [PubMed] [Google Scholar]
- 18.Kile BT, Schulman BA, Alexander WS, Nicola NA, Martin HM, Hilton DJ. The SOCS box: a tale of destruction and degradation. Trends Biochem. Sci. 2002;27:235–241. doi: 10.1016/s0968-0004(02)02085-6. [DOI] [PubMed] [Google Scholar]
- 19.Kamizono S, Hanada T, Yasukawa H, Minoguchi S, Kato R, Minoguchi M, et al. The SOCS box of SOCS-1 accelerates ubiquitin-dependent proteolysis of TEL-JAK2. J. Biol. Chem. 2001;276:12530–12538. doi: 10.1074/jbc.M010074200. [DOI] [PubMed] [Google Scholar]
- 20.Kamura T, Maenaka K, Kotoshiba S, Matsumoto M, Kohda D, Conaway RC, et al. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 2004;18:3055–3065. doi: 10.1101/gad.1252404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willems AR, Schwab M, Tyers M. A hitchhiker’s guide to the cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta. 2004;1695:133–170. doi: 10.1016/j.bbamcr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 22.Boyle K, Egan P, Rakar S, Willson TA, Wicks IP, Metcalf D, et al. The SOCS box of suppressor of cytokine signaling-3 contributes to the control of G-CSF responsiveness in vivo. Blood. 2007;110:1466–1474. doi: 10.1182/blood-2007-03-079178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Babon JJ, McManus EJ, Yao S, DeSouza DP, Mielke LA, Sprigg NS, et al. The structure of SOCS3 reveals the basis of the extended SH2 domain function and identifies an unstructured insertion that regulates stability. Mol. Cell. 2006;22:205–216. doi: 10.1016/j.molcel.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 24.Bergamin E, Wu J, Hubbard SR. Structural basis for phosphotyrosine recognition by suppressor of cytokine signaling-3. Structure. 2006;14:1285–1292. doi: 10.1016/j.str.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 25.Buchberger A, Howard MJ, Freund SM, Proctor M, Butler PJ, Fersht AR, Bycroft M. Biophysical characterization of elongin C from Saccharomyces cerevisiae. Biochemistry. 2000;39:11137–11146. doi: 10.1021/bi0008231. [DOI] [PubMed] [Google Scholar]
- 26.Stebbins CE, Kaelin WG, Jr, Pavletich NP. Structure of the VHL–ElonginC–ElonginB complex: implications for VHL tumor suppressor function. Science. 1999;284:455–461. doi: 10.1126/science.284.5413.455. [DOI] [PubMed] [Google Scholar]
- 27.Bullock AN, Debreczeni JE, Edwards AM, Sundstrom M, Knapp S. Crystal structure of the SOCS2–elongin C–elongin B complex defines a prototypical SOCS box ubiquitin ligase. Proc. Natl Acad. Sci. USA. 2006;103:7637–7642. doi: 10.1073/pnas.0601638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lavens D, Ulrichts P, Catteeuw D, Gevaert K, Vandekerckhove J, Peelman F, et al. The C-terminus of CIS defines its interaction pattern. Biochem. J. 2007;401:257–267. doi: 10.1042/BJ20060242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Botuyan MV, Koth CM, Mer G, Chakrabartty A, Conaway JW, Conaway RC, et al. Binding of elongin A or a von Hippel–Lindau peptide stabilizes the structure of yeast elongin C. Proc. Natl Acad. Sci. USA. 1999;96:9033–9038. doi: 10.1073/pnas.96.16.9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bullock AN, Rodriguez MC, Debreczeni JE, Songyang Z, Knapp S. Structure of the SOCS4–elonginB/C complex reveals a distinct SOCS box interface and the molecular basis for SOCS-dependent EGFR degradation. Structure. 2007;15:1493–1504. doi: 10.1016/j.str.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gunasekaran K, Tsai CJ, Kumar S, Zanuy D, Nussinov R. Extended disordered proteins: targeting function with less scaffold. Trends Biochem. Sci. 2003;28:81–85. doi: 10.1016/S0968-0004(03)00003-3. [DOI] [PubMed] [Google Scholar]
- 32.Tompa P, Szasz C, Buday L. Structural disorder throws new light on moonlighting. Trends Biochem. Sci. 2005;30:484–489. doi: 10.1016/j.tibs.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 33.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell. Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 34.Sitko JC, Guevara CI, Cacalano NA. Tyrosine-phosphorylated SOCS3 interacts with the Nck and Crk-L adapter proteins and regulates Nck activation. J. Biol. Chem. 2004;279:37662–37669. doi: 10.1074/jbc.M404007200. [DOI] [PubMed] [Google Scholar]
- 35.Mahrour N, Redwine WB, Florens L, Swanson SK, Martin-Brown S, Bradford WD, et al. Characterization of Cullin-box sequences that direct recruitment of Cul2-Rbx1 and Cul5-Rbx2 modules to Elongin BC-based ubiquitin ligases. J. Biol. Chem. 2008;283:8005–8013. doi: 10.1074/jbc.M706987200. [DOI] [PubMed] [Google Scholar]
- 36.Duhrsen U, Villeval JL, Boyd J, Kannourakis G, Morstyn G, Metcalf D. Effects of recombinant human granulocyte colony-stimulating factor on hematopoietic progenitor cells in cancer patients. Blood. 1988;72:2074–2081. [PubMed] [Google Scholar]
- 37.Morstyn G, Campbell L, Duhrsen U, Souza LM, Alton NK, Villeval JL, et al. Clinical studies with granulocyte colony stimulating factor (G-CSF) in patients receiving cytotoxic chemotherapy. Behring Inst. Mitt. 1988:234–239. [PubMed] [Google Scholar]
- 38.Visakorpi T. The molecular genetics of prostate cancer. Urology. 2003;62:3–10. doi: 10.1016/s0090-4295(03)00776-3. [DOI] [PubMed] [Google Scholar]
- 39.Yu Y, Xiao Z, Ehrlich ES, Yu X, Yu XF. Selective assembly of HIV-1 Vif–Cul5–ElonginB–ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 2004;18:2867–2872. doi: 10.1101/gad.1250204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shirakawa K, Takaori-Kondo A, Kobayashi M, Tomonaga M, Izumi T, Fukunaga K, et al. Ubiquitination of APOBEC3 proteins by the Vif–Cullin5–ElonginB–ElonginC complex. Virology. 2006;344:263–266. doi: 10.1016/j.virol.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 41.Kobayashi M, Takaori-Kondo A, Miyauchi Y, Iwai K, Uchiyama T. Ubiquitination of APO-BEC3G by an HIV-1 Vif–Cullin5–Elongin B–Elongin C complex is essential for Vif function. J. Biol. Chem. 2005;280:18573–18578. doi: 10.1074/jbc.C500082200. [DOI] [PubMed] [Google Scholar]
- 42.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif–Cul5–SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 43.Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 44.Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J. Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Babon JJ, Yao S, DeSouza DP, Harrison CF, Fabri LJ, Liepinsh E, et al. Secondary structure assignment of mouse SOCS3 by NMR defines the domain boundaries and identifies an unstructured insertion in the SH2 domain. FEBS J. 2005;272:6120–6130. doi: 10.1111/j.1742-4658.2005.05010.x. [DOI] [PubMed] [Google Scholar]
- 46.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, et al. Structure of the Cul1–Rbx1–Skp1–F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 47.Vistica J, Dam J, Balbo A, Yikilmaz E, Mariuzza RA, Rouault TA, Schuck P. Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal. Biochem. 2004;326:234–256. doi: 10.1016/j.ab.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 48.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding SE, Rowe AJ, Horton JC, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Cambridge, UK: The Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- 49.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 50.DeLano WL. Use of PYMOL as a communications tool for molecular science. Abstr. Pap.-Am. Chem. Soc. 2004;228:U313–U314. [Google Scholar]
- 51.Koradi R, Billeter M, Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics. 1996;14:51. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 52.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.