SUMMARY
Ring NTPases of the ASCE superfamily perform a variety of cellular functions. An important question about the operation of these molecular machines is how the ring subunits coordinate their chemical and mechanical transitions. Here, we present a comprehensive mechanochemical characterization of a homomeric ring ATPase—the bacteriophage φ29 packaging motor—a homopentamer that translocates double-stranded DNA in cycles composed of alternating dwells and bursts. We use high-resolution optical tweezers to determine the effect of nucleotide analogs on the cycle. We find that ATP hydrolysis occurs sequentially during the burst and that ADP release is interlaced with ATP binding during the dwell, revealing a high degree of coordination among ring subunits. Moreover, we show that the motor displays an unexpected division of labor: although all subunits of the homopentamer bind and hydrolyze ATP during each cycle, only four participate in translocation, whereas the remaining subunit plays an ATP-dependent regulatory role.
INTRODUCTION
Multi-subunit, ring-shaped nucleoside triphosphatases (NTPases) are a group of enzymes that drive translocation or rotation of their substrates, such as nucleic acids and polypeptides, by coupling nucleotide binding and hydrolysis to conformational changes in the substrate-binding motif (Bustamante et al., 2004). A large number of these oligomeric ring NTPases belong to the additional strand conserved E (glutamate) (ASCE) superfamily, and are involved in a multitude of cellular tasks, including DNA replication, transcription regulation, protein degradation, cargo transport, chromosome segregation, and viral genome packaging (Erzberger and Berger, 2006; Iyer et al., 2004).
A central question about the operating principles of ring NTPases is how these motors couple and coordinate chemical events to processive mechanical movement (Lyubimov et al., 2011; Singleton et al., 2007). It is difficult to answer this question using traditional biochemical methods which necessarily involve ensemble averaging. Although crystal structures containing the motor, the substrate, and the bound nucleotides often possess sufficient resolution to relate chemical states to mechanical states of the motor, they do not provide dynamic information about the overall mechanochemical cycle. The advent of single-molecule techniques has made it possible to follow the trajectories of individual molecules in real time. By directly imaging the angular movement of a heteromeric rotary motor, F1-ATPase, Adachi et al. (2007) achieved the most comprehensive mechanochemical characterization of an ASCE ring NTPase to date. However, a similar level of description has not yet been accomplished for any homomeric nucleic acid or polypeptide translocase, which comprises a major fraction of ASCE ring NTPases. This is largely due to the lack of robust and sensitive assays to monitor nanometer-scale substrate translocation. We have recently developed high-resolution optical tweezers that enable us to observe the discrete translocation steps of the DNA packaging motor of the Bacillus subtilis bacteriophage φ29 (Moffitt et al., 2009). Thus this double-stranded DNA (dsDNA) translocase is an ideal model system to study the mechanochemical coupling and coordination mechanism of homomeric ring ATPases.
The functional core of the φ29 packaging motor is a homopentameric ATPase, gene product 16 (gp16) (Morais et al., 2008), that belongs to the HerA/FtsK clade of the ASCE superfamily (Burroughs et al., 2007). The biochemical and biophysical properties of gp16 have been studied extensively by bulk and single-molecule experiments (Casjens, 2011; Hetherington et al., 2012; Rao and Feiss, 2008). Previously we reported that the φ29 packaging motor translocates DNA by cycling through two phases: a stationary or “dwell” phase during which ATPs are loaded to the ring, and a translocation or “burst” phase during which 10 base pairs (bp) of DNA are packaged in four 2.5 bp steps (Figures 1A–1D; Figure S1 available online) (Moffitt et al., 2009). We note that the step size was measured based upon changes in the length of the B-form DNA tether outside of the motor complex, and thus does not depend on the DNA structure within the motor. Each 2.5 bp translocation step is powered by the release of inorganic phosphate (Pi) from one subunit (Figure 1D) (Chemla et al., 2005), indicating that only four of the five ring subunits are involved in DNA translocation in every 10 bp cycle. We have also shown that the motor makes a specific electrostatic contact with a pair of adjacent backbone phosphates on the 5′-3′ DNA strand every 10 bp and that this phosphate contact plays an important regulatory role in the motor’s cycle (Aathavan et al., 2009). These results set the stage to pursue a full description of the mechanochemical coupling in the φ29 packaging motor. Specifically, we asked: Where in the cycle, burst or dwell, does ATP hydrolysis occur? Where in the cycle does ADP release happen? How do individual ATPase subunits coordinate their catalytic cycles? Does the nontranslocating subunit bind and hydrolyze nucleotide each cycle? Does the nontranslocating subunit play any role in the motor operation?
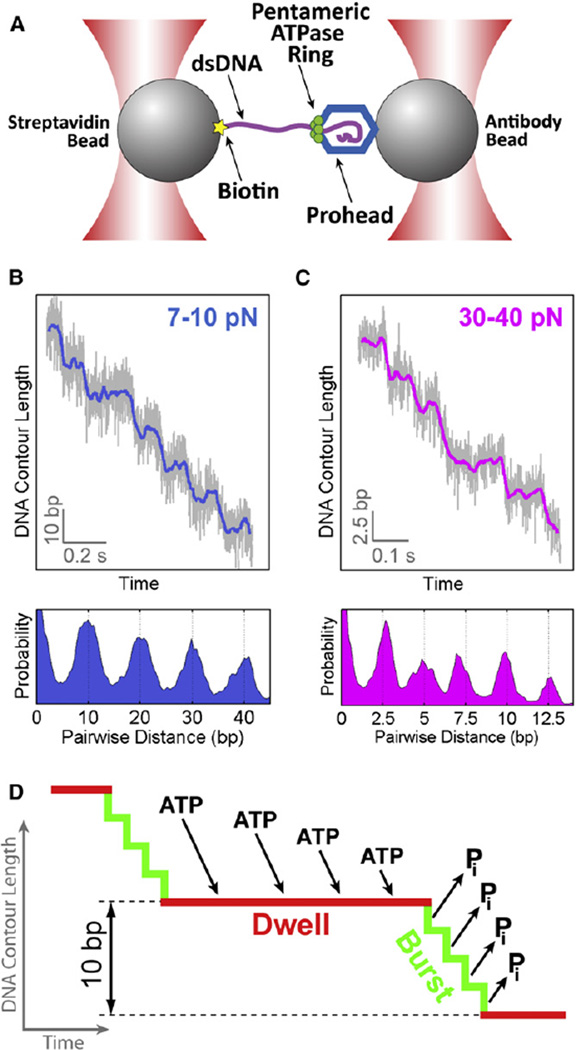
Figure 1. φ29 Motor Packages DNA via a Dwell-Burst Mechanism.
(A) Dual-trap optical tweezers used to study the φ29 packaging motor.
(B) A sample packaging trace collected at an external force of 7–10 pN (top) and the corresponding pairwise distance distribution (PWD) (bottom). Raw data (2500 Hz) (light gray) were filtered and decimated to 250 Hz (blue). In this force regime, DNA is translocated in nearly-instantaneous 10 bp increments.
(C) A sample packaging trace collected at an external force of 30–40 pN (top) and its corresponding PWD (bottom). In this force regime the 10 bp bursts break up into four 2.5 bp steps.
(D) Diagram of the dwell-burst cycle of the φ29 motor.
See also Figure S1.
To determine how chemical transitions are coupled to mechanical motion and how subunits are coordinated in the φ29 ATPase, we use various nucleotide analogs to perturb specific chemical transitions of the catalytic cycle. We follow the translocation dynamics of single packaging motors with high-resolution optical tweezers (Figure 1A). By monitoring the mechanical response of the motor to these chemical perturbations we show that ATP hydrolysis occurs in the burst phase and that ADP release occurs in the dwell phase. Furthermore, we establish that ATP binding and ADP release are interlaced and strictly coordinated during the dwell phase. Moreover, our data indicate that the special nontranslocating subunit binds and hydrolyzes ATP at a well-defined stage in the dwell-burst cycle, in order to play an essential regulatory role in the motor’s operation. This finding reveals an unprecedented division of labor among subunits in a homomeric ring ATPase. Finally, we present a complete mechanochemical model for the operation of a homomeric ring ATPase based on the dynamic information obtained from single-molecule data.
RESULTS
ATP Hydrolysis Occurs in the Burst Phase
To determine where in the dwell-burst cycle ATP hydrolysis occurs and how the individual hydrolysis events are coordinated among subunits, we monitored DNA translocation by φ29 packaging complexes in the presence of ATPγS, a nonhydrolyzable ATP analog. It was previously shown that ATPγS binds to the motor and induces long pauses (Chemla et al., 2005). We analyzed single-molecule trajectories at base pair resolution to determine what phase of the cycle (dwell or burst) is affected by ATPγS. Figure 2A shows individual packaging traces at various ATPγS concentrations and a fixed, saturating ATP concentration. ATPγS-induced events appeared as pauses in DNA translocation that became more frequent with increasing nucleotide analog concentration (Figures 2A and 2B). The majority (~75%) of all ATPγS-induced pausing events consisted of a single pause (Figure 2C). Surprisingly, a significant portion (~25%) of all ATPγS-induced pausing events consisted of two or more consecutive pauses separated by 10 bp (Figure 2C). We refer to the latter fraction of pausing events as pause clusters.
Figure 2. Nonhydrolyzable ATP Analog Induces Pausing Events.
(A) Packaging traces at saturating [ATP] (250 µM) and various amounts of ATPγS.
(B) The density of ATPγS-induced pausing events (number of pausing events per kilobase of DNA translocated) is linearly proportional to [ATPγS]. Error bars represent 95% confidence intervals.
(C) ATPγS-induced pausing events consist of one or more pauses separated by 10 bp bursts. Pausing events are characterized by their duration (orange bar) and span (the length of DNA translocated during an event, green bar). Pausing events consisting of two or more pauses are referred to as pause clusters.
(D) The duration of pausing events pooled from all ATPγS concentrations is well fit by a single exponential (n = 516). Inset: the mean pausing event duration is independent of [ATPγS]. Error bars represent 95% confidence intervals.
(E) The span of pausing events pooled from all [ATPγS] comprises three distinct groups: single pauses with ~0 bp span, two-pause-clusters with ~10 bp span, and three-pause clusters with ~20 bp span. The width of the peaks is due to noise in single-molecule data. Inset: the fraction of ATPγS-induced pausing events that are pause clusters is independent of [ATPγS]. Error bars represent 95% confidence intervals.
See also Figure S2.
We then asked whether the multiple pauses observed within a pause cluster were caused by the successive binding of multiple ATPγS molecules to the motor. At the highest [ATPγS] sampled (2.0 µM), we observed an average of one ATPγS-induced pausing event for every 170 bp of DNA packaged (Figure 2B). Therefore the probability of observing two consecutive pauses caused by two independent ATPγS binding events within a cluster should be <1/17 (~6%). This analysis indicates that each pause cluster is caused by the binding of a single ATPγS to the motor. This conclusion is reinforced by the following observations: (1) the number of ATPγS-induced pausing events per kilobase of DNA packaged—including both single pauses and pause clusters—increases linearly with [ATPγS] (Figure 2B), indicating that each pausing event is caused by the binding of a single nucleotide analog; (2) the distribution of pausing event durations is well fit by a single-exponential, indicating that a single stochastic process—most likely the dissociation of a single ATPγS—is responsible for the termination of the pausing event (Figure 2D); in contrast, the distribution of durations of individual pauses (including single pauses and pauses within a pause cluster) cannot be fit by a single-exponential, further suggesting that the dissociation of ATPγS from the motor determines the lifetime of the entire pausing event but not the duration of individual pauses that make up a cluster (Figure S2); (3) the average duration of pausing events is independent of [ATPγS] (Figure S2); and (4) the fraction of pause clusters among all pausing events is also independent of [ATPγS] (Figures 2E, inset, and S2).
Taken together, our results support a mechanism in which all pausing events are caused by a single nonhydrolyzable ATP analog binding to the ring. While the analog remains bound, the motor can stochastically take a few 10 bp bursts, resulting in a pause cluster. As will be discussed later, the observation of pause clusters provides insight about the division of labor among subunits and their coordination in the ring.
Having established that all pausing events are caused by the binding of a single ATP analog to the motor, we can now determine where ATP hydrolysis occurs in the dwell-burst cycle. Previous work indicated that Pi release precedes or coincides with each 2.5 bp power stroke during the burst phase (Chemla et al., 2005; Moffitt et al., 2009). Therefore, within a single subunit, ATP hydrolysis should happen after nucleotide binding and before the power stroke. However, this constraint does not uniquely determine the timing or the coordination of hydrolysis with respect to other chemical events in the motor’s cycle. ATP hydrolysis can occur during the dwell phase, either interlaced with nucleotide binding (Figure 3A, H-Scenario 1) or temporally segregated from binding (Figure 3A, H-Scenario 2). Alternatively, ATP hydrolysis could take place immediately before Pi release during the burst phase (Figure 3A, H-Scenario 3).
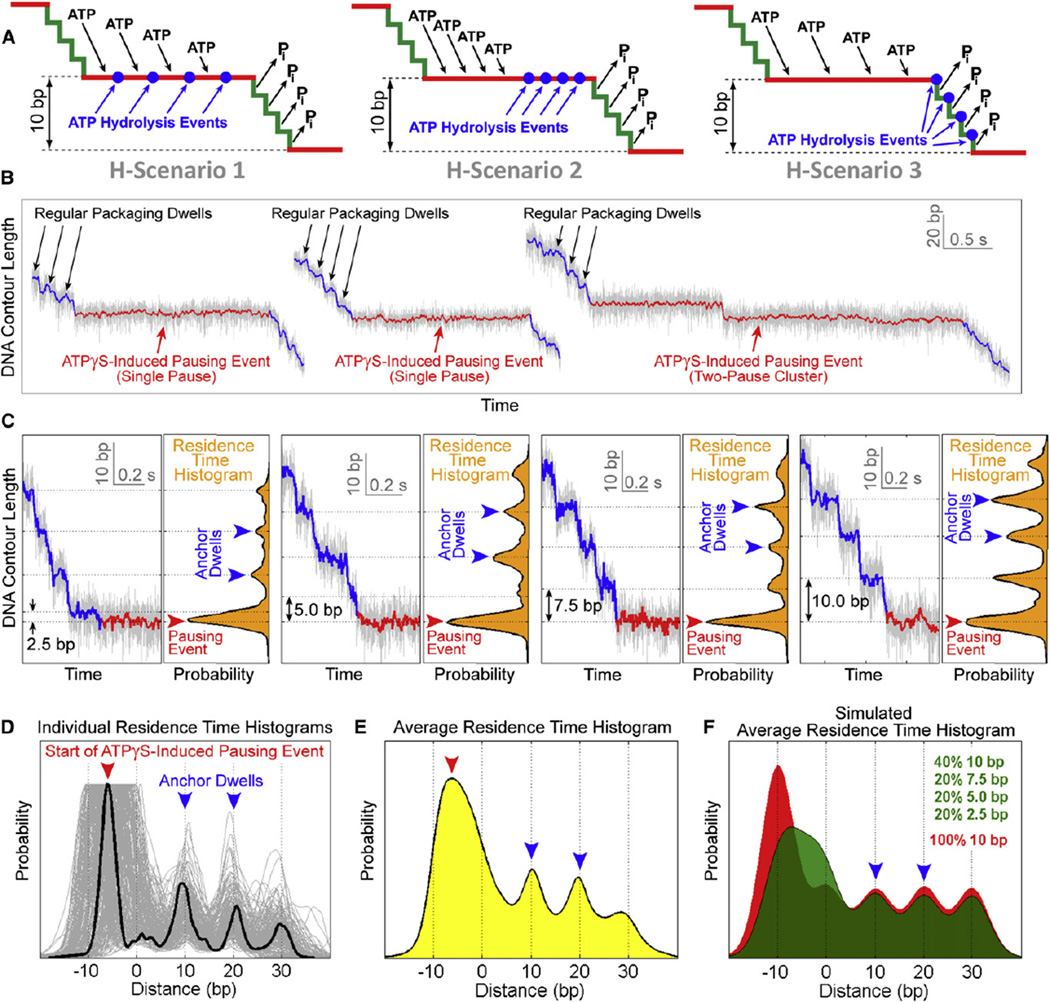
Figure 3. Determining the Timing of ATP Hydrolysis in the Dwell-Burst Cycle.
(A) Possible ATP-hydrolysis scenarios. H-Scenario 1: ATP hydrolysis is interlaced with ATP binding during the dwell. H-Scenario 2: ATP hydrolysis occurs during the dwell after all subunits have bound ATP. H-Scenario 3: ATP hydrolysis is interlaced with Pi release during the burst.
(B) Overview of ATPγS-induced pausing events highlighting the difference in duration between regular packaging dwells (black arrows) and pauses (red arrows).
(C) Detailed view of four sample packaging traces containing an ATPγS-induced pausing event, from 40 bp upstream of the pausing event to 0.5 s after the start of the pausing event. Regular packaging is shown in blue and the start of the pausing event is shown in red. The large peak (red arrow) in the residence time histogram corresponds to the start of the pausing event. Two regular dwells (blue arrows) were used as anchors for aligning different residence time histograms.
(D) Residence time histograms superimposed and aligned using their two anchor dwells, which are located at 10 bp and 20 bp marks (blue arrows). The 0 bp mark denotes the position of the regular dwell immediately before a pausing event. The burst size before an ATPγS-induced pausing event is given by the distance between the 0 bp mark and the large peak in the histogram (red arrow).
(E) The residence time histogram obtained by averaging all histograms from (D) (n = 209), using experimental data from all [ATPγS].
(F) The average residence time histogram constructed from simulated data for scenarios where hydrolysis occurs during the dwell (red) or during the burst (green).
See also Figure S3.
Using high-resolution optical tweezers, we can distinguish among these alternative scenarios by determining what mechanical phase of the cycle is interrupted by the nonhydrolyzable analog. If all ATP hydrolysis events occur during the dwell (H-Scenarios 1 and 2), the analog-induced pausing event should always start during a dwell and should always be preceded by a complete 10 bp burst, regardless of which subunit binds the analog (Figure S3A). In contrast, if hydrolysis occurs during the burst (H-Scenario 3), we expect the motor to pause after it takes zero, one, two, or three 2.5 bp steps depending on the order of the analog-bound subunit in the hydrolysis sequence, resulting in a 10 bp, 2.5 bp, 5.0 bp, or 7.5 bp prepause burst, respectively (Figure S3B). Note that when the motor takes zero 2.5 bp steps before pausing, the prepause burst appears as 10 bp in size.
Analysis of individual molecular trajectories revealed a broad distribution containing both complete (10 bp) and incomplete (2.5 bp, 5.0 bp, and 7.5 bp) prepause bursts (Figures 3B and 3C). To objectively analyze the distribution of prepause burst sizes without biasing against smaller bursts, we constructed residence time histograms, which represent the amount of time the motor resides at a particular location along the DNA (Figure 3C, orange histograms). On these histograms, regular packaging dwells appeared as small peaks, whereas ATPγS-induced pauses appeared as large local maxima (Figure 3C, red arrows). Two regular packaging dwells (Figure 3C, blue arrows) upstream of the pause served as anchor points. The distance between the beginning of a pausing event and its anchor dwells provides a direct measurement of the prepause burst size. Furthermore, the anchor dwells enabled us to align all traces and construct an average residence time histogram (Figures 3D and 3E). Note that the large peak corresponding to the pause is broad (Figure 3E, red arrow) and centered away from the 10 bp lattice defined by the anchor points (Figure 3E, dotted vertical lines). For comparison, Figure 3F shows simulated average residence time histograms corresponding to different hydrolysis scenarios. If all prepause bursts are 10 bp in size (H-Scenarios 1 and 2), the pause peak should fall on the 10 bp lattice (Figure 3F, red histogram). If instead a significant fraction of prepause bursts is 2.5 bp, 5.0 bp, or 7.5 bp in size (H-Scenario 3), the pause peak should be broad and centered off the 10 bp lattice (Figure 3F, green histogram). The average residence time histogram constructed from experimental data is clearly consistent with the latter case (Figures 3E and 3F), indicating that ATP is hydrolyzed during the burst.
Our data suggest that ATPγS is able to temporarily stall the motor after it takes zero, one, two, or three 2.5 bp steps, depending on the position of the analog-bound subunit relative to the first subunit performing hydrolysis. This observation requires that hydrolysis events be strictly coordinated and sequential; otherwise the ATP-bound subunits would always hydrolyze first, resulting in the same burst size prior to each pausing event. To summarize, our results demonstrate that hydrolysis occurs during the burst phase in a strictly coordinated fashion, with each subunit hydrolyzing ATP immediately before taking a 2.5 bp step (Figure 3A, H-Scenario 3).
ADP Release Occurs in the Dwell Phase
To determine the timing and coordination of ADP release within the dwell-burst cycle, we perturbed DNA packaging using orthovanadate (VO43−), a Pi analog that forms stable complexes with ADP, delaying the dissociation of ADP from the binding pocket (Baird et al., 1999; Sharma and Davidson, 2000; Yang and Catalano, 2004). Within a single subunit, ADP release must occur after nucleotide hydrolysis and before a new ATP molecule docks. These requirements allow only three possible ADP release scenarios: all ADP release events are temporally segregated from ATP binding at the beginning of the dwell (Figure 4A, R-Scenario 1), ADP release is interlaced with ATP binding during the dwell (Figure 4A, R-Scenario 2), or ADP release is interlaced with Pi release during the burst (Figure 4A, R-Scenario 3).
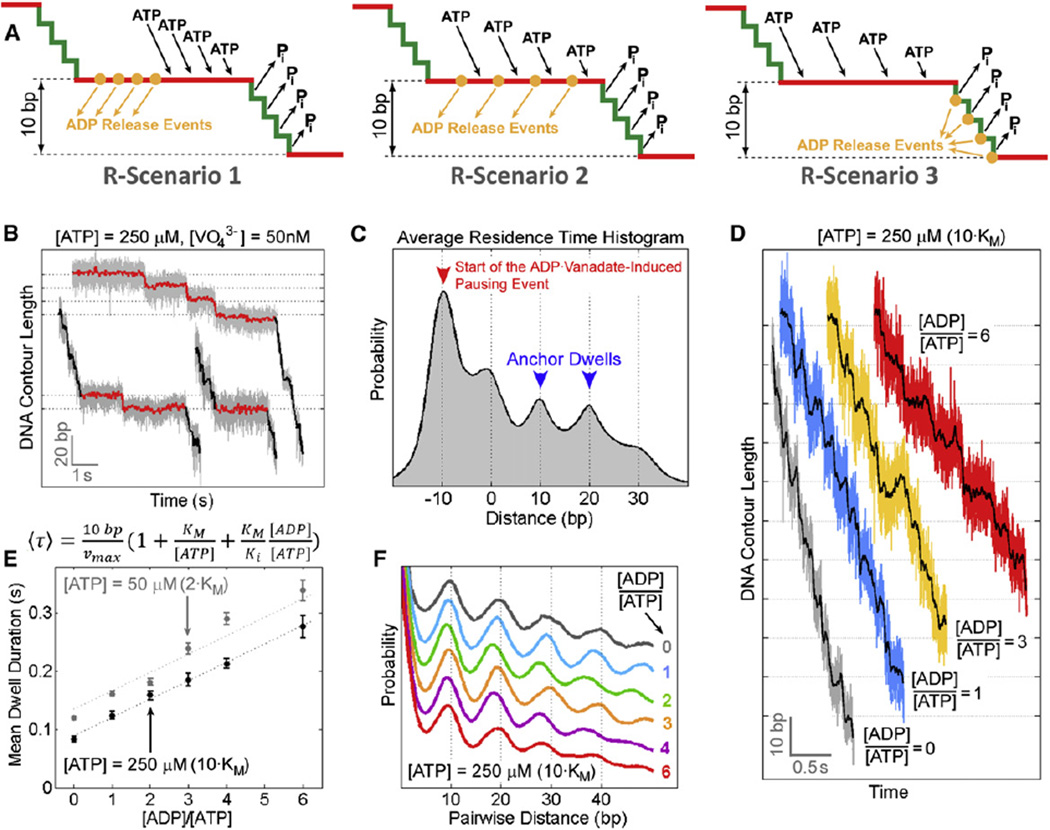
Figure 4. Determining the Timing and Coordination of ADP Release.
(A) Possible ADP-release scenarios. R-Scenario 1: ADP release is temporally segregated from ATP binding during the dwell. R-Scenario 2: ADP release is interlaced with ATP binding during the dwell. R-Scenario 3: ADP release is interlaced with hydrolysis and Pi release during the burst.
(B) Sample packaging traces in an ADP-free packaging buffer containing saturating [ATP] and 50 nM of sodium orthovanadate. ADP-vanadate-induced pausing events (red) consist of single pauses or pause clusters containing multiple pauses separated by 10 bp bursts.
(C) The average residence time histogram for ADP-vanadate-induced pausing events (n = 37). The blue arrows denote the anchor dwells, and the red arrow indicates the start of the pausing event.
(D) Sample packaging traces at saturating [ATP] (250 µM) and various ADP concentrations.
(E) The mean dwell duration as a function of [ADP] at [ATP] = 250 µM(black) and 50 µM(gray). Data were fit to a competitive inhibition model. Error bars represent 95% confidence intervals.
(F) PWD plots computed from translocation traces at [ATP] = 250 µM and various [ADP]. The peaks at integer multiples of 10 bp indicate that the burst size is 10 bp, and is not affected by [ADP].
See also Figure S4, Table S1, and Extended Experimental Procedures.
Small amounts of Na3VO4 (50 nM) were added to an ADP-free packaging buffer, such that vanadate could form complexes only in situ with ADP that remained bound to the ATPase after the last round of hydrolysis. We found that ADP-vanadate also induces pausing events (Figure 4B). However, in contrast to the ATPγS results (Figure 3E), the bursts preceding ADP-vanadate-induced pausing events are overwhelmingly 10 bp in size, and the pause peak in the average residence time histogram falls on the 10 bp lattice (Figure 4C). This result is only consistent with scenarios where ADP release occurs during the dwell phase (Figure 4A, R-Scenarios 1 and 2).
ADP Release Is Interlaced with ATP Binding
We have previously reported that all ATPs are loaded to the gp16 ring during the dwell phase and that at any given time during the dwell only one ATPase subunit is capable of binding ATP (Moffitt et al., 2009). As shown above, ADP release also occurs during the dwell phase. The question then arises: how do individual gp16 subunits coordinate their ADP release and ATP binding transitions? Are ADP release and ATP binding temporally segregated (Figure 4A, R-Scenario 1), or are they interlaced (Figure 4A, R-Scenario 2)? To discriminate between these two scenarios, we sought to probe the effect of ADP on the motor dynamics. High-resolution packaging traces were collected at saturating [ATP] and various [ADP]. Increasing [ADP] from 0 to 6 × [ATP] gradually lengthens the duration of all dwells in a linear fashion (Figures 4D and 4E). The effect of ADP is clearly distinct from that of ATPγS or ADP-vanadate, which cause long-lived pausing events once bound. Thus, the dissociation rate of ADP must be sufficiently fast so that binding of one single ADP does not induce distinct pauses. The increase in the dwell duration at high [ADP] is most likely due to multiple rounds of ADP binding and release events from an ATPase subunit before that subunit finally docks ATP. These experiments also show that ADP does not affect the burst size or burst duration (Figures 4D and 4F).
The dependence of the mean dwell duration on [ADP] is well described by a competitive-inhibition model (Figure 4E), consistent with our previous results (Chemla et al., 2005). If ADP release and ATP binding occur in an alternating fashion (Figure 4A, R-Scenario 2), ADP and ATP always compete for the same state of the ATPase, making this scenario consistent with competitive inhibition. Moreover, in this case, the mean dwell duration should be linearly dependent on [ADP] because at any given time only one binding site is available. On the contrary, if ADP release and ATP binding were temporally segregated (Figure 4A, R-Scenario 1), ADP would act as a noncompetitive inhibitor, and inhibition by ADP could not be offset with increasing [ATP], in contradiction to our previous observations (Chemla et al., 2005). Moreover, R-Scenario 1 predicts a nonlinear dependence of the mean dwell duration on [ADP], because multiple binding sites would be simultaneously available for ADP binding (Segel, 1975). Therefore, our results and those from previous studies (Chemla et al., 2005; Moffitt et al., 2009) are consistent with a model in which ADP release events are interlaced with ATP binding during the dwell phase (Figure 4A, R-Scenario 2). An alternative model in which ADP inhibition occurs through an off-pathway state can be shown to be invalid (Figure S4; Table S1; Extended Experimental Procedures).
ADP Release Events Are Rate-Limiting at Saturating [ATP]
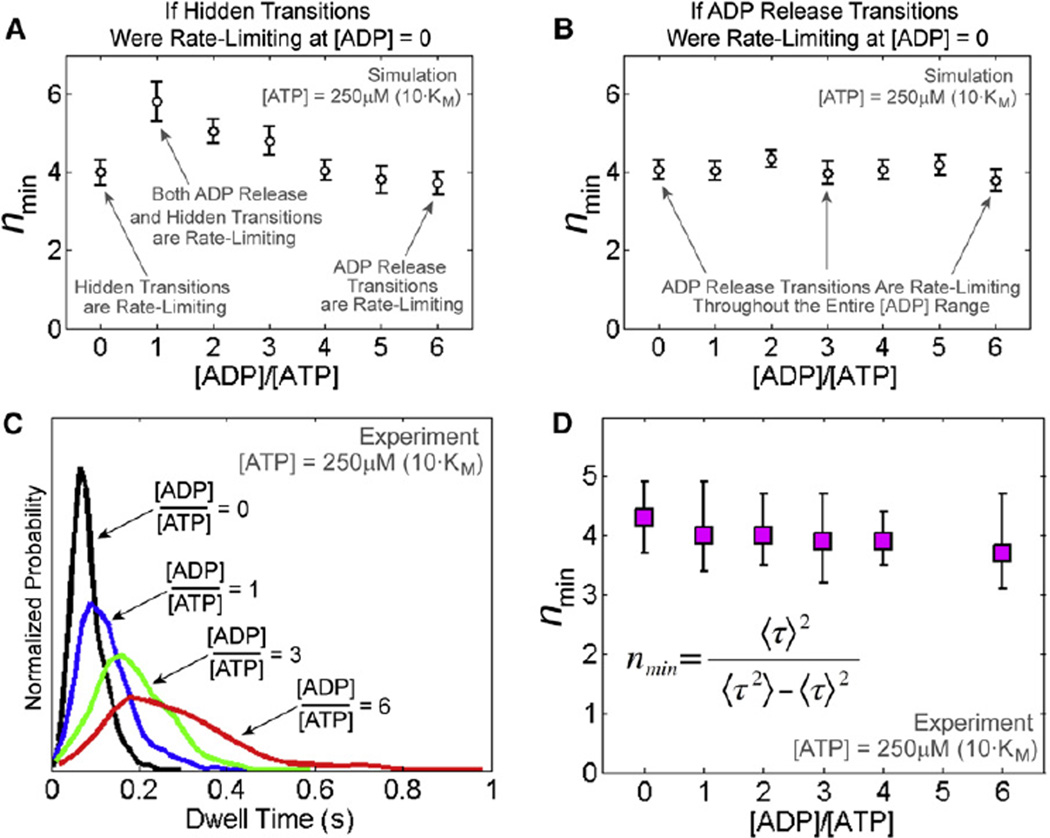
As shown previously (Moffitt et al., 2010; Schnitzer and Block, 1995), the ratio of the squared mean of dwell times to their variance, nmin = 〈τ〉2/(〈τ〉2 − 〈τ〉2), constitutes a lower bound for the number of rate-limiting events in a dwell: the larger the number of rate-limiting events the higher the nmin value. Using nmin analysis, we previously established that, at saturating [ATP], the dwell phase contains at least four rate-limiting transitions of unknown identity, which are distinct from ATP binding (Moffitt et al., 2009). By determining how nmin varies with [ADP], it is possible to establish the identity of those rate-limiting events.
As shown in Figures 4D and 4E, high concentrations of ADP prolong the amount of time the motor spends in the dwell phase, most likely by delaying the replacement of ADP with ATP in the catalytic pockets, and effectively rendering ADP release rate-limiting. Let us now imagine that the dwell phase contains at least four hidden transitions that are neither ATP binding nor ADP release, and that these hidden transitions are rate-limiting at saturating [ATP] and zero [ADP]. As [ADP] is raised, the ADP release transition effectively becomes slower. When the average duration of one ADP release event becomes comparable to the average duration of one hidden transition, the number of rate-limiting events within the dwell phase will necessarily increase (nmin should increase). As [ADP] is raised further, ADP release transitions become slower than the hidden transitions, and eventually ADP release events become the only rate-limiting steps (nmin should decrease). In other words, if processes other than ADP release were rate-limiting at zero [ADP], the identity of the rate-limiting transitions should change with increasing [ADP], and the value of nmin should first rise, reach a peak, and then decrease asymptotically. Monte Carlo simulations of different rate-limiting scenarios corroborate this prediction (Figures 5A and 5B; Table S2; Extended Experimental Procedures).
Figure 5. ADP Release Events Are Rate Limiting in the Dwell at Saturating [ATP].
(A) Simulated nmin dependence on [ADP] if four hidden transitions were rate-limiting during the dwell at saturating [ATP] and zero [ADP]. nmin is computed from the dwell time distribution and represents a lower limit for the number of rate-limiting transitions during the dwell. Error bars represent 95% confidence intervals.
(B) Simulated nmin dependence on [ADP] if four ADP release transitions were rate-limiting at saturating [ATP] and zero [ADP]. Error bars represent 95% confidence intervals.
(C) Experimental dwell time distributions at saturating [ATP] (250 µM) and various [ADP].
(D) nmin computed from experimental dwell time distributions at various [ADP]. Error bars represent 95% confidence intervals estimated via bootstrapping.
We compiled dwell time distributions for packaging traces in buffers containing saturating [ATP] (250 µM) and [ADP] ranging from 0 to 1,500 µM (Figure 5C). Remarkably, nmin remained constant even though the mean dwell duration increased ~3-fold across the entire range of ADP concentrations tested (Figures 5D and 4E). This result can only be explained if, at saturating [ATP], ADP release events are already rate-limiting at zero [ADP] and remain so at all ADP concentrations. Therefore we conclude that under saturating [ATP] conditions at least four ADP release events rate-limit the duration of the dwell.
The Special Nontranslocating Subunit Binds and Hydrolyzes ATP Every Cycle
The results presented above reveal that the φ29 packaging motor operates as a highly coordinated machine in which four of its five subunits release ADP, bind ATP, hydrolyze ATP, and translocate DNA in a precisely timed fashion throughout its mechanochemical cycle. Several questions then arise naturally: What is the role of the special nontranslocating subunit? Does it bind ATP? Does it hydrolyze ATP? Three observations provide clues to answer these questions.
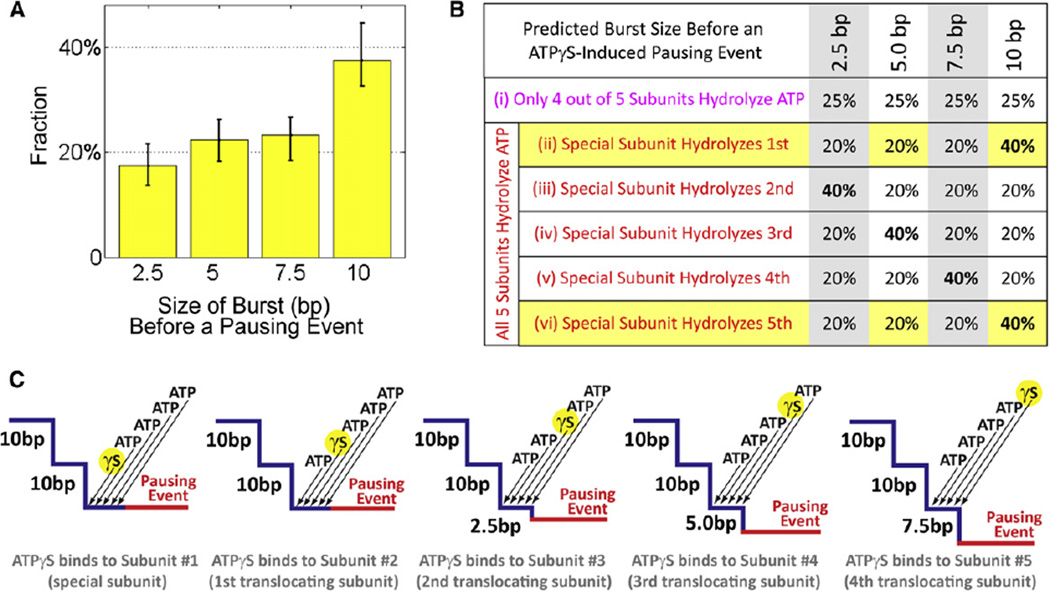
(1) Approximately 40% of all bursts preceding ATPγS-induced pausing events are 10 bp in size, whereas the remaining 60% are evenly split among 2.5 bp, 5.0 bp, and 7.5 bp bursts (Figure 6A). This observation can be most naturally explained by a mechanism in which all five motor subunits, including the special one, bind and hydrolyze nucleotide in every dwell-burst cycle. To demonstrate this point, let us consider the contrary, namely that only the four translocating subunits bind and hydrolyze ATP every dwell-burst cycle. If so the prepause burst size should be evenly distributed among 2.5 bp, 5.0 bp, 7.5 bp, and 10 bp (Figure 6B, Case (i)). In contrast, if instead all five gp16 subunits bind and hydrolyze ATP during every dwell-burst cycle, we should expect a nonuniform prepause burst size distribution (Figure 6B, Cases (ii)–(vi)). Our observation that the prepause burst sizes are not uniformly distributed indicates that all five subunits bind and hydrolyze ATP in each cycle. Moreover, the shape of the prepause burst size distribution can be used to determine the temporal order of hydrolysis of the special subunit with respect to the other four translocating subunits. As shown in Figures 6B and 6C, the experimentally observed 2:1:1:1 ratio of 10 bp, 7.5 bp, 5.0 bp, and 2.5 bp prepause bursts, respectively, requires that the special subunit be either the first or the last to hydrolyze nucleotide.
Figure 6. Nucleotide State of the Special Subunit.
(A) Distribution of the burst size before an ATPγS-induced pausing event, measured from residence time histograms (Figure 3D). Bursts of 10 bp occur roughly twice as often as 2.5 bp, 5.0 bp, or 7.5 bp bursts (n = 209). Error bars represent 95% confidence intervals estimated via bootstrapping.
(B) Predicted distribution of 2.5 bp, 5.0 bp, 7.5 bp, and 10 bp bursts before a pausing event for various cases of the nucleotide state of the special subunit. Cases (ii) and (vi) are consistent with experimental observations (A).
(C) Diagrams for the prepause burst size distribution given Case (ii) from (B). In this case, all five subunits, including the special one, bind and hydrolyze nucleotide during every cycle and the special subunit is always the first to undergo hydrolysis. Arrows indicate the temporal order of nucleotide binding events.
See also Figure S5.
(2) The pausing event span histogram displays peaks at 0, 10, and 20 bp (Figure 2E), indicating that during an ATPγS-induced pause cluster, the motor predominantly takes 10 bp bursts. We obtained similar results when the experiments were repeated with a different nonhydrolyzable ATP analog, AMP-PNP (Figure S5). These observations show that, while one ATPase subunit is occupied by a nonhydrolyzable nucleotide analog, the motor is capable of packaging DNA in nearly-instantaneous 10 bp bursts separated by pauses that are much longer than regular dwells. This phenomenon requires that: (i) during the first pause of a pause cluster the motor resets, and the ATPγS-bound subunit adopts the identity of the special nontranslocating subunit, whether or not it fulfilled this role in the previous cycle, and (ii) while the special subunit remains bound to ATPγS, the remaining four subunits bind ATP, hydrolyze ATP, and translocate DNA by 2.5 bp each. In this mechanism, the long pauses observed within a pause cluster reflect the response of the motor to the inability of the newly assigned special subunit to hydrolyze the analog nucleotide.
(3) ADP-vanadate can also induce pause clusters that contain 10 bp bursts (Figure 4B), indicating that, during an ADP-vanadate-induced pause cluster, only one subunit is bound to ADP-vanadate, and that this subunit assumes the special identity. This observation implies that, besides hydrolysis, timely product release by the special subunit is also required for the normal activity of the ring.
In conclusion, our observations support a coordination mechanism in which all five subunits, including the special one, bind and hydrolyze ATP every dwell-burst cycle. We have already shown that hydrolysis events must be strictly coordinated and sequential among the four translocating subunits. The prepause burst size distribution now indicates that hydrolysis of the special subunit is also strictly timed with respect to those of the other subunits—it either initiates or concludes the hydrolysis cascade. This mechanism naturally reconciles the outstanding discrepancy between the bulk measurement of five ATP molecules consumed for every 10 bp of DNA packaged (Chemla et al., 2005; Guo et al., 1987), and the single-molecule measurement of four 2.5 bp power strokes per dwell-burst cycle. The strict timing and coordination of the catalytic activity of the special subunit lead us to speculate that the hydrolysis by the special subunit is coupled to an essential regulatory process, such as triggering or resetting the motor’s mechanochemical cycle.
Note that the role of the special subunit is inferred assuming that nucleotide analogs trap the ATPase at a certain on-pathway state without dramatically altering the kinetic mechanism of the motor, and that ATPγS is equally likely to bind to any one of the five catalytic sites. This is a reasonable assumption, and it has been widely used to dissect the reaction pathway of other NTPases. We acknowledge that alternative models for the operation of the motor can be envisioned; here we have chosen the most parsimonious one to explain the data.
DISCUSSION
Symmetry Breaking in the φ29 Homomeric Ring ATPase
It has been shown that functional asymmetry exists in heteromeric ring NTPases such as dynein (Carter et al., 2011), the bacterial clamp loader (Johnson and O’Donnell, 2003), and the eukaryotic MCM helicase (Bochman et al., 2008). In these enzymes, a subset of the subunits is responsible for the mechanical task, whereas the others either are inactive or play regulatory roles. On the other hand, subunits from homomeric ring NTPases are generally believed to undertake the same mechanical task, although they could be in different nucleotide states at a particular stage in the mechanochemical cycle (Lyubimov et al., 2011). Here we show that a homomeric ring ATPase can also display a division of labor: only four of the five gp16 subunits translocate the substrate, whereas the special one appears to play a critical regulatory role.
Although structural differences between subunits in a heteromeric ring NTPase naturally provide the basis for functional asymmetry, it is less clear how such division of labor arises in a homomeric ring motor. It is possible that the functional asymmetry observed in gp16 is caused by the cracking of its ring into a lock-washer shape as has been seen in the crystal structures of Rho, DnaA, DnaC, and recently DnaB (Duderstadt et al., 2011; Mott et al., 2008; Skordalakes and Berger, 2003; Itsathitphaisarn et al., 2012), or by an asymmetric closed planar ring as seen for ClpX (Glynn et al., 2009). However, any asymmetric structure must always involve an underlying symmetry-breaking mechanism. For example, in the case of F1-ATPase, the 3-fold symmetry of the trimer of αβ dimers is most likely broken by the successive interaction of each β subunit with the central γ subunit that acts as an external coordination agent of the cycle (Abrahams et al., 1994). In the φ29 packaging motor, it has been shown that the electrostatic contact made by the motor with a pair of adjacent DNA phosphates every 10 bp plays an important regulatory role in the motor’s operation (Aathavan et al., 2009). Our results here suggest that one of the five ATPase subunits also plays a critical regulatory function. Hence, it is sensible to propose that the subunit making the electrostatic contact with DNA is the one that adopts the identity of the special, nontranslocating subunit. In other words, the symmetry breaking in the φ29 motor could arise during every dwell from the crucial electrostatic contact the motor makes every 10 bp with the DNA backbone.
What is the role of this nontranslocating subunit? We have inferred that the special subunit either initiates or concludes the hydrolysis cascade (Figures 6A and 6B). Therefore, the role of this subunit could be to time the motor’s cycle by starting or terminating the burst. Of these two possibilities, we favor the one in which the special subunit is the first to hydrolyze ATP, using this energy to break its strong electrostatic contact with the DNA backbone and to trigger the burst phase. When the special subunit is occupied by a nucleotide analog (ATPγS, AMP-PNP, ADP-vanadate), the breaking of the motor-DNA contact at the beginning of the burst and the release of the first ADP molecule at the beginning of the dwell must take place spontaneously. Hence the progression of the cycle slows down due to the absence of the timing signal arising from the ATP turnover by the special subunit.
Is the regulatory role always performed by the same subunit, or is that task passed around the ring in subsequent cycles? We have argued that, within a pause cluster, the analog-bound subunit assumes the special role and retains it throughout successive cycles in a cluster. One could argue that a model in which the identity of the special subunit does not change is only valid when one of the motor subunits is bound to ATPγS. However, this model is also favored by symmetry considerations. Because the helical pitch of dsDNA is 10.4 bp, the subunit best positioned to make the crucial regulatory phosphate interaction after a 10 bp burst is the same subunit that made that contact in the previous dwell. We note that this model does not prevent the motor from reassigning the identity of the special subunit under certain circumstances when regular packaging is disrupted, such as after slips or upon analog-induced pausing events, nor does it exclude the small probability of special subunit reassignment during normal packaging due to the intrinsic stochasticity of the system.
Complete Mechanochemical Characterization of the φ29 Packaging Motor Reveals a Highly Coordinated Ring ATPase
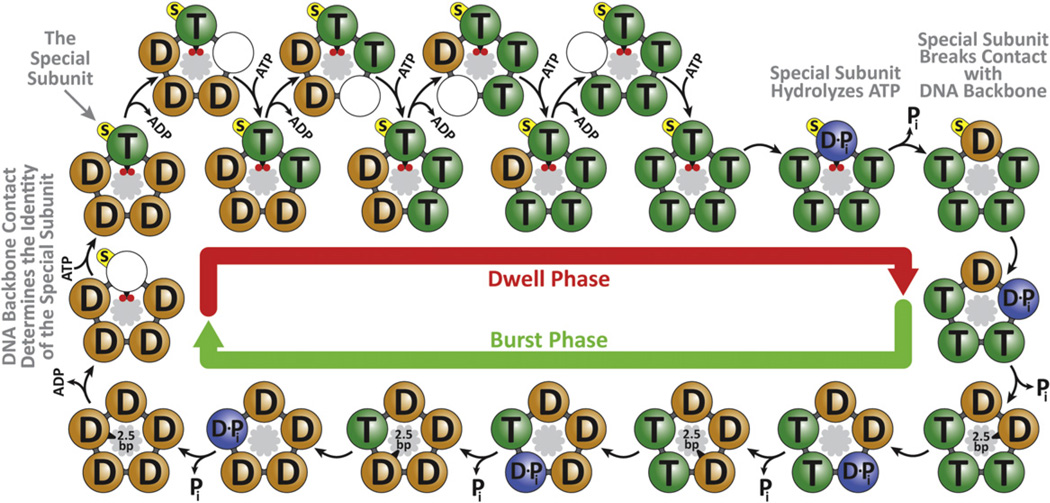
Based on our previous results and current findings, we can now propose a complete model of the mechanochemical cycle of the φ29 packaging motor, as depicted in Figure 7. At the end of the burst phase, all five motor subunits contain ADPs from the previous hydrolysis cycle. At the beginning of the new dwell phase, the special subunit makes the crucial phosphate contact, causing the release of its ADP. The subsequent docking and tight binding of ATP triggers the release of ADP from the next subunit. Coordinated release and binding events then repeat around the ring until all five subunits have replaced ADP with ATP. Next, the special subunit is the first to hydrolyze ATP, breaking its electrostatic contact with the DNA and triggering the sequential hydrolysis, Pi release, and 2.5 bp DNA translocation by the remaining four subunits.
Figure 7. Complete Mechanochemical Model of the φ29 ATPase.
At the end of the burst, all subunits are ADP-bound (“D” label). At the beginning of the dwell, the motor makes an electrostatic contact with two backbone phosphates (small red circles) on the dsDNA substrate (inside the ATPase ring). This unique contact determines the identity of the special subunit (yellow label “s”). The formation of the electrostatic contact facilitates ADP release by the special subunit. Subsequent ATP (“T” label) binding and ADP release events are interlaced, with ATP binding to one subunit enabling ADP release from its neighbor. After all five subunits have bound ATP, the special subunit hydrolyzes ATP (“D•Pi” label), releases Pi, and uses the hydrolysis free-energy to break the electrostatic contact with DNA, triggering the burst phase. During the burst, the remaining four ATP-bound subunits sequentially hydrolyze ATP, release Pi, and translocate DNA by 2.5 bp. The motor-DNA geometry (10.0 bp burst size versus 10.4 bp dsDNA helical pitch) favors a mechanism in which the same subunit is special in consecutive cycles.
The model above depicts a ring ATPase with multiple levels of coordination among the catalytic cycles of individual subunits. First, ADP release and ATP binding occur in an interlaced and sequential fashion. It is intuitive to see how binding and release events can be kinetically coupled between nearest neighbors in a ring structure: ATP tight binding in one subunit may provide the strain energy to open the binding pocket of the following subunit, facilitating ADP release (Wang and Oster, 1998; Yu et al., 2010). Significantly, our finding that ATP binding and ADP release are interlaced during the dwell explains why we experimentally determined an ATP-binding Hill coefficient of n = 1 (Chemla et al., 2005; Moffitt et al., 2009), in apparent contradiction with a highly coordinated motor operation, because only one site is vacant and available for ATP docking at any given time during the dwell and tight-binding represents a largely irreversible step (Moffitt et al., 2009).
Second, hydrolysis also occurs sequentially around the ring. In related ring NTPases such as the T7 gp4 helicase and the φ12 P4 packaging motor, it has been suggested that hydrolysis of the preceding subunit drives the insertion of its arginine finger into the next catalytic site, accelerating the next hydrolysis event (Kainov et al., 2008; Singleton et al., 2000). It is conceivable that the same mechanism is employed by φ29 gp16, as comparative genomic studies have identified a highly conserved arginine finger (R146) (Burroughs et al., 2007).
Finally, the Pi release events could either be strictly interlaced with hydrolysis or take place stochastically following hydrolysis and preceding ADP release. The great processivity of the motor and intricate coordination of other chemical events make the stochastic Pi release model less likely.
Implications for Other Ring Motors
The operation of ASCE ring NTPases has been proposed to occur through either a rotary (Adachi et al., 2007; Adelman et al., 2006; Crampton et al., 2006; Enemark and Joshua-Tor, 2006; Mancini et al., 2004; Massey et al., 2006; Singleton et al., 2000; Sun et al., 2008; Thomsen and Berger, 2009), a concerted (Gai et al., 2004), or a stochastic mechanism (Martin et al., 2005), which have been seen as mutually exclusive of each other. In the model we present here for the φ29 motor, nucleotide binding, hydrolysis, and release events all proceed from one subunit to the next in an ordinal fashion, consistent with a canonical rotary mechanism. However, the level of detail attained in our study also reveals elements that are typically associated with either the concerted or the stochastic mechanism. First, observations of an “all-ATP-bound” and an “all-ADP-bound” state in the crystal structures of the SV40 LTag helicase have been used to support a concerted mechanism for this motor in which all subunits turnover nucleotides simultaneously (Gai et al., 2004). We note that in our model, the motor also adopts an “all-ATP-bound” and an “all-ADP-bound” state at certain points in the cycle, even though nucleotide turnover is sequential. Second, several motors, such as ClpX and MCM helicases, have been shown to tolerate multiple inactive subunits, a phenomenon that has been rationalized by complete or partial stochastic models (Ilves et al., 2010; Martin et al., 2005; Moreau et al., 2007). Here we show that the φ29 motor, although highly coordinated, exhibits flexibility reminiscent of a stochastic mechanism, bypassing the usual coordination scheme and allowing alternative subunits to translocate DNA when one subunit is temporarily inactivated. A mechanism that allows such flexibility might be particularly useful for overcoming mechanical or chemical obstacles. The combination of coordination and adaptability may be a general design feature shared by ring NTPases, and may serve as a cautionary tale against strict canonical mechanisms which need not be mutually exclusive: motors may exhibit features from multiple mechanisms to better suit their unique biological functions.
EXPERIMENTAL PROCEDURES
Sample Preparation
Fiberless proheads, ATPase gp16, and genomic DNA were isolated as described previously (Zhao et al., 2008). Genomic DNA was digested with ClaI or XbaI (New England Biolabs) and biotinylated using Klenow exo-(New England Biolabs) to fill in the overhang with biotinylated nucleotides (Invitrogen). Proheads were partially packaged with biotinylated genomic DNA and stalled with ATPγS. The packaging buffer contained 25 mM Tris-HCl, pH 7.8, 50 mM NaCl, and 5 mM MgCl2. Sodium orthovanadate stocks (100 mM) were prepared in H2O, adjusted to pH 10 with 6 N HCl, and diluted to desired concentration prior to use (Goodno, 1982).
Optical Trapping
High-resolution packaging measurements were conducted on a dual-trap optical tweezers instrument as described previously (Bustamante et al., 2008). Tethers were formed between a 0.90-µm-diameter streptavidin-coated bead and a 0.88-µm-diameter anti-capsid-antibody-coated bead (Spherotech) held in separate optical traps. Packaging was restarted in an ATP-containing buffer, and DNA translocation by the motor was determined from the decrease in the bead-to-bead distance. All packaging experiments were conducted in a semi-passive mode in which the distance between the two traps was adjusted periodically so that the tension applied to the motor was kept within 7–12 pN. An oxygen scavenging system (100 µg ml−1 glucose oxidase, 20 µg ml−1 catalase, and 5 mg ml−1 dextrose; Sigma-Aldrich) was included in the buffer to prevent the formation of reactive singlet oxygen.
Residence Time Histogram Analysis
Raw 2,500 Hz data were filtered and decimated to 250 Hz. Each filtered point was represented as a Gaussian centered at the mean of the data, with a width equal to the local standard error of the unfiltered data. The residence time histogram was constructed by summing up the Gaussian representations of all filtered data points. Simulated packaging traces for residence time histograms were generated using the experimentally measured dwell-time distribution and signal-to-noise ratio.
Step Finding Algorithm
High-resolution packaging traces were selected based on their pairwise distance distribution (PWD) (Figures S1A and S1B). A modified Schwartz Information Criterion (SIC) method was used to find steps (Kalafut and Visscher, 2008) (Figure S1C). At saturating [ATP] and in the absence of analogs, >99% of all dwells were shorter than 500 ms. Thus dwells longer than 500 ms were considered as analog-induced pauses.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Dangkulwanich, T. Ishibashi, C. Kaiser, N. Liu, R. Maillard, M. Morais, and M. Sen for critical discussions. This work was supported in part by grants from the National Institutes of Health (GM071552 to C.B., GM059604 and DE003606 to S.G.) and the U.S. Department of Energy, Office of Basic Energy Sciences, Division of Materials Sciences and Engineering under Contract No. DE-AC02-05CH11231 (single molecule force measurement) to C.B.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, five figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2012.10.031.
REFERENCES
- Aathavan K, Politzer AT, Kaplan A, Moffitt JR, Chemla YR, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Substrate interactions and promiscuity in a viral DNA packaging motor. Nature. 2009;461:669–673. doi: 10.1038/nature08443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- Adachi K, Oiwa K, Nishizaka T, Furuike S, Noji H, Itoh H, Yoshida M, Kinosita K., Jr Coupling of rotation and catalysis in F(1)-ATPase revealed by single-molecule imaging and manipulation. Cell. 2007;130:309–321. doi: 10.1016/j.cell.2007.05.020. [DOI] [PubMed] [Google Scholar]
- Adelman JL, Jeong YJ, Liao JC, Patel G, Kim DE, Oster G, Patel SS. Mechanochemistry of transcription termination factor Rho. Mol. Cell. 2006;22:611–621. doi: 10.1016/j.molcel.2006.04.022. [DOI] [PubMed] [Google Scholar]
- Baird CL, Harkins TT, Morris SK, Lindsley JE. Topoisomerase II drives DNA transport by hydrolyzing one ATP. Proc. Natl. Acad. Sci. USA. 1999;96:13685–13690. doi: 10.1073/pnas.96.24.13685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman ML, Bell SP, Schwacha A. Subunit organization of Mcm2–7 and the unequal role of active sites in ATP hydrolysis and viability. Mol. Cell. Biol. 2008;28:5865–5873. doi: 10.1128/MCB.00161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Iyer LM, Aravind L. Comparative genomics and evolutionary trajectories of viral ATP dependent DNA-packaging systems. Genome Dyn. 2007;3:48–65. doi: 10.1159/000107603. [DOI] [PubMed] [Google Scholar]
- Bustamante C, Chemla YR, Forde NR, Izhaky D. Mechanical processes in biochemistry. Annu. Rev. Biochem. 2004;73:705–748. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- Bustamante C, Chemla YR, Moffitt JR. High-resolution dual-trap optical tweezers with differential detection. In: Selvin PR, Ha T, editors. Single-Molecule Techniques: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2008. pp. 297–324. [Google Scholar]
- Carter AP, Cho C, Jin L, Vale RD. Crystal structure of the dynein motor domain. Science. 2011;331:1159–1165. doi: 10.1126/science.1202393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens SR. The DNA-packaging nanomotor of tailed bacteriophages. Nat. Rev. Microbiol. 2011;9:647–657. doi: 10.1038/nrmicro2632. [DOI] [PubMed] [Google Scholar]
- Chemla YR, Aathavan K, Michaelis J, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Mechanism of force generation of a viral DNA packaging motor. Cell. 2005;122:683–692. doi: 10.1016/j.cell.2005.06.024. [DOI] [PubMed] [Google Scholar]
- Crampton DJ, Mukherjee S, Richardson CC. DNA-induced switch from independent to sequential dTTP hydrolysis in the bacteriophage T7 DNA helicase. Mol. Cell. 2006;21:165–174. doi: 10.1016/j.molcel.2005.11.027. [DOI] [PubMed] [Google Scholar]
- Duderstadt KE, Chuang K, Berger JM. DNA stretching by bacterial initiators promotes replication origin opening. Nature. 2011;478:209–213. doi: 10.1038/nature10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- Gai D, Zhao R, Li D, Finkielstein CV, Chen XS. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell. 2004;119:47–60. doi: 10.1016/j.cell.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell. 2009;139:744–756. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodno CC. Myosin active-site trapping with vanadate ion. Methods Enzymol. 1982;85(Pt B):116–123. doi: 10.1016/0076-6879(82)85014-3. [DOI] [PubMed] [Google Scholar]
- Guo P, Peterson C, Anderson D. Prohead and DNA-gp3-dependent ATPase activity of the DNA packaging protein gp16 of bacteriophage phi 29. J. Mol. Biol. 1987;197:229–236. doi: 10.1016/0022-2836(87)90121-5. [DOI] [PubMed] [Google Scholar]
- Hetherington CL, Moffitt JR, Jardine PJ, Bustamante C. Viral DNA packaging motors. In: Egelman E, editor. Comprehensive Biophysics. San Diego, CA: Academic Press; 2012. pp. 420–446. [Google Scholar]
- Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2–7 helicase by association with Cdc45 and GINS proteins. Mol. Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsathitphaisarn O, Wing RA, Eliason WK, Wang J, Steitz TA. The hexameric helicase DnaB adopts a nonplanar conformation during translocation. Cell. 2012;151:267–277. doi: 10.1016/j.cell.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Johnson A, O’Donnell M. Ordered ATP hydrolysis in the gamma complex clamp loader AAA+ machine. J. Biol. Chem. 2003;278:14406–14413. doi: 10.1074/jbc.M212708200. [DOI] [PubMed] [Google Scholar]
- Kainov DE, Mancini EJ, Telenius J, Lísal J, Grimes JM, Bamford DH, Stuart DI, Tuma R. Structural basis of mechanochemical coupling in a hexameric molecular motor. J. Biol. Chem. 2008;283:3607–3617. doi: 10.1074/jbc.M706366200. [DOI] [PubMed] [Google Scholar]
- Kalafut B, Visscher K. An objective, model-independent method for detection of non-uniform steps in noisy signals. Comput. Phys. Commun. 2008;179:716–723. [Google Scholar]
- Lyubimov AY, Strycharska M, Berger JM. The nuts and bolts of ring-translocase structure and mechanism. Curr. Opin. Struct. Biol. 2011;21:240–248. doi: 10.1016/j.sbi.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini EJ, Kainov DE, Grimes JM, Tuma R, Bamford DH, Stuart DI. Atomic snapshots of an RNA packaging motor reveal conformational changes linking ATP hydrolysis to RNA translocation. Cell. 2004;118:743–755. doi: 10.1016/j.cell.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Rebuilt AAA + motors reveal operating principles for ATP-fuelled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- Massey TH, Mercogliano CP, Yates J, Sherratt DJ, Löwe J. Double-stranded DNA translocation: structure and mechanism of hexameric FtsK. Mol. Cell. 2006;23:457–469. doi: 10.1016/j.molcel.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Moffitt JR, Chemla YR, Aathavan K, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Chemla YR, Bustamante C. Methods in statistical kinetics. Methods Enzymol. 2010;475:221–257. doi: 10.1016/S0076-6879(10)75010-2. [DOI] [PubMed] [Google Scholar]
- Morais MC, Koti JS, Bowman VD, Reyes-Aldrete E, Anderson DL, Rossmann MG. Defining molecular and domain boundaries in the bacteriophage phi29 DNA packaging motor. Structure. 2008;16:1267–1274. doi: 10.1016/j.str.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau MJ, McGeoch AT, Lowe AR, Itzhaki LS, Bell SD. ATPase site architecture and helicase mechanism of an archaeal MCM. Mol. Cell. 2007;28:304–314. doi: 10.1016/j.molcel.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Mott ML, Erzberger JP, Coons MM, Berger JM. Structural synergy and molecular crosstalk between bacterial helicase loaders and replication initiators. Cell. 2008;135:623–634. doi: 10.1016/j.cell.2008.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu. Rev. Genet. 2008;42:647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- Schnitzer MJ, Block SM. Statistical kinetics of processive enzymes. Cold Spring Harb. Symp. Quant. Biol. 1995;60:793–802. doi: 10.1101/sqb.1995.060.01.085. [DOI] [PubMed] [Google Scholar]
- Segel IH. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems. New York, NY: John Wiley & Sons, Inc.; 1975. [Google Scholar]
- Sharma S, Davidson AL. Vanadate-induced trapping of nucleotides by purified maltose transport complex requires ATP hydrolysis. J. Bacteriol. 2000;182:6570–6576. doi: 10.1128/jb.182.23.6570-6576.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- Skordalakes E, Berger JM. Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading. Cell. 2003;114:135–146. doi: 10.1016/s0092-8674(03)00512-9. [DOI] [PubMed] [Google Scholar]
- Sun S, Kondabagil K, Draper B, Alam TI, Bowman VD, Zhang Z, Hegde S, Fokine A, Rossmann MG, Rao VB. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135:1251–1262. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Thomsen ND, Berger JM. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell. 2009;139:523–534. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Oster G. Energy transduction in the F1 motor of ATP synthase. Nature. 1998;396:279–282. doi: 10.1038/24409. [DOI] [PubMed] [Google Scholar]
- Yang Q, Catalano CE. A minimal kinetic model for a viral DNA packaging machine. Biochemistry. 2004;43:289–299. doi: 10.1021/bi035532h. [DOI] [PubMed] [Google Scholar]
- Yu J, Moffitt J, Hetherington CL, Bustamante C, Oster G. Mechanochemistry of a viral DNA packaging motor. J. Mol. Biol. 2010;400:186–203. doi: 10.1016/j.jmb.2010.05.002. [DOI] [PubMed] [Google Scholar]
- Zhao W, Morais MC, Anderson DL, Jardine PJ, Grimes S. Role of the CCA bulge of prohead RNA of bacteriophage ø29 in DNA packaging. J. Mol. Biol. 2008;383:520–528. doi: 10.1016/j.jmb.2008.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.