Abstract
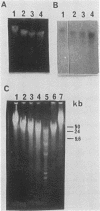
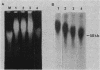
The active beta-globin genes in chicken erythrocytes, like all active genes, reside in large chromatin domains which are preferentially sensitive to digestion by DNase I. We have recently proposed that the special structure of chromatin in active domains is maintained by torsional stress in the DNA (Villeponteau et al., Cell 39:469-478, 1984). This hypothesis predicts that nicking of the DNA within any such chromosomal domain in vivo will relax the DNA and lead to loss of the special DNase I-sensitive state. Here we have tested this prediction by using gamma irradiation and bleomycin treatment to cleave DNA within intact chicken embryo erythrocytes. Both treatments cause reversal of DNase I sensitivity. Moreover, reversal occurs at approximately one nick per 150 kilobase pairs for both agents despite their entirely unrelated modes of cell penetration and DNA attack. These results suggest that the domain of DNase I sensitivity surrounding the beta-globin genes comprises 150 kilobase pairs of chromatin under torsional stress and that a single DNA nick in this region is sufficient to reverse the DNase I sensitivity throughout the entire domain.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alevy M. C., Tsai M. J., O'Malley B. W. DNase I sensitive domain of the gene coding for the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase. Biochemistry. 1984 May 8;23(10):2309–2314. doi: 10.1021/bi00305a034. [DOI] [PubMed] [Google Scholar]
- Bakayev V. V., Yugai A. A., Luchnik A. N. Effect of X-ray induced DNA damage on DNAase I hypersensitivity of SV40 chromatin: relation to elastic torsional strain in DNA. Nucleic Acids Res. 1985 Oct 11;13(19):7079–7093. doi: 10.1093/nar/13.19.7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsoum J., Berg P. Simian virus 40 minichromosomes contain torsionally strained DNA molecules. Mol Cell Biol. 1985 Nov;5(11):3048–3057. doi: 10.1128/mcb.5.11.3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin R. C., Gill D. M. ADP-ribosylation in mammalian cell ghosts. Dependence of poly(ADP-ribose) synthesis on strand breakage in DNA. J Biol Chem. 1980 Nov 10;255(21):10493–10501. [PubMed] [Google Scholar]
- Benyajati C., Worcel A. Isolation, characterization, and structure of the folded interphase genome of Drosophila melanogaster. Cell. 1976 Nov;9(3):393–407. doi: 10.1016/0092-8674(76)90084-2. [DOI] [PubMed] [Google Scholar]
- Berry D. E., Kilkuskie R. E., Hecht S. M. DNA damage induced by bleomycin in the presence of dibucaine is not predictive of cell growth inhibition. Biochemistry. 1985 Jun 18;24(13):3214–3219. doi: 10.1021/bi00334a021. [DOI] [PubMed] [Google Scholar]
- Certa U., Colavito-Shepanski M., Grunstein M. Yeast may not contain histone H1: the only known 'histone H1-like' protein in Saccharomyces cerevisiae is a mitochondrial protein. Nucleic Acids Res. 1984 Nov 12;12(21):7975–7985. doi: 10.1093/nar/12.21.7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook P. R., Brazell I. A. Supercoils in human DNA. J Cell Sci. 1975 Nov;19(2):261–279. doi: 10.1242/jcs.19.2.261. [DOI] [PubMed] [Google Scholar]
- Cotten M., Bresnahan D., Thompson S., Sealy L., Chalkley R. Novobiocin precipitates histones at concentrations normally used to inhibit eukaryotic type II topoisomerase. Nucleic Acids Res. 1986 May 12;14(9):3671–3686. doi: 10.1093/nar/14.9.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R. Structural specificities of five commonly used DNA nucleases. J Mol Biol. 1984 Jul 15;176(4):535–557. doi: 10.1016/0022-2836(84)90176-1. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA structural variations in the E. coli tyrT promoter. Cell. 1984 Jun;37(2):491–502. doi: 10.1016/0092-8674(84)90379-9. [DOI] [PubMed] [Google Scholar]
- Fox K. R., Waring M. J. DNA structural variations produced by actinomycin and distamycin as revealed by DNAase I footprinting. Nucleic Acids Res. 1984 Dec 21;12(24):9271–9285. doi: 10.1093/nar/12.24.9271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher M., Weinberg R., Simpson M. V. Effect of the bacterial DNA gyrase inhibitors, novobiocin, nalidixic acid, and oxolinic acid, on oxidative phosphorylation. J Biol Chem. 1986 Jul 5;261(19):8604–8607. [PubMed] [Google Scholar]
- Gazit B., Cedar H., Lerer I., Voss R. Active genes are sensitive to deoxyribonuclease I during metaphase. Science. 1982 Aug 13;217(4560):648–650. doi: 10.1126/science.6283640. [DOI] [PubMed] [Google Scholar]
- Glikin G. C., Ruberti I., Worcel A. Chromatin assembly in Xenopus oocytes: in vitro studies. Cell. 1984 May;37(1):33–41. doi: 10.1016/0092-8674(84)90298-8. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J. M. Novobiocin inhibits RNA polymerase III transcription in vitro by a mechanism distinct from DNA topoisomerase II. Nucleic Acids Res. 1986 Mar 11;14(5):2075–2088. doi: 10.1093/nar/14.5.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groudine M., Peretz M., Weintraub H. Transcriptional regulation of hemoglobin switching in chicken embryos. Mol Cell Biol. 1981 Mar;1(3):281–288. doi: 10.1128/mcb.1.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson F. Chemical changes induced in DNA by ionizing radiation. Prog Nucleic Acid Res Mol Biol. 1985;32:115–154. doi: 10.1016/s0079-6603(08)60347-5. [DOI] [PubMed] [Google Scholar]
- Jantzen K., Fritton H. P., Igo-Kemenes T. The DNase I sensitive domain of the chicken lysozyme gene spans 24 kb. Nucleic Acids Res. 1986 Aug 11;14(15):6085–6099. doi: 10.1093/nar/14.15.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiec E. B., Ryoji M., Worcel A. Gyration is required for 5S RNA transcription from a chromatin template. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1305–1309. doi: 10.1073/pnas.83.5.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiec E. B., Worcel A. The positive transcription factor of the 5S RNA gene induces a 5S DNA-specific gyration in Xenopus oocyte extracts. Cell. 1985 Jul;41(3):945–953. doi: 10.1016/s0092-8674(85)80075-1. [DOI] [PubMed] [Google Scholar]
- Kuo M. T. Preferential damage of active chromatin by bleomycin. Cancer Res. 1981 Jun;41(6):2439–2443. [PubMed] [Google Scholar]
- Landes G. M., Villeponteau B., Pribyl T. M., Martinson H. G. Hemoglobin switching in chickens. Is the switch initiated post-transcriptionally? J Biol Chem. 1982 Sep 25;257(18):11008–11014. [PubMed] [Google Scholar]
- Lawson G. M., Knoll B. J., March C. J., Woo S. L., Tsai M. J., O'Malley B. W. Definition of 5' and 3' structural boundaries of the chromatin domain containing the ovalbumin multigene family. J Biol Chem. 1982 Feb 10;257(3):1501–1507. [PubMed] [Google Scholar]
- Lohr D., Hereford L. Yeast chromatin is uniformly digested by DNase-I. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4285–4288. doi: 10.1073/pnas.76.9.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low C. M., Drew H. R., Waring M. J. Sequence-specific binding of echinomycin to DNA: evidence for conformational changes affecting flanking sequences. Nucleic Acids Res. 1984 Jun 25;12(12):4865–4879. doi: 10.1093/nar/12.12.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchnik A. N., Bakayev V. V., Zbarsky I. B., Georgiev G. P. Elastic torsional strain in DNA within a fraction of SV40 minichromosomes: relation to transcriptionally active chromatin. EMBO J. 1982;1(11):1353–1358. doi: 10.1002/j.1460-2075.1982.tb01322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydersen B. K., Pettijohn D. E. Interactions stabilizing DNA tertiary structure in the Escherichia coli chromosome investigated with ionizing radiation. Chromosoma. 1977 Jul 8;62(3):199–215. doi: 10.1007/BF00286044. [DOI] [PubMed] [Google Scholar]
- Reeves R. Transcriptionally active chromatin. Biochim Biophys Acta. 1984 Sep 10;782(4):343–393. doi: 10.1016/0167-4781(84)90044-7. [DOI] [PubMed] [Google Scholar]
- Richmond T. J., Finch J. T., Rushton B., Rhodes D., Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984 Oct 11;311(5986):532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- Ryoji M., Worcel A. Chromatin assembly in Xenopus oocytes: in vivo studies. Cell. 1984 May;37(1):21–32. doi: 10.1016/0092-8674(84)90297-6. [DOI] [PubMed] [Google Scholar]
- Saavedra R. A., Huberman J. A. Both DNA topoisomerases I and II relax 2 micron plasmid DNA in living yeast cells. Cell. 1986 Apr 11;45(1):65–70. doi: 10.1016/0092-8674(86)90538-6. [DOI] [PubMed] [Google Scholar]
- Sealy L., Cotten M., Chalkley R. Novobiocin inhibits passive chromatin assembly in vitro. EMBO J. 1986 Dec 1;5(12):3305–3311. doi: 10.1002/j.1460-2075.1986.tb04644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden R. R., Carlson J. O., Pettijohn D. E. Torsional tension in the DNA double helix measured with trimethylpsoralen in living E. coli cells: analogous measurements in insect and human cells. Cell. 1980 Oct;21(3):773–783. doi: 10.1016/0092-8674(80)90440-7. [DOI] [PubMed] [Google Scholar]
- Sinden R. R., Pettijohn D. E. Chromosomes in living Escherichia coli cells are segregated into domains of supercoiling. Proc Natl Acad Sci U S A. 1981 Jan;78(1):224–228. doi: 10.1073/pnas.78.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stalder J., Larsen A., Engel J. D., Dolan M., Groudine M., Weintraub H. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell. 1980 Jun;20(2):451–460. doi: 10.1016/0092-8674(80)90631-5. [DOI] [PubMed] [Google Scholar]
- Villeponteau B., Landes G. M., Pankratz M. J., Martinson H. G. The chicken beta globin gene region. Delineation of transcription units and developmental regulation of interspersed DNA repeats. J Biol Chem. 1982 Sep 25;257(18):11015–11023. [PubMed] [Google Scholar]
- Villeponteau B., Lundell M., Martinson H. Torsional stress promotes the DNAase I sensitivity of active genes. Cell. 1984 Dec;39(3 Pt 2):469–478. doi: 10.1016/0092-8674(84)90454-9. [DOI] [PubMed] [Google Scholar]
- Villeponteau B., Pribyl T. M., Grant M. H., Martinson H. G. Novobiocin induces the in vivo cleavage of active gene sequences in intact cells. J Biol Chem. 1986 Aug 5;261(22):10359–10365. [PubMed] [Google Scholar]
- Vogelstein B., Pardoll D. M., Coffey D. S. Supercoiled loops and eucaryotic DNA replicaton. Cell. 1980 Nov;22(1 Pt 1):79–85. doi: 10.1016/0092-8674(80)90156-7. [DOI] [PubMed] [Google Scholar]
- Wang J. C. DNA topoisomerases. Annu Rev Biochem. 1985;54:665–697. doi: 10.1146/annurev.bi.54.070185.003313. [DOI] [PubMed] [Google Scholar]
- Weintraub H. A dominant role for DNA secondary structure in forming hypersensitive structures in chromatin. Cell. 1983 Apr;32(4):1191–1203. doi: 10.1016/0092-8674(83)90302-1. [DOI] [PubMed] [Google Scholar]
- Weintraub H. Assembly and propagation of repressed and depressed chromosomal states. Cell. 1985 Oct;42(3):705–711. doi: 10.1016/0092-8674(85)90267-3. [DOI] [PubMed] [Google Scholar]
- Weintraub H. Assembly of an active chromatin structure during replication. Nucleic Acids Res. 1979 Oct 10;7(3):781–792. doi: 10.1093/nar/7.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Cheng P. F., Conrad K. Expression of transfected DNA depends on DNA topology. Cell. 1986 Jul 4;46(1):115–122. doi: 10.1016/0092-8674(86)90865-2. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weintraub H. Histone-H1-dependent chromatin superstructures and the suppression of gene activity. Cell. 1984 Aug;38(1):17–27. doi: 10.1016/0092-8674(84)90522-1. [DOI] [PubMed] [Google Scholar]