Abstract
Objectives
This study sought to ascertain the relationship of 9p21 locus with: 1) angiographic coronary artery disease (CAD) burden; and 2) myocardial infarction (MI) in individuals with underlying CAD.
Background
Chromosome 9p21 variants have been robustly associated with coronary heart disease, but questions remain on the mechanism of risk, specifically whether the locus contributes to coronary atheroma burden or plaque instability.
Methods
We established a collaboration of 21 studies consisting of 33,673 subjects with information on both CAD (clinical or angiographic) and MI status along with 9p21 genotype. Tabular data are provided for each cohort on the presence and burden of angiographic CAD, MI cases with underlying CAD, and the diabetic status of all subjects.
Results
We first confirmed an association between 9p21 and CAD with angiographically defined cases and control subjects (pooled odds ratio [OR]: 1.31, 95% confidence interval [CI]: 1.20 to 1.43). Among subjects with angiographic CAD (n = 20,987), random-effects model identified an association with multivessel CAD, compared with those with single-vessel disease (OR: 1.10, 95% CI: 1.04 to 1.17)/copy of risk allele). Genotypic models showed an OR of 1.15, 95% CI: 1.04 to 1.26 for heterozygous carrier and OR: 1.23, 95% CI: 1.08 to 1.39 for homozygous carrier. Finally, there was no significant association between 9p21 and prevalent MI when both cases (n = 17,791) and control subjects (n = 15,882) had underlying CAD (OR: 0.99, 95% CI: 0.95 to 1.03)/risk allele.
Conclusions
The 9p21 locus shows convincing association with greater burden of CAD but not with MI in the presence of underlying CAD. This adds further weight to the hypothesis that 9p21 locus primarily mediates an atherosclerotic phenotype.
Keywords: 9p21, angiography, coronary artery disease, meta-analysis, myocardial infarction, single nucleotide polymorphism
Genome-wide association studies (GWAS) first identified the 9p21 locus as associating with coronary heart disease (CHD) in 2007 (1-3). A plethora of replication studies have since confirmed and validated this association in a series of different ethnic populations, making this the most robust genetic finding for CHD to date. The need for large study samples in many of these studies has led to phenotypic heterogeneity with inclusion of cases with acute or stable clinical presentations with presumed healthy control populations and varying definitions of CHD, including clinical or noninvasive diagnosis of coronary artery disease (CAD), angiographic CAD, validated myocardial infarction (MI), or a combination of these (4,5). This lack of phenotypic clarity has resulted in uncertainty about the primary phenotype mediated by the 9p21 locus, specifically whether it predisposes to atherosclerosis or promotes a more abrupt plaque rupture or thrombotic process leading to MI. This in turn has hampered contextualization of early functional studies attempting to resolve the underlying mechanism (6).
There have been several attempts to tease apart the closely related phenotypes of CAD and MI (7-9). In the most comprehensive analysis to date, Reilly et al. (8) demonstrated that, although 11 variants had shown robust association with MI in GWAS when compared with healthy control subjects, they did not associate with MI when both cases and control subjects had underlying CAD. It was thus proposed that the primary association for these variants was likely to be with development of CAD rather than predisposition to plaque rupture or thrombosis per se. It follows then that carriers of the risk allele at the 9p21 locus should demonstrate a greater burden of coronary atherosclerosis compared with non-risk carriers. Although studies with computerized tomography have demonstrated a correlation between 9p21 and greater coronary artery calcification, indicating a role in predisposing to atheroma formation (10,11), studies using invasive coronary angiography as a more direct and widely available means of visualizing plaque have demonstrated discrepant results, leading to ongoing uncertainty with regard to the mechanism of risk (12-15).
To address this lack of consistency we sought to establish a collaboration of genetic studies and perform a comprehensive meta-analysis of the association between 9p21 and angiographically defined CAD burden as well as to replicate the lack of association with superimposed MI in subjects with underlying CAD.
Methods
Search strategy and selection criteria
We performed a systematic published data search for studies of 9p21 variation in relation to CAD/MI, published before June 2011 on MEDLINE and EMBASE, combined with cross references and manual searches. Search terms included “coronary artery disease,” “myocardial infarction,” or “atherosclerosis,” in combination with “9p21.” No language restrictions were used. A hand-search of articles and cited reference search were also performed to identify all articles that cited the index publication. Experts contributed to identification of cohorts with published and unpublished data (M.R./N.J.S.). For each included cohort, data were tabulated for angiographic presence or absence of CAD, burden of CAD, number of MI cases in subjects with underlying CAD, and the diabetic status of all subjects. Published data search, data collection/abstraction, and entry were performed independently and reconciled by 2 trained investigators (K.C. and R.P.).
For the analysis of 9p21 association with atherosclerosis burden, only studies with coronary angiographic data in relation to 9p21 single nucleotide polymorphisms (SNPs) were included. For the analysis of 9p21 association with MI in subjects with underlying CAD, studies were included if they had information for both CAD status (angiographic or clinical diagnosis) and MI sufficient to classify individuals as MI+/CAD+ or MI−/CAD+. Myocardial infarction was defined with standard criteria on the basis of clinical symptoms, electrocardiography, and raised troponin (16). Coronary artery disease was defined as angiographically documented CAD (≥50% or ≥70% luminal stenosis on visual estimation) or clinical CAD on the basis of physician diagnosis with history of prior coronary revascularization. Studies that used only the single phenotypes of CAD or MI or an indistinguishable admixture of phenotypes in primary comparisons with healthy control populations were not included.
Data sources
We contacted the principal investigators of the identified studies/cohorts meeting inclusion criteria to obtain complete published and unpublished data for the prevalence of MI in subjects with CAD as well as angiographic data where available. For 2 of the eligible studies, sufficient data were unavailable in the published reports and could not be obtained on request (Online Table 1). Details of all included study cohorts are provided in the Online Appendix. From the individual studies, we received (or abstracted) information about: 1) angiographic CAD, specifically on the number of diseased major epicardial vessels and criteria used to define disease (≥50% or ≥70% luminal stenosis on visual estimation); and 2) prevalence of MI in the presence or absence of CAD by 9p21 genotype. We also collected details on race, sex, age, diabetes status (documented history of diabetes), and the identification (rs number) of the particular SNP genotyped.
Statistical analysis
Statistical analysis followed guidelines from the Cochrane Handbook for Systematic Reviews and HuGE Review Handbook for meta-analysis of genetic association studies (17,18). Random effect models (DerSimonian and Laird) were used to estimate the pooled per-allele and per-genotype odds ratios (ORs) and 95% confidence intervals (CIs) according to the 9p21 polymorphism genotypes (RR, homozygous risk allele carrier; RW, heterozygous risk allele carrier; WW, homozygous reference group) with results presented as Forest plots (19).
Meta-analysis was first performed for association between 9p21 and angiographic CAD against angiographic control subjects with the cohorts that had data available for both. Angiographic control subjects were defined as having completely smooth unobstructed vessels or <10% luminal stenosis anywhere in the coronary tree. Subsequently we examined association with angiographic CAD burden, by comparing subjects with coronary atherosclerosis/stenosis in a single-vessel with those with 2-vessel, 3-vessel, or multi-vessel (≥1 vessels) disease. Subgroup analyses were planned a priori to investigate the association with multivessel disease by study size, race, and diabetes status. Finally, we estimated a pooled estimate for odds of prevalent MI in the presence of underlying CAD (all CAD definitions), by comparing those with MI and CAD (MI+/CAD+) with those without MI but with CAD (MI−/CAD+). For the latter we also performed a priori subgroup analysis for alternate classifications of CAD, including: 1) clinical diagnosis alone; 2) ≥50% stenosis on coronary angiography; or 3) ≥70% stenosis on coronary angiography.
In all meta-analyses we used Cochran’s Q test to evaluate the degree of heterogeneity between studies and the I2 statistic to measure the proportion of total variation in study estimates attribute to heterogeneity (20). This was further investigated through subgroup analyses on the basis of cohort size, ethnicity, and diabetic status of individual patients. In addition to subgroup analyses by cohort size, possible publication bias was also explored by analysis of a funnel plot and Egger’s test (21). We identified a priori that a major source of between-study heterogeneity is likely to arise due to use of different 9p21 markers in different studies. The markers used in this series of studies included rs1333049 (C), rs10757278 (G), and rs2383206 (G), which are all in tight linkage disequilibrium (LD) (Online Fig. S1). Therefore, we used a random effects model to permit heterogeneity in the association being measured between studies. This delivers more conservative CIs than a fixed effects model. However, to further model individual 9p21 SNP effects we also applied a Bayesian method, developed to address the problem of meta-analyzing gene-disease association studies that report different SNPs. Rather than including all SNPs in a single univariate test under an assumption of exchangeability (as in the aforementioned random effects analysis), separate effects are modeled for each SNP as part of an adjusted multivariate model. This is made possible by imputing the unobserved SNPs in each study according to LD information from HapMap. Further details are available in the Online Appendix Methods section.
The random effects meta-analyses were performed with a STATA software package (version 11.0, College Station, Texas). Bayesian analysis was performed with the Java package “BayesMeta,” described in Newcombe et al. (22). Post hoc power calculations were performed with G*power software package. Figures were created with STATA.
Results
Characteristics of the study cohorts
We identified 114 studies matching our database search terms, for association between 9p21 and CAD and/or MI with a further 2 identified through manual searching of references. Of these, 89 were excluded, because they were reviews, letters, or repeat studies on the same cohorts. A further 4 studies were excluded, because they only examined associations between CAD/MI with healthy control subjects. Of the remaining 23 studies, 2 had sufficient data published for this meta-analysis. We contacted the PIs for the 21 other studies and received information from 19. In total, sufficient data for meta-analysis were obtained for 21 studies consisting of 29 unique cohorts (Fig. 1).
Figure 1. Published Data Search Results Identifying Association Studies for 9p21 and CAD/MI.
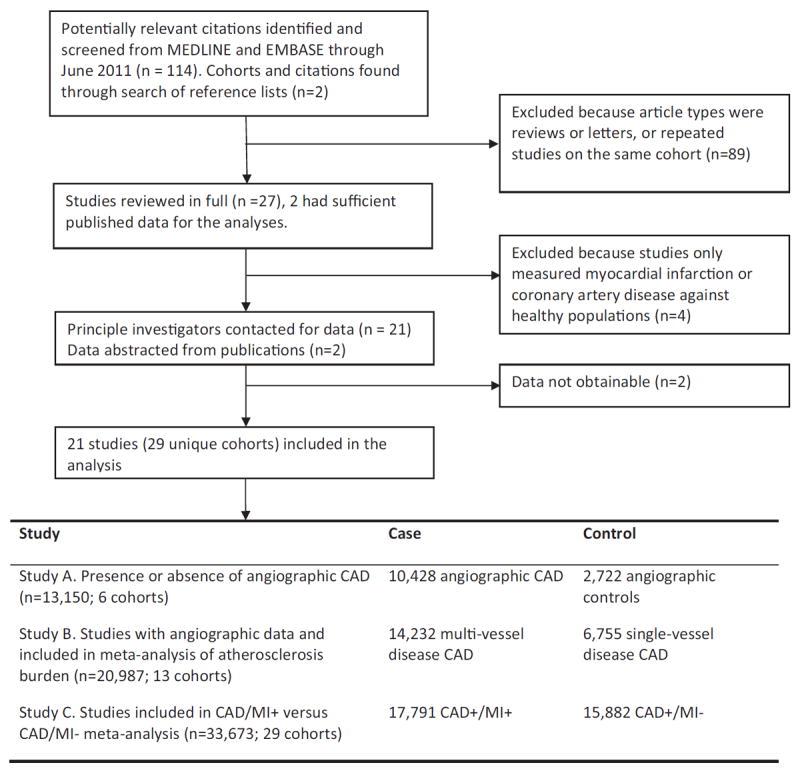
CAD = coronary artery disease; MI = myocardial infarction.
Of the 21 studies included, there were 6 cohorts (n = 13,150) available for study A (presence/absence angiographic CAD), 13 cohorts (n = 20,987) for study B (burden of angiographic CAD), and 29 cohorts (n = 33,673) for study C (prevalent MI in CAD subjects) (Fig. 1). Table 1 lists age, sex, and ethnicity distributions of the different cohorts. The mean age for the populations ranged from 47.6 to 70.0 years; 73.1% were male, with 86.1% of European ancestry, 11.1% Asians, 1.5% Hispanics, and 0.9% Africans; 52.8% of the pooled population had a history of MI (Table 1). Genotype frequency data were obtained for all studies except 1, for which only summary statistics were available (23). The genotype distributions for all 9p21 markers were in agreement with Hardy-Weinberg equilibrium, with overall risk allele frequencies similar to HapMap and published reports.
Table 1.
Description of Studies Included in Meta-Analysis
| First Author (Ref. #) | Cohort | Country | SNP (effect allele) |
Risk Allele Frequency |
Mean or Cutoff Age (yrs) | Ethnicity | Male% | Coronary Angiography | No. of Subjects | CAD Definitions | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1-Vessel Disease |
2-Vessel Disease |
3-Vessel Disease |
CAD/MI+ (nondiabetic subjects) |
CAD/MI− (nondiabetic subjects) |
CAD Definition | ||||||||
| Assimes et al. (11) | ADVANCE | U.S. | rs10757278 (G) | 0.53 | Mean age 58.2. Early onset CAD were 18 to 45 yrs (men) and 18 to 55 women, older onset were any age >18 but also not eligible to be included in early onset group | European ancestry/African/East Asian/South Asian | 67.7% | — | — | — | 1,005 (809) | 547 (432) | Clinical/symptomatic |
| Broadbent et al. (23)* | PROCARDIS | Germany, Italy, Sweden, and U.K. | rs1333049 (C) | 0.52 | Mean age (at date of recruitment) 62.2 yrs | European ancestry | 72.7% | — | — | — | 2,890 (2449) | 1361 (1158) | Clinical/symptomatic |
| Chen et al. (15) | LCAS | U.S. | rs1333049 (C) | 0.55 | Mean age 59.3±7.7, all <75 yrs | European ancestry | 84.0% | — | — | — | 141 | 191 | Angiography (30%–75% stenosis) |
| Dandona et al. (12) | OHGS (Early onset) | U.S. | rs1333049 (C) | 0.55 | Mean age 56.1±9.6, male <55 yrs and female <65 yrs | European ancestry | 70.8% | 518 | 563 | 633 | 517 (517) | 433 (433) | Angiography (≥50% stenosis) |
| OHGS (Late onset) | U.S. | rs1333049 (C) | 0.55 | Mean age 70.0±8.0 | European ancestry | 82.6% | — | — | — | 271 (271) | 493 (493) | Angiography (≥50% stenosis) | |
| Ellis et al. (9) | CDCS | New Zealand | rs1333049 (C) | 0.51 | Mean age 67.1±12.5 | European ancestry/Asian/ Pacific Island ancestry | 69.2% | 201 | 205 | 272 | 854 (703) | 162 (142) | Angiography (≥50% stenosis) |
| Gong et al. (25) | INVEST | U.S. | rs1333049 (C) | 0.54 | Mean age 66±10 | European ancestry/African/Hispanic | 44.2% | — | — | — | 443 (344) | 997 (713) | Clinical/symptomatic |
| Hauser et al. | CATHGEN | U.S. | rs10757278 (G) | 0.52 | Mean age 64.4±11.0 (CAD/MI+ group), 61.02±10.65 (CAD/MI− group) | European ancestry | 73.0% | 1,049 | 792 | 1180 | 1,357 (941) | 1664 (1,159) | Angiography (≥50% stenosis) |
| Hazen et al. | Cleveland GB | USA | rs1333049 (C) | 0.54 | Mean age 61.7.9±11.1 | European ancestry | 74.0% | 745 | 672 | 1,054 | 1,332 (1039) | 1079 (806) | Angiography (≥50% stenosis) |
| Hinohara et al. (36) | — | Japan | rs1333049 (C) | 0.56 | Mean age 59.3±10.1 | Japanese | 83.3% | 245 | 170 | 181 | 572 (395) | 47 (29) | Angiography (≥50% stenosis) |
| Hinohara et al. (36) | — | Korea | rs1333049 (C) | 0.52 | Mean age 61.2±11.1 | Korean | 76.4% | 243 | 162 | 117 | 433 (222) | 260 (51) | Angiography (≥50% stenosis) |
| Hoppmann et al. (37) | — | Germany | rs1333049 (C) | 0.51 | Mean age 66.3±10.5 | European ancestry | 78.2% | 364 | 579 | 1,085 | 733 | 1295 | Angiography (≥50% stenosis) |
| Horne et al. (7) | IHCS (Sample set) | U.S. | rs2383206 (C) | 0.54 | Mean age 63±10 yrs (range 21–78 yrs) | European ancestry | 77.0% | 666 | 473 | 609 | 869 (613) | 879 (705) | Angiography (≥70% stenosis) |
| IHCS (Replication set) | U.S. | rs2383206 (C) | 0.53 | Mean age 61±10 yrs (range 24–75 yrs) | European ancestry | 73.0% | 429 | 300 | 285 | 507 (347) | 507 (368) | Angiography (≥70% stenosis) | |
| Krawczak et al. (38) | PopGen | Germany | rs1333049 (C) | 0.51 | Mean age 47.6±6.3 | European ancestry | 79.4% | — | — | — | 1,366 (1,111) | 718 (584) | Angiography (≥70% stenosis) |
| Muendlein et al. (39) | — | Austria | rs1333049 (C) | 0.55 | Mean age 65.1± 10.6; male 64.1 ± 10.8; female 68.6 ± 9.2 | European ancestry | 77.1% | 362 | 290 | 262 | 357 (248) | 576 (429) | Angiography (≥50% stenosis) |
| Patel et al. (13) | EmCB | U.S. | rs10757278 (G) | 0.51 | Mean age 63.9±11 | European ancestry | 67.8% | 498 | 545 | 1,314 | 891 (611) | 864 (596) | Angiography (≥50% stenosis) |
| Reilly et al. (8) | MEDSTAR | U.S. | rs1333049 (C) | 0.54 | All aged <61, male <55 and female <60 | European ancestry | 71.9% | 330 | 290 | 204 | 421 (294) | 454 (308) | Angiography (≥50% stenosis) |
| Reilly et al. (8) | PENNCATH | U.S. | rs1333049 (C) | 0.57 | All aged <66, male <60 and female <65 | European ancestry | 75.0% | 247 | 258 | 336 | 470 (123) | 463 (111) | Angiography (≥50% stenosis) |
| Samani et al. (1) | WTCCC | U.K. | rs1333049 (C) | 0.55 | All Aged <66 | European ancestry | 79.5% | — | — | — | 1,376 (1214) | 532 (467) | Clinical/symptomatic |
| Shen et al. (40) | SK-CAD | Korea | rs10757278 (G) | 0.53 | Mean age 63.7±10.1 | Korean | 70.9% | — | — | — | 603 (410) | 485 (332) | Angiography (≥70% stenosis) |
| Wang et al. (41) | — | China | rs1333049 (C) | 0.54 | Mean age 66.81±9.8 | Chinese | 56.1% | 419 | 389 | 357 | 555 (300) | 610 (245) | Angiography (≥50% stenosis) |
| Ye et al. (42) | SAS | U.K. | rs1333049 (C) | 0.52 | Mean age 63.2±9.95 | European ancestry | 76.6% | 439 | 373 | 282 | 592 (513) | 501 (434) | Angiography (≥50% stenosis) |
Allele frequencies were estimated from computed odds ratio.
CDCS = Coronary Disease Cohort Study; Cleveland GB = Cleveland GeneBank Study; EmCB = Emory Cardiovascular Biobank; IHCS = Utah Intermountain Heart Collaborative Study; LCAS = Lipoprotein and Coronary Atherosclerosis Study; INVEST = International Verapamil SR Trandolapril Study Genetic Substudy; OHGS = Ottawa Heart Genomics Study; PROCARDIS = Precocious Coronary Artery Disease; SAS = Southampton Atherosclerosis Study; WTCCC = Wellcome Trust Case-Control Consortium.
Study A: 9p21 association with angiographic CAD presence/absence
We first confirmed an association between the 9p21 risk locus and presence or absence of angiographic CAD, with a subset of studies in which genotype data were also available for those with normal coronary arteries on angiography (n = 10,428 cases, n = 2,722 control subjects). Meta-analysis revealed an overall pooled allelic OR of 1.31 (95% CI: 1.20 to 1.43) for association with angiographic CAD (Online Fig. S2A). Heterozygotes for the risk allele had 39% greater odds of having CAD (OR: 1.39, 95% CI: 1.21 to 1.59), compared with those without any risk alleles, whereas homozygotes for the risk allele had 73% greater odds (OR: 1.73, 95% CI: 1.45 to 2.05) (Online Figs. S2B and S2C).
Study B: 9p21 association with atherosclerosis burden on angiographic CAD
In the combined analysis on all those with angiographic CAD (n = 20,987), the 9p21 risk variant was associated with a greater odds of multivessel disease (n = 14,232) compared with single-vessel disease (n = 6755), with a summary OR of 1.10 (95% CI: 1.04 to 1.17)/risk allele (Fig. 2A). Under genotypic models, heterozygotes had 15% greater odds of having multivessel disease (OR: 1.15, 95% CI 1.04 to 1.26), compared with non-risk allele carriers, whereas homozygotes for the risk allele had 23% greater odds (OR: 1.23, 95% CI: 1.08 to 1.39) (Figs. 2B and 2C). The 9p21 risk allele carriers also showed significantly increased odds of 2-vessel disease over 1-vessel disease (OR: 1.08, 95% CI: 1.02 to 1.13) as well as 3-vessel disease over 2-vessel disease (OR: 1.07, 95% CI: 1.01 to 1.13) (Online Figs. S3 and S4).
Figure 2. Association Between 9p21 and Multivessel CAD as Compared With Single-Vessel CAD.
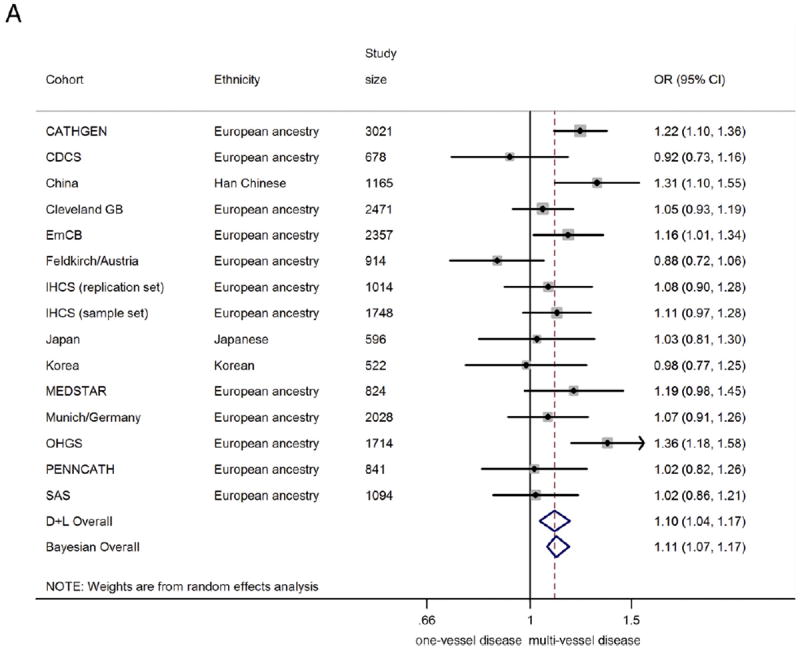
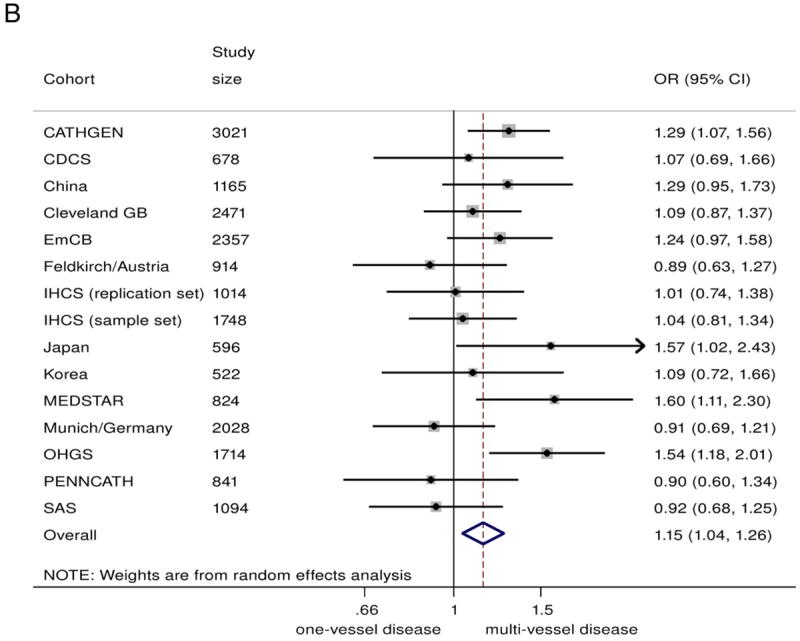
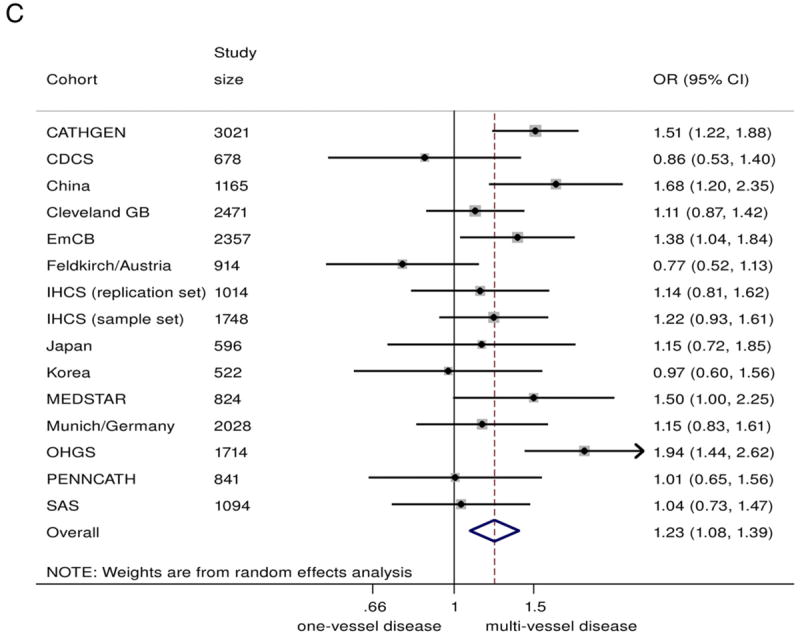
(A) Odds ratio (OR)/copy of risk allele (allelic model).
(B) heterozygote risk versus homozygote non-risk (genotypic model), random-effects analysis;
(C) homozygote risk versus homozygotes non-risk (genotypic model), random-effects analysis. Total n = 20,987; 14,232 multivessel disease, 6,755 single-vessel disease. Non-European ancestry cohorts were not included in the Bayesian analysis. Bayesian analysis showed no distinguishable differences between rs10757278/rs1333049 and rs2383206, for which the marginal effect estimate was nearly identical. CI = confidence interval; Bayesian = Bayesian model; CDCS = Coronary Disease Cohort Study; Cleveland GB = Cleveland GeneBank Study; D+L = DerSimonian and Laird random effects model; EmCB = Emory Cardiovascular Biobank; IHCS = Utah Intermountain Heart Collaborative Study; OHGS = Ottawa Heart Genomics Study; SAS = Southampton Atherosclerosis Study.
There was some evidence of heterogeneity between studies (I2 = 49.5%, Q = 27.7, p = 0.015, Egger test p = 0.058). However, upon exclusion of smaller studies (n < 1,000), the evidence of heterogeneity substantially reduced (I2= 43.2%, Q = 14.1, p = 0.08, Egger test p = 0.69), suggesting that part of the heterogeneity was caused by publication bias. The funnel plot across all studies was also consistent with possible publication bias (Online Fig. S8). Encouragingly, the effect estimate was robust to exclusion of the smaller (n < 1,000) studies: Among the 9 remaining studies (total sample size of n = 16,612) the meta-analyzed OR was 1.15 (95% CI: 1.08 to 1.23).
Although studies of subjects of European ancestry and studies of Asians showed similar effect sizes, the association was nonsignificant for the studies of Asians, likely due to smaller overall sample size (OR: 1.12, 95% CI 0.92 to 1.35, n = 2,283). Diabetes status did not significantly alter the association of 9p21 with greater CAD burden (Fig. 3).
Figure 3. Summary ORs/Copy of 9p21 Risk Allele for Multivessel Versus Single-Vessel CAD.
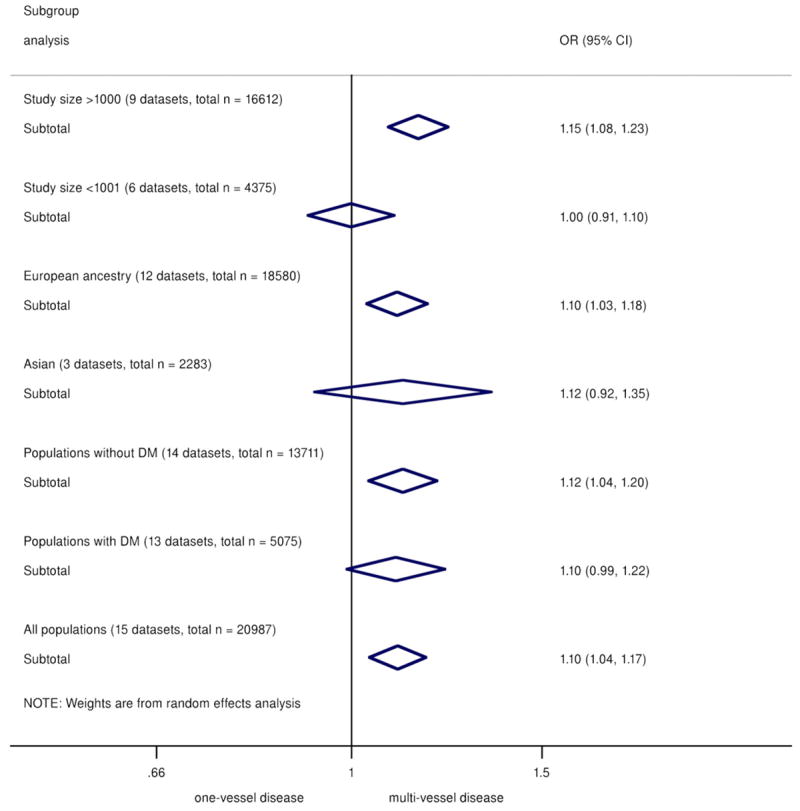
Categorized by subgroups of study size, ethnicity, and diabetes status. CI = confidence interval; DM = diabetes mellitus; OR = odds ratio.
Finally, in the Bayesian analysis that formally models different SNPs as different variables, the SNPs were largely indistinguishable under model selection, suggesting that all tested SNPs are good markers for association with CAD burden. This is perhaps not surprising, given the high levels of LD observed in HapMap (r2= 1 for rs1333049 and rs10757278; and r2= 0.87 for rs1333049 and rs2383206).
Study C: No association between 9p21 and MI in subjects with CAD
The 9p21 SNPs did not show a significant association with MI when comparing MI cases with control subjects consisting of subjects without MI but with underlying CAD (Fig. 4). The summary effect size was estimated at 0.99 (95% CI: 0.95 to 1.03)/risk allele, with no evidence of publication bias (Egger p = 0.843) and little heterogeneity between studies (I2 = 20.9%) (Online Fig. S9). Similar findings were noted in genotypic analysis, with an OR of 0.98 (95% CI: 0.92 to 1.04) for heterozygotes and 0.99 (95% CI: 0.91 to 1.07) for homozygotes (Online Figs. S5 and S6). Subgroup analysis revealed that the findings were not significantly altered by CAD definition (on the basis of clinical definitions or confirmed angiographic stenosis of ≥50% or ≥70%), ethnicity, study size, and diabetes status of the population (Online Fig. S7). Results from the Bayesian meta-analysis were entirely consistent with those described in the preceding text (Online Table 2).
Figure 4. Association Between 9p21 and MI in Patients With Underlying CAD.
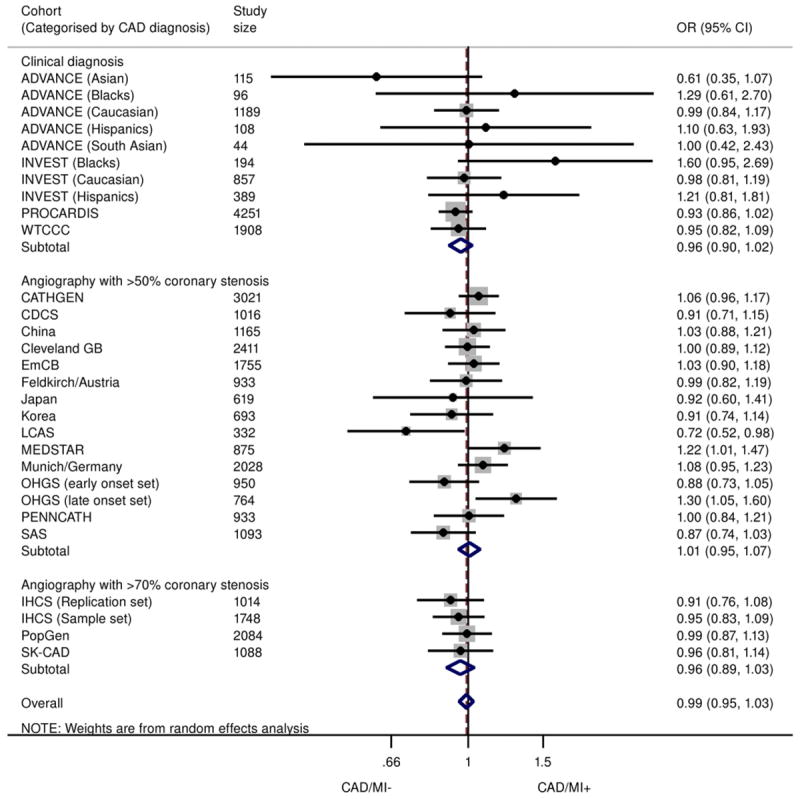
Odds ratios/copy of 9p21 risk allele for MI in subjects with CAD (allelic model), stratified according to CAD definition (clinical, angiographic stenosis ≥50% or ≥70%). Total n = 33,673; 17,791 CAD+/MI+, 15,882 CAD+/MI−. ADVANCE = Atherosclerotic Disease, Vascular Function, and Genetic Epidemiology; CAD/MI+ = cases with history of MI and underlying CAD; CAD/M− = control subjects without history of MI but with underlying CAD; LCAS = Lipoprotein and Coronary Atherosclerosis Study; PROCARDIS = Precocious Coronary Artery Disease; WTCCC = Wellcome Trust Case-Control Consortium; other abbreviations as in Figures 1, 2, and 3.
Discussion
We have demonstrated, through a comprehensive and collaborative meta-analysis, that the 9p21 risk locus is associated with a greater burden of angiographically defined CAD. Homozygotes for the risk allele are 23% more likely to have multivessel disease than single-vessel disease when compared with homozygotes for the non-risk allele. Furthermore, we confirmed that there is no association between 9p21 and MI in the presence of background CAD. These results support the hypothesis that the 9p21 locus primarily promotes atherosclerotic lesion development and functionally significant coronary stenosis rather than precipitating plaque rupture and thrombosis.
Early GWAS demonstrated significant association between 9p21 and both MI and CAD (1-3). Given that MI predominantly occurs in those with CAD, it was initially unclear whether 9p21 promoted the upstream phenotype of atherosclerosis or the later complication of MI. Subsequent studies attempted to refine this phenotype and demonstrated robust association with CAD, including angiographically defined atherosclerosis (7). Three large studies have demonstrated further association between 9p21 and CAD burden with a number of diseased vessels or semi-quantitative methods such as the Gensini score, suggesting that 9p21 promotes progressive atherosclerosis (12,13,24). However, other studies have not confirmed this association, and this lack of consistency has led to difficulties in reconciling association with presence but not extent of CAD (14,15). However, this meta-analysis, which includes almost all published and unpublished reports on 9p21 and angiographic CAD, convincingly demonstrates that 9p21 is associated with greater CAD burden, overall.
Interestingly, subgroup analysis showed that study size significantly influenced the observed effect. Association with multivessel disease was observed in large studies (sample size >1,000) with similar effect size as the overall pooled results but not in smaller studies. Several of the smaller studies reported on association with CAD burden as secondary analysis, not suitably powered to detect differences in a crude phenotype (number of vessels with significant stenoses) combined with a modest genetic effect. Ethnicity meanwhile did not seem to influence the overall association, although the smaller Asian population size rendered a wide CI. Finally, diabetes status showed little modification on the results, which is relevant given the exclusion of diabetic cases in some of the reported studies (12).
Additionally, we demonstrated in a sample size of 33,673 that the 9p21 locus was not associated with prevalent MI when both cases and control subjects had documented CAD. This analysis has been proposed by others as a means to tease out the effect of 9p21 on CAD and MI. Given that MI primarily occurs in the presence of CAD, comparing MI cases with healthy control subjects might also be testing the hypothesis of CAD versus no CAD. Thus, by examining control subjects without MI but with underlying CAD, one can determine whether 9p21 truly associates with an MI phenotype, such as tendency to plaque instability or thrombosis, or not (7,8,14,25). However, if the locus promotes atherosclerosis, then no association would be observed, because the frequency of risk allele would be similar in both cases and control subjects. In the largest such analysis to date, Reilly et al. (8) demonstrated in 9,427 subjects that 9p21 along with other novel genomic variants did not reach statistically significant associations with MI when all subjects had angiographic CAD. Our analysis adds to these findings by including almost 4 times as many subjects. Furthermore, we also ascertained that the various definitions of CAD do not influence this observation, and neither does race, diabetes, or study size.
Therefore, our results support the hypothesis that the 9p21 risk locus primarily promotes an atherosclerotic process and is not associated with a thrombotic or plaque rupture phenotype. These findings are consistent with several lines of evidence, including significant associations with: 1) subclinical atherosclerosis (26); 2) coronary calcification (10,11); 3) carotid atherosclerosis (27); and 4) peripheral arterial disease (28,29). Additionally, these data are also in line with the observations that 9p21 variants are associated with earlier age of CAD onset as well as emerging evidence that 9p21 does not predict risk of incident MI events in those with existing CAD (9,25,30).
Biological studies have also yielded important data suggesting an atherogenic mechanism. The nearest genes to the non-coding 9p21 region include cyclin-dependent kinase inhibitors associated with cell cycling and death, which are relevant to the atherosclerotic process (31). Visel et al. (32) observed, in a mouse knockout model for the orthologous 9p21 region, increased expression of these cyclin-dependent kinase inhibitors (CDKN2A and CDKN2B), with excessive proliferation and diminished senescence of vascular smooth muscle cells (VSMCs). More recently, Kim et al. (33) reported that CDKN2A knockout mice demonstrate smaller aortic atheromatous lesions compared with wild-type mice. Furthermore, the 9p21 variant has been shown to be associated with CDKN2A and CDKN2B expression in VSMCs, VSMC proliferation, and VSMC content in atherosclerotic plaque (34), whereas proliferation of neointimal content of macrophages has also been described (35). These are all key pathological changes implicated in the early development and progression of atherosclerotic plaques. The exact mechanistic pathway leading to these events remains to be elucidated, although the regulatory role of the 9p21 region is likely to involve antisense non-coding ribonucleic acid (ANRIL) at a transcription level (34) and extensive compensatory regulation of gene expression (33). Thus, our finding of enhanced atherosclerosis in humans provides robust proof of concept for emerging mechanistic and murine studies demonstrating a possible proliferative phenotype for the 9p21 risk locus.
Implications
By refining the underlying phenotype, this study has significant implications for our understanding of the 9p21 risk locus. Firstly, by demonstrating that the mechanism of risk is likely through development of atherosclerosis, our work helps translate and contextualize functional studies, which in turn has potential implications for developing novel therapies to reduce atherosclerotic burden and cardiovascular risk. Secondly, these findings might help explain why 9p21 does not enhance risk prediction for MI, given that this is a more downstream phenotype. With expanding use of direct-to-consumer and physician-ordered testing for 9p21, this work will have greater relevance and will better inform those interpreting these tests about the actual underlying genetic risk.
Strengths and limitations
The major strength of this study is the collaborative collection of cohorts combining published and unpublished data, including 14 that had not previously reported on data with regard to presence of MI in subjects with CAD. We also present the use of a Bayesian analysis to account for potential heterogeneity from use of different 9p21 markers and confirm from a statistical perspective that the several 9p21 markers (rs1333049, rs10757278, and rs2383206) can be used interchangeably in meta-analyses such as this. There are also some limitations to our study. First, the cohorts with angiographic data are inherently subject to selection bias, but because our primary objective was to determine differences in disease burden, this is not a major concern. Second, we noted significant heterogeneity between the studies, and although it is possible that there were some clinical differences between studies, such as referral bias in different countries, these are likely to be small, and we devised a priori subgroup analyses to investigate these further. Third, our phenotype was relatively crude for assessing CAD burden, and we did not have more details on, for example, presence of more proximal prognostic disease, but this was unavoidable given that few cohorts collect extensive CAD scores such as the SYNTAX score (13). Finally, it is possible that we might have missed a small effect of risk of MI in the context of CAD, but post hoc power calculations suggested the analysis had >0.95 power to detect an OR of 1.1 for MI among subjects with CAD. This observation combined with the consistency of the data from different groups makes it unlikely that the finding is spurious.
Conclusions
Our analysis reveals that the 9p21 risk locus is associated with greater CAD burden but not with prevalent MI when both cases and control subjects have documented angiographic CAD. Taken together our meta-analysis confirms that the primary phenotype mediated by the 9p21 locus is one of predisposition to greater atherosclerosis.
Supplementary Material
Acknowledgments
Work in the laboratory of author S.Y. is supported by the British Heart Foundation and forms part of the research themes contributing to the translational research portfolio of Barts Cardiovascular Biomedical Research Unit, which is supported and funded by the National Institute of Health Research.
The Japan and Korean study was supported in part by Grant-in-Aids from the Ministry of Education, Science, Sports, Culture and Technology (MEXT) of Japan, a grant for Japan-Korea collaboration research program from Japan Society for the Promotion of Science (JSPS), the National Research Foundation of Korea (NRF) Grant (NRF-616-2010-2-E00), and a follow-up grants provided from the Tokyo Medical and Dental University.
The SK-CAD research was supported by the American Heart Association National Innovative Research Grant 11IRG5570046 to Q.K.W.
The Utah Intermountain Heart Collaborative Study was funded in part by the Intermountain Research and Medical Foundation.
The Precocious Coronary Artery Disease work was supported by the European Community Sixth Framework Program [LSHM-CT- 2007-037273], the British Heart foundation, the Oxford BHF Centre of Research Excellence, the Wellcome Trust [090532/Z/09/Z], and AstraZeneca.
In Munich, Germany, the study was funded by an institutional grant from the German Heart Centre Munich, Munich, Germany.
The Coronary Disease Cohort Study was supported by Christchurch Cardioendocrine Research Group, National Heart Foundation of New Zealand, and Health Research Council of New Zealand.
The Austrian study was supported by grants from the “Land Vorarlberg” and the “Europaeischer Fonds fuer regionale Entwicklung.”
The International Verapamil SR Trandolapril Study Genetic Substudy was supported by grants from the National Institutes of Health (NIH) (HL074730 and GM074492), Abbott Laboratories, and University of Florida Opportunity Fund and in part by the NIH/National Center for Research Resources Clinical and Translational Science Award to the University of Florida UL1RR029890.
The Atherosclerotic Disease, Vascular Function, and Genetic Epidemiology study was supported by a grant from the Donald W. Reynolds Foundation, Las Vegas, Nevada, and the Stanford Cardiovascular Institute, Stanford, California.
Recruitment of the PennCATH cohort was supported by the Cardiovascular Institute of the University of Pennsylvania. Recruitment of the MedStar cohort was supported by a research grant from GlaxoSmithKline. Genotyping was performed at the Center for Applied Genomics at the Children’s Hospital of Philadelphia and supported by GlaxoSmithKline through an Alternate Drug Discovery Initiative research alliance award to the University of Pennsylvania School of Medicine (M.P.R. and D.J.R.).
The Cleveland Clinic GeneBank study is supported by P01 HL076491, P01 HL098055, and R01HL103866 to S.L.H. and R01HL103931 to W.H.W.T.
This work at the Emory Cardiovascular Biobank was supported by the American Heart Association (Postdoctoral Fellowship for R.S.P.), NIH R01 HL89650-01, Robert W. Woodruff Health Sciences Center Fund, and Emory Heart and Vascular Center Funds and supported in part by NIH Grant UL1 RR025008 from the Clinical and Translational Science Award Program.
The authors acknowledge Drs. Taishi Sasaoka, Shigeru Hohda, and Toru Izumi for their contributions in collecting blood samples and clinical information from Japanese cases, in the Japan and Korean study.
Abbreviations and Acronyms
- CAD
coronary artery disease
- CHD
coronary heart disease
- CI
confidence interval
- GWAS
genome-wide association studies
- LD
linkage disequilibrium
- MI
myocardial infarction
- OR
odds ratio
- SNP
single nucleotide polymorphism
- VSMC
vascular smooth muscle cell
Footnotes
APPENDIX
For supplementary figures, tables, and text, please see the online version of this article.
References
- 1.Samani NJ, Erdmann J, Hall AS, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–53. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helgadottir A, Thorleifsson G, Manolescu A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–3. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 3.McPherson R, Pertsemlidis A, Kavaslar N, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–91. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schunkert H, Gotz A, Braund P, et al. Repeated replication and a prospective meta-analysis of the association between chromosome 9p21.3 and coronary artery disease. Circulation. 2008;117:1675–84. doi: 10.1161/CIRCULATIONAHA.107.730614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palomaki GE, Melillo S, Bradley LA. Association between 9p21 genomic markers and heart disease: a meta-analysis. JAMA. 2010;303:648–56. doi: 10.1001/jama.2010.118. [DOI] [PubMed] [Google Scholar]
- 6.Kitsios GD, Dahabreh IJ, Trikalinos TA, Schmid CH, Huggins GS, Kent DM. Heterogeneity of the phenotypic definition of coronary artery disease and its impact on genetic association studies. Circ Cardiovasc Genet. 2011;4:58–67. doi: 10.1161/CIRCGENETICS.110.957738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horne BD, Carlquist JF, Muhlestein JB, Bair TL, Anderson JL. Association of variation in the chromosome 9p21 locus with myocardial infarction versus chronic coronary artery disease. Circ Cardiovasc Genet. 2008;1:85–92. doi: 10.1161/CIRCGENETICS.108.793158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reilly MP, Li M, He J, et al. Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: two genome-wide association studies. Lancet. 2011;377:383–92. doi: 10.1016/S0140-6736(10)61996-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis KL, Pilbrow AP, Frampton CM, et al. A common variant at chromosome 9P21.3 is associated with age of onset of coronary disease but not subsequent mortality. Circ Cardiovasc Genet. 2010;3:286–93. doi: 10.1161/CIRCGENETICS.109.917443. [DOI] [PubMed] [Google Scholar]
- 10.O’Donnell CJ, Kavousi M, Smith AV, et al. Genome-wide association study for coronary artery calcification with follow-up in myocardial infarction. Circulation. 2011;124:2855–64. doi: 10.1161/CIRCULATIONAHA.110.974899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Assimes TL, Knowles JW, Basu A, et al. Susceptibility locus for clinical and subclinical coronary artery disease at chromosome 9p21 in the multi-ethnic ADVANCE study. Hum Mol Genet. 2008;17:2320–8. doi: 10.1093/hmg/ddn132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dandona S, Stewart AF, Chen L, et al. Gene dosage of the common variant 9p21 predicts severity of coronary artery disease. J Am Coll Cardiol. 2010;56:479–86. doi: 10.1016/j.jacc.2009.10.092. [DOI] [PubMed] [Google Scholar]
- 13.Patel RS, Su S, Neeland IJ, et al. The chromosome 9p21 risk locus is associated with angiographic severity and progression of coronary artery disease. Eur Heart J. 2010;31:3017–23. doi: 10.1093/eurheartj/ehq272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson JL, Horne BD, Kolek MJ, et al. Genetic variation at the 9p21 locus predicts angiographic coronary artery disease prevalence but not extent and has clinical utility. Am Heart J. 2008;156:1155–62.e2. doi: 10.1016/j.ahj.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 15.Chen SN, Ballantyne CM, Gotto AM, Jr, Marian AJ. The 9p21 susceptibility locus for coronary artery disease and the severity of coronary atherosclerosis. BMC Cardiovasc Disord. 2009;9:3. doi: 10.1186/1471-2261-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alpert JS, Thygesen K, Antman E, Bassand JP. Myocardial infarction redefined—a consensus document of The Joint European Society of Cardiology/American College of Cardiology Committee for the re-definition of myocardial infarction. J Am Coll Cardiol. 2000;36:959–69. doi: 10.1016/s0735-1097(00)00804-4. [DOI] [PubMed] [Google Scholar]
- 17.Higgins JPT, Green S, editors. The Cochrane Collaboration. [March 21, 2012];Cochrane Handbook for Systematic Reviews of Interventions, Version 5.1.0. 2011 Available at: http://www.cochrane.org/training/cochrane-handbook.
- 18.Little J, Higgins JPT, editors. [March 21, 2012];The HuGENetTM HuGE Review Handbook, Version 1.0. Available at: http://www.med.uottawa.ca/public-health-genomics/web/assets/documents/huge_review_handbook_v1_0.pdf.
- 19.DerSimonian R, Kacker R. Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials. 2007;28:105–14. doi: 10.1016/j.cct.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newcombe PJ, Verzilli C, Casas JP, Hingorani AD, Smeeth L, Whittaker JC. Multilocus Bayesian meta-analysis of gene-disease associations. Am J Hum Genet. 2009;84:567–80. doi: 10.1016/j.ajhg.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broadbent HM, Peden JF, Lorkowski S, et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet. 2008;17:806–14. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- 24.Chan K, Motterle A, Laxton RC, Ye S. Common variant on chromosome 9p21 predicts severity of coronary artery disease. J Am Coll Cardiol. 2011;57:1497–8. doi: 10.1016/j.jacc.2010.09.078. [DOI] [PubMed] [Google Scholar]
- 25.Gong Y, Beitelshees AL, Cooper-DeHoff RM, et al. Chromosome 9p21 haplotypes and prognosis in white and black patients with coronary artery disease. Circ Cardiovasc Genet. 2011;4:169–78. doi: 10.1161/CIRCGENETICS.110.959296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Donnell CJ, Cupples LA, D’Agostino RB, et al. Genome-wide association study for subclinical atherosclerosis in major arterial territories in the NHLBI’s Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S4. doi: 10.1186/1471-2350-8-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye S, Willeit J, Kronenberg F, Xu Q, Kiechl S. Association of genetic variation on chromosome 9p21 with susceptibility and progression of atherosclerosis: a population-based, prospective study. J Am Coll Cardiol. 2008;52:378–84. doi: 10.1016/j.jacc.2007.11.087. [DOI] [PubMed] [Google Scholar]
- 28.Cluett C, McDermott MM, Guralnik J, et al. The 9p21 myocardial infarction risk allele increases risk of peripheral artery disease in older people. Circ Cardiovasc Genet. 2009;2:347–53. doi: 10.1161/CIRCGENETICS.108.825935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murabito JM, White CC, Kavousi M, et al. Association between chromosome 9p21 variants and the ankle-brachial index identified by a meta-analysis of 21 genome-wide association studies. Circ Cardiovasc Genet. 2012;5:100–12. doi: 10.1161/CIRCGENETICS.111.961292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel RS, Eapen DJ, Zafari AM, Vaccarino V, Quyyumi AA. Letter by Patel et al regarding article “Chromosome 9p21 haplotypes and prognosis in white and black patients with coronary artery disease”. Circ Cardiovasc Genet. 2011;4:e11. doi: 10.1161/CIRCGENETICS.111.960641. [DOI] [PubMed] [Google Scholar]
- 31.Jarinova O, Stewart AF, Roberts R, et al. Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arterioscler Thromb Vasc Biol. 2009;29:1671–7. doi: 10.1161/ATVBAHA.109.189522. [DOI] [PubMed] [Google Scholar]
- 32.Visel A, Zhu Y, May D, et al. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010;464:409–12. doi: 10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim JB, Deluna A, Mungrue IN, et al. Effect of 9p21.3 coronary artery disease locus neighboring genes on atherosclerosis in mice. Circulation. 2012;126:1896–906. doi: 10.1161/CIRCULATIONAHA.111.064881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Motterle A, Pu X, Wood H, et al. Functional analyses of coronary artery disease associated variation on chromosome 9p21 in vascular smooth muscle cells. Hum Mol Genet. 2012;21:4021–9. doi: 10.1093/hmg/dds224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonzalez-Navarro H, Abu Nabah YN, Vinue A, et al. p19(ARF) deficiency reduces macrophage and vascular smooth muscle cell apoptosis and aggravates atherosclerosis. J Am Coll Cardiol. 2010;55:2258–68. doi: 10.1016/j.jacc.2010.01.026. [DOI] [PubMed] [Google Scholar]
- 36.Hinohara K, Nakajima T, Takahashi M, et al. Replication of the association between a chromosome 9p21 polymorphism and coronary artery disease in Japanese and Korean populations. J Hum Genet. 2008;53:357–9. doi: 10.1007/s10038-008-0248-4. [DOI] [PubMed] [Google Scholar]
- 37.Hoppmann P, Erl A, Turk S, et al. No association of chromosome 9p21.3 variation with clinical and angiographic outcomes after placement of drug-eluting stents. J Am Coll Cardiol Intv. 2009;2:1149–55. doi: 10.1016/j.jcin.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 38.Krawczak M, Nikolaus S, von Eberstein H, Croucher PJ, El Mokhtari NE, Schreiber S. PopGen: population-based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Community Genet. 2006;9:55–61. doi: 10.1159/000090694. [DOI] [PubMed] [Google Scholar]
- 39.Muendlein A, Saely CH, Rhomberg S, et al. Evaluation of the association of genetic variants on the chromosomal loci 9p21.3, 6q25.1, and 2q36.3 with angiographically characterized coronary artery disease. Atherosclerosis. 2009;205:174–80. doi: 10.1016/j.atherosclerosis.2008.10.035. [DOI] [PubMed] [Google Scholar]
- 40.Shen GQ, Li L, Rao S, et al. Four SNPs on chromosome 9p21 in a South Korean population implicate a genetic locus that confers high cross-race risk for development of coronary artery disease. Arterioscler Thromb Vasc Biol. 2008;28:360–5. doi: 10.1161/ATVBAHA.107.157248. [DOI] [PubMed] [Google Scholar]
- 41.Wang W, Peng WH, Lu L, et al. Polymorphism on chromosome 9p21.3 contributes to early-onset and severity of coronary artery disease in non-diabetic and type 2 diabetic patients. Chin Med J (Engl) 2011;124:66–71. doi: 10.3901/jme.2011.08.066. [DOI] [PubMed] [Google Scholar]
- 42.Ye S, Dunleavey L, Bannister W, et al. Independent effects of the -219 G>T and epsilon 2/ epsilon 3/ epsilon 4 polymorphisms in the apolipoprotein E gene on coronary artery disease: the Southampton Atherosclerosis Study. Eur J Hum Genet. 2003;11:437–43. doi: 10.1038/sj.ejhg.5200983. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


