Abstract
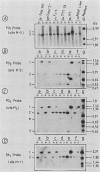
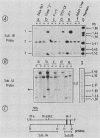
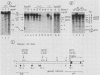
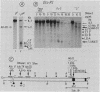
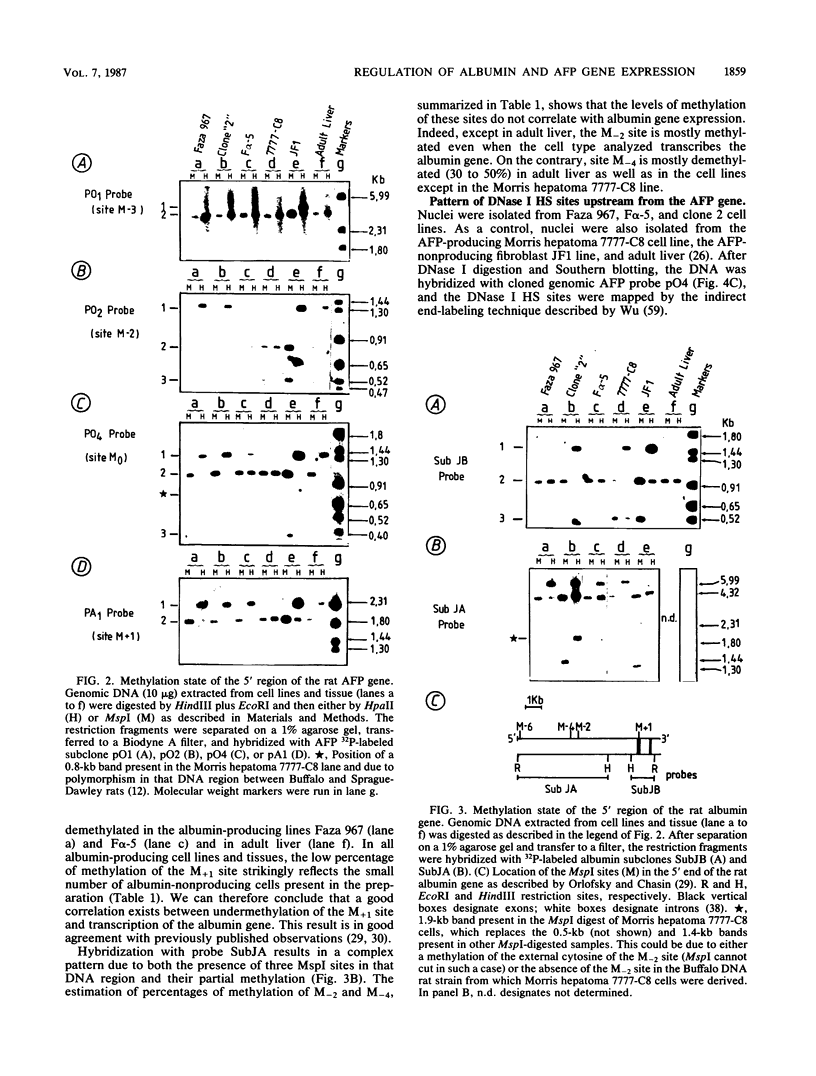
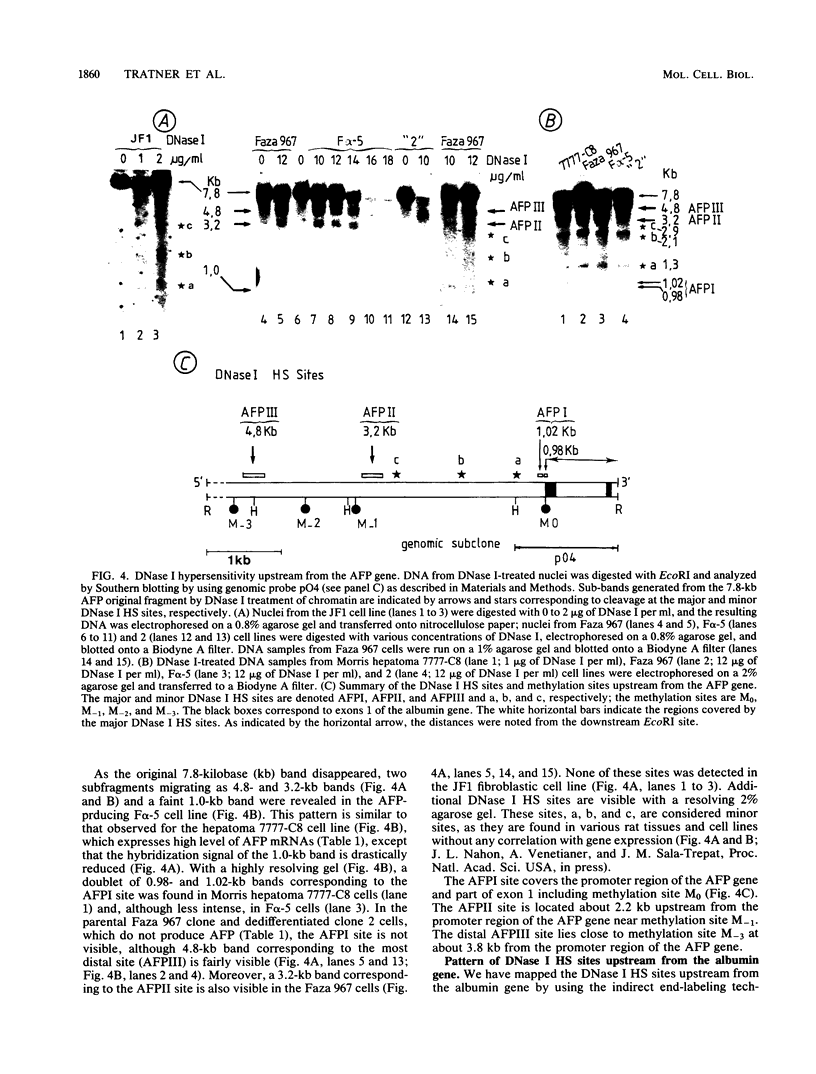
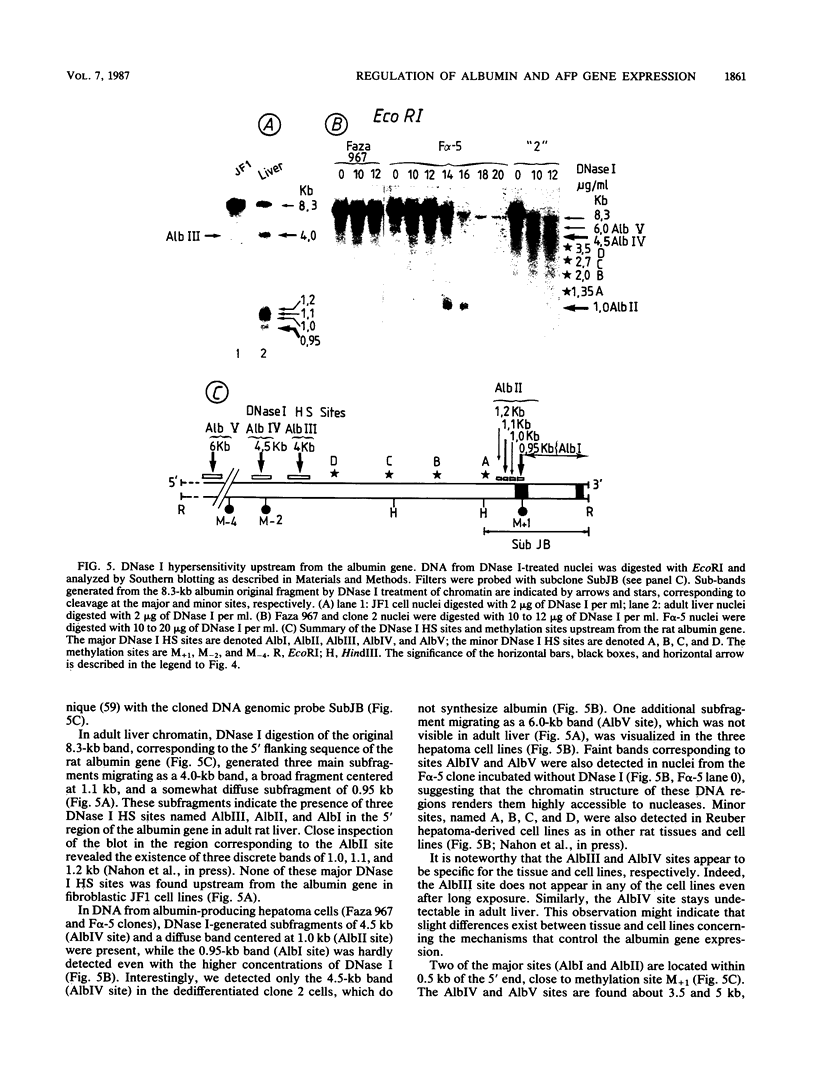
We examined DNA methylation and DNase I hypersensitivity of the alpha-fetoprotein (AFP) and albumin gene region in hepatoma cell lines which showed drastic differences in the level of expression of these genes. We assayed for methylation of the CCGG sequences by using the restriction enzyme isoschizomers HpaII and MspI. We found two methylation sites located in the 5' region of the AFP gene and one in exon 1 of the albumin gene for which hypomethylation is correlated with gene expression. Another such site, located about 4,000 base pairs upstream from the AFP gene, seems to be correlated with the tissue specificity of the cells. DNase I-hypersensitive sites were mapped by using the indirect end-labeling technique with cloned genomic DNA probes. Three tissue-specific DNase I-hypersensitive sites were mapped in the 5' flanking region of the AFP gene when this gene was transcribed. Similarly, three tissue-specific DNase I-hypersensitive sites were detected upstream from the albumin gene in producing cell lines. In both cases, the most distal sites were maintained after cessation of gene activity and appear to be correlated with the potential expression of the gene. Interestingly, specific methylation sites are localized in the same DNA region as DNase I hypersensitive sites. This suggests that specific alterations of chromatin structure and changes in methylation pattern occur in specific critical regulatory regions upstream from the albumin and AFP genes in rat hepatoma cell lines.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abelev G. I. Alpha-fetoprotein in ontogenesis and its association with malignant tumors. Adv Cancer Res. 1971;14:295–358. doi: 10.1016/s0065-230x(08)60523-0. [DOI] [PubMed] [Google Scholar]
- Anderson J. N., Vanderbilt J. N., Lawson G. M., Tsai M. J., O'Malley B. W. Chromatin structure of the ovalbumin gene family in the chicken oviduct. Biochemistry. 1983 Jan 4;22(1):21–30. doi: 10.1021/bi00270a004. [DOI] [PubMed] [Google Scholar]
- Bernuau D., Poliard A., Tournier I., Sala-Trepat J., Feldmann G. All hepatocytes are involved in the expression of the albumin gene in the normal adult rat: a demonstration by in situ hybridization and immunoperoxidase techniques. Cell Biol Int Rep. 1985 Jan;9(1):31–42. doi: 10.1016/0309-1651(85)90139-0. [DOI] [PubMed] [Google Scholar]
- Blin N., Stafford D. W. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976 Sep;3(9):2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busslinger M., Hurst J., Flavell R. A. DNA methylation and the regulation of globin gene expression. Cell. 1983 Aug;34(1):197–206. doi: 10.1016/0092-8674(83)90150-2. [DOI] [PubMed] [Google Scholar]
- Bélanger L., Frain M., Baril P., Gingras M. C., Bartkowiak J., Sala-Trepat J. M. Glucocorticosteroid suppression of alpha1-fetoprotein synthesis in developing rat liver. Evidence for selective gene repression at the transcriptional level. Biochemistry. 1981 Nov 10;20(23):6665–6672. doi: 10.1021/bi00526a022. [DOI] [PubMed] [Google Scholar]
- Chiu J. F., Massari R. J., Schwartz C. E., Meisler N. T., Thanassi J. W. Hormonal modulation of alpha-fetoprotein gene expression in newborn rat livers. Nucleic Acids Res. 1981 Dec 21;9(24):6917–6933. doi: 10.1093/nar/9.24.6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschatrette J., Weiss M. C. Characterization of differentiated and dedifferentiated clones from a rat hepatoma. Biochimie. 1974;56(11-12):1603–1611. doi: 10.1016/s0300-9084(75)80286-0. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Control of eukaryotic messenger RNA synthesis by sequence-specific DNA-binding proteins. 1985 Aug 29-Sep 4Nature. 316(6031):774–778. doi: 10.1038/316774a0. [DOI] [PubMed] [Google Scholar]
- Elgin S. C. DNAase I-hypersensitive sites of chromatin. Cell. 1981 Dec;27(3 Pt 2):413–415. doi: 10.1016/0092-8674(81)90381-0. [DOI] [PubMed] [Google Scholar]
- Gal A., Nahon J. L., Gomez-Garcia M., Tratner I., Sala-Trepat J. M. Organization of the albumin and alpha-fetoprotein genes in fetal and adult rat tissues, and rat hepatomas. Differentiation. 1985;29(3):238–242. doi: 10.1111/j.1432-0436.1985.tb00322.x. [DOI] [PubMed] [Google Scholar]
- Gal A., Nahon J. L., Lucotte G., Sala-Trepat J. M. Structural variants of the alpha-fetoprotein gene in different inbred strains of rat. Mol Gen Genet. 1984;195(1-2):153–158. doi: 10.1007/BF00332738. [DOI] [PubMed] [Google Scholar]
- Gerber-Huber S., May F. E., Westley B. R., Felber B. K., Hosbach H. A., Andres A. C., Ryffel G. U. In contrast to other Xenopus genes the estrogen-inducible vitellogenin genes are expressed when totally methylated. Cell. 1983 May;33(1):43–51. doi: 10.1016/0092-8674(83)90333-1. [DOI] [PubMed] [Google Scholar]
- Godbout R., Ingram R., Tilghman S. M. Multiple regulatory elements in the intergenic region between the alpha-fetoprotein and albumin genes. Mol Cell Biol. 1986 Feb;6(2):477–487. doi: 10.1128/mcb.6.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groudine M., Eisenman R., Weintraub H. Chromatin structure of endogenous retroviral genes and activation by an inhibitor of DNA methylation. Nature. 1981 Jul 23;292(5821):311–317. doi: 10.1038/292311a0. [DOI] [PubMed] [Google Scholar]
- Keshet I., Lieman-Hurwitz J., Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986 Feb 28;44(4):535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- Keshet I., Yisraeli J., Cedar H. Effect of regional DNA methylation on gene expression. Proc Natl Acad Sci U S A. 1985 May;82(9):2560–2564. doi: 10.1073/pnas.82.9.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumlauf R., Hammer R. E., Tilghman S. M., Brinster R. L. Developmental regulation of alpha-fetoprotein genes in transgenic mice. Mol Cell Biol. 1985 Jul;5(7):1639–1648. doi: 10.1128/mcb.5.7.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunnath L., Locker J. Developmental changes in the methylation of the rat albumin and alpha-fetoprotein genes. EMBO J. 1983;2(3):317–324. doi: 10.1002/j.1460-2075.1983.tb01425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M. T., Iyer B., Wu J. R., Lapeyre J. N., Becker F. F. Methylation of the alpha-fetoprotein gene in productive and nonproductive rat hepatocellular carcinomas. Cancer Res. 1984 Apr;44(4):1642–1647. [PubMed] [Google Scholar]
- Lucotte G., Gal A., Nahon J. L., Sala-Trepat J. M. Eco RI restriction-site polymorphism of the albumin gene in different inbred strains of rat. Biochem Genet. 1982 Dec;20(11-12):1105–1115. doi: 10.1007/BF00498935. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Ginder G. D. Specific DNA methylation sites in the vicinity of the chicken beta-globin genes. Nature. 1979 Aug 2;280(5721):419–420. doi: 10.1038/280419a0. [DOI] [PubMed] [Google Scholar]
- McKeon C., Ohkubo H., Pastan I., de Crombrugghe B. Unusual methylation pattern of the alpha 2 (l) collagen gene. Cell. 1982 May;29(1):203–210. doi: 10.1016/0092-8674(82)90104-0. [DOI] [PubMed] [Google Scholar]
- Muglia L., Rothman-Denes L. B. Cell type-specific negative regulatory element in the control region of the rat alpha-fetoprotein gene. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7653–7657. doi: 10.1073/pnas.83.20.7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahon J. L., Gal A., Erdos T., Sala-Trepat J. M. Differential DNase I sensitivity of the albumin and alpha-fetoprotein genes in chromatin from rat tissues and cell lines. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5031–5035. doi: 10.1073/pnas.81.16.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi H., Taketa K., Yamane T., Oda M., Sato J. Hormonal control of alpha-fetoprotein secretion in human hepatoma cell lines proliferating in chemically defined medium. Cancer Res. 1985 Dec;45(12 Pt 1):6379–6383. [PubMed] [Google Scholar]
- Nelson C., Crenshaw E. B., 3rd, Franco R., Lira S. A., Albert V. R., Evans R. M., Rosenfeld M. G. Discrete cis-active genomic sequences dictate the pituitary cell type-specific expression of rat prolactin and growth hormone genes. Nature. 1986 Aug 7;322(6079):557–562. doi: 10.1038/322557a0. [DOI] [PubMed] [Google Scholar]
- Orlofsky A., Chasin L. A. A domain of methylation change at the albumin locus in rat hepatoma cell variants. Mol Cell Biol. 1985 Jan;5(1):214–225. doi: 10.1128/mcb.5.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M. O., Sperling L., Cassio D., Levilliers J., Sala-Trepat J., Weiss M. C. Undermethylation at the 5' end of the albumin gene is necessary but not sufficient for albumin production by rat hepatoma cells in culture. Cell. 1982 Oct;30(3):825–833. doi: 10.1016/0092-8674(82)90287-2. [DOI] [PubMed] [Google Scholar]
- Ott M. O., Sperling L., Herbomel P., Yaniv M., Weiss M. C. Tissue-specific expression is conferred by a sequence from the 5' end of the rat albumin gene. EMBO J. 1984 Nov;3(11):2505–2510. doi: 10.1002/j.1460-2075.1984.tb02164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Szyf M. DNA methylation patterns. Formation and function. Biochim Biophys Acta. 1984 Sep 10;782(4):331–342. doi: 10.1016/0167-4781(84)90043-5. [DOI] [PubMed] [Google Scholar]
- Reeves R. Transcriptionally active chromatin. Biochim Biophys Acta. 1984 Sep 10;782(4):343–393. doi: 10.1016/0167-4781(84)90044-7. [DOI] [PubMed] [Google Scholar]
- Sargent T. D., Wu J. R., Sala-Trepat J. M., Wallace R. B., Reyes A. A., Bonner J. The rat serum albumin gene: analysis of cloned sequences. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3256–3260. doi: 10.1073/pnas.76.7.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent T. D., Yang M., Bonner J. Nucleotide sequence of cloned rat serum albumin messenger RNA. Proc Natl Acad Sci U S A. 1981 Jan;78(1):243–246. doi: 10.1073/pnas.78.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffery M., Rifkind R. A., Marks P. A. Murine erythroleukemia cell differentiation: DNase I hypersensitivity and DNA methylation near the globin genes. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1180–1184. doi: 10.1073/pnas.79.4.1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenlist U., Durand D. B., Bressler P., Holbrook N. J., Norris C. A., Kamoun M., Kant J. A., Crabtree G. R. Promoter region of interleukin-2 gene undergoes chromatin structure changes and confers inducibility on chloramphenicol acetyltransferase gene during activation of T cells. Mol Cell Biol. 1986 Sep;6(9):3042–3049. doi: 10.1128/mcb.6.9.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein R., Razin A., Cedar H. In vitro methylation of the hamster adenine phosphoribosyltransferase gene inhibits its expression in mouse L cells. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3418–3422. doi: 10.1073/pnas.79.11.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein R., Sciaky-Gallili N., Razin A., Cedar H. Pattern of methylation of two genes coding for housekeeping functions. Proc Natl Acad Sci U S A. 1983 May;80(9):2422–2426. doi: 10.1073/pnas.80.9.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumph W. E., Baez M., Beattie W. G., Tsai M. J., O'Malley B. W. Characterization of deoxyribonucleic acid sequences at the 5' and 3' borders of the 100 kilobase pair ovalbumin gene domain. Biochemistry. 1983 Jan 18;22(2):306–315. doi: 10.1021/bi00271a012. [DOI] [PubMed] [Google Scholar]
- Szpirer J., Szpirer C. The control of serum protein synthesis in hepatoma-fibroblast hybrids. Cell. 1975 Sep;6(1):53–60. doi: 10.1016/0092-8674(75)90073-2. [DOI] [PubMed] [Google Scholar]
- Theisen M., Stief A., Sippel A. E. The lysozyme enhancer: cell-specific activation of the chicken lysozyme gene by a far-upstream DNA element. EMBO J. 1986 Apr;5(4):719–724. doi: 10.1002/j.1460-2075.1986.tb04273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedel M., Gomez-Garcia M., Sala M., Sala-Trepat J. M. Changes in methylation pattern of albumin and alpha-fetoprotein genes in developing rat liver and neoplasia. Nucleic Acids Res. 1983 Jul 11;11(13):4335–4354. doi: 10.1093/nar/11.13.4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venetianer A., Bösze Z. Expression of differentiated functions in dexamethasone-resistant hepatoma cells. Differentiation. 1983;25(1):70–78. doi: 10.1111/j.1432-0436.1984.tb01340.x. [DOI] [PubMed] [Google Scholar]
- Venetianer A., Pintér Z., Gál A. Examination of glucocorticoid sensitivity and receptor content of hepatoma cell lines. Cytogenet Cell Genet. 1980;28(4):280–283. doi: 10.1159/000131541. [DOI] [PubMed] [Google Scholar]
- Venetianer A., Poliard A., Poiret M., Erdös T., Hermesz E., Sala-Trepat J. M. Activation of alpha-fetoprotein synthesis in rat hepatoma cells with reduced sensitivity to dexamethasone. Differentiation. 1986;32(2):148–156. doi: 10.1111/j.1432-0436.1986.tb00567.x. [DOI] [PubMed] [Google Scholar]
- Venetianer A., Schiller D. L., Magin T., Franke W. W. Cessation of cytokeratin expression in a rat hepatoma cell line lacking differentiated functions. Nature. 1983 Oct 20;305(5936):730–733. doi: 10.1038/305730a0. [DOI] [PubMed] [Google Scholar]
- Walker M. D., Edlund T., Boulet A. M., Rutter W. J. Cell-specific expression controlled by the 5'-flanking region of insulin and chymotrypsin genes. Nature. 1983 Dec 8;306(5943):557–561. doi: 10.1038/306557a0. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Larsen A., Groudine M. Alpha-Globin-gene switching during the development of chicken embryos: expression and chromosome structure. Cell. 1981 May;24(2):333–344. doi: 10.1016/0092-8674(81)90323-8. [DOI] [PubMed] [Google Scholar]
- Widen S. G., Papaconstantinou J. Liver-specific expression of the mouse alpha-fetoprotein gene is mediated by cis-acting DNA elements. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8196–8200. doi: 10.1073/pnas.83.21.8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilks A. F., Cozens P. J., Mattaj I. W., Jost J. P. Estrogen induces a demethylation at the 5' end region of the chicken vitellogenin gene. Proc Natl Acad Sci U S A. 1982 Jul;79(14):4252–4255. doi: 10.1073/pnas.79.14.4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilks A., Seldran M., Jost J. P. An estrogen-dependent demethylation at the 5' end of the chicken vitellogenin gene is independent of DNA synthesis. Nucleic Acids Res. 1984 Jan 25;12(2):1163–1177. doi: 10.1093/nar/12.2.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W. I., Felsenfeld G. Chromatin structure of the chicken beta-globin gene region. Sensitivity to DNase I, micrococcal nuclease, and DNase II. J Biol Chem. 1982 Jul 10;257(13):7730–7736. [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Yaniv M. Regulation of eukaryotic gene expression by transactivating proteins and cis acting DNA elements. Biol Cell. 1984;50(3):203–216. doi: 10.1111/j.1768-322x.1984.tb00268.x. [DOI] [PubMed] [Google Scholar]
- van der Ploeg L. H., Flavell R. A. DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroid tissues. Cell. 1980 Apr;19(4):947–958. doi: 10.1016/0092-8674(80)90086-0. [DOI] [PubMed] [Google Scholar]