Abstract
To determine if ethanol consumption and alcoholism cause global DNA methylation disturbances, we examined alcoholics and controls using methylation specific microarrays to detect all annotated gene and non-coding micro-RNA promoters and their CpG islands. DNA was isolated and immunoprecipitated from the frontal cortex of 10 alcoholics and 10 age and gender-matched controls then labeled prior to co-hybridization. A modified Kolmogorov–Smirnov test was used to predict differentially enriched regions (peaks) from log-ratio estimates of amplified vs input DNA. More than 180,000 targets were identified for each subject which correlated with >30,000 distinct, integrated peaks or high probability methylation loci. Peaks were mapped to regions near 17,810 separate annotated genes per subject representing hypothetical methylation targets. No global methylation differences were observed between the two subject groups with 80% genetic overlap, but extreme methylation was observed in both groups at specific loci corresponding with known methylated genes (e.g., H19) and potentially other genes of unknown methylation status. Methylation density patterns targeting CpG islands visually correlated with recognized chromosome banding. Our study provides insight into global epigenetic regulation in the human brain in relationship to controls and potentially novel targets for hypothesis generation and follow-up studies of alcoholism.
Keywords: Alcoholism, Epigenetics, Brain, Frontal cortex, DNA methylation, Global methylation
1. Introduction
Alcoholism is a chronic, relapsing illness associated with significant psychosocial, behavioral and physical dysfunction and broad sociological impact. Decades of scientific study have implicated events and characteristics associated with social, biological and interpersonal influences as risk factors for the development of alcoholism (Goodwin, 1979; Goodwin et al., 1974; Vaillant, 1975, 1976). Genetic and biological influences are the most consistent predictors; however, strong interactions exist across multiple predictors over the life of an individual. Today it is widely assumed that genes account for 40 to 60% of the variance associated with alcoholic drinking (Prescott and Kendler, 1999; Prescott et al., 1999). The search for specific genes has identified several genetic variants with strong linkage to alcoholism (Agrawal and Lynskey, 2008; Ducci and Goldman, 2008; Mayfield et al., 2008). Specific ways in which genes interact with environmental factors to influence alcoholism are presently unknown and the subject of intense clinical interest.
Ethanol is not only an addictive drug, but alters the activities and function of multiple organ systems including brain, liver, gastrointestinal and cardiovascular affecting human health. Alcohol consumption causes disruption in gene expression regulating cellular signaling pathways impacting on transcriptional factors and gene regulation specifically for stress response, metabolism, olfaction, cytoskeletal organization, and nucleic acid binding and epigenetics (Awofala, 2011; Miranda et al., 2010).
1.1. Epigenetics of alcoholism
Epigenetics is an emerging field in the study of mental illness. Disturbances in epigenetic regulation have been implicated in several psychiatric disorders including schizophrenia, depression and the addictions (Abdolmaleky et al., 2005; Grayson et al., 2005; Maze and Nestler, 2011; Polesskaya et al., 2006). Epigenetic mechanisms regulate gene expression through non-permanent genetic alterations such as histone modification or chromatin restructuring as well as DNA methylation occurring in promoter regions and “CpG islands”. CpG islands are defined as DNA regions of more than 200 bases with G + C content ≥50% and a ratio of observed to statistically expected CpG frequencies of ≥0.6. In mammals, CpG islands are typically 300–3000 base pairs in size. CpG dinucleotides are relatively rare within the genome (~1%) (Flatscher-Bader et al., 2005) while approximately 60% of genes have CpG islands upstream of their promoters (Bird, 2002; Portela and Esteller, 2010). Computational analyses have predicted roughly 29,000 CpG islands in the human genome (Bird, 2002).
The majority of genes are unmethylated (activated) in all tissues during developmental stages (Bird, 2002). In normal adult cells 70–80% of CpG islands are methylated (inactivated) (Wilson, 2008). However, mechanisms have been reported for ethanol exposure and reduction of DNA methylation levels. DNA methylation in mammals is controlled by five proteins within the DNA methyltransferase (DNMT) family while ethanol use is associated with reduced DNMT levels (Ouko et al., 2009). DNMT enzymes catalyze the transfer of methyl groups from S-adenosyl methionine (SAM) to DNA. Ethanol consumption impairs 1-carbon metabolism lowering the availability of SAM, the methyl donor for both DNA and histone methylation (Hamid et al., 2009). Aberrant gene methylation is thought to play a role in a variety of disorders including autoimmune diseases and inflammation (e.g., rheumatoid arthritis and multiple sclerosis), cancer, genetic disorders (e.g., Prader–Willi, Angelman and Beckwith–Wiedemann syndromes) and neurological disorders (e.g., Alzheimer disease) (Butler, 2009; Portela and Esteller, 2010).
Epigenetic changes are influenced by external factors such as nutrition, smoking and alcohol use. Epigenetic disturbances have been identified in blood samples taken from alcohol dependent patients (Bleich et al., 2006; Heberlein et al., 2011; Hillemacher et al., 2009; Muschler et al., 2010) and have correlated with measureable effects on gene expression, level of alcohol intoxication and withdrawal as well as psychological assessments of alcohol craving. Subjective effects of alcohol lend support that epigenetic disturbances measured in peripheral blood can accurately reflect central changes. However, a critical link necessary to validate future study of dynamic responses to alcohol exposure should include tissue specific studies of epigenetic effects using human brain tissue. For example, Taqi et al., 2011 used human prefrontal cortex to identify differential methylation of CpG dinucleotides which overlapped with three prodynorphin single nucleotide polymorphisms (SNPs) in alcoholics relative to controls. Methylation was also increased in alcoholics and positively correlated with dynorphin expression. These early studies highlight the importance of detailed investigations of epigenetic influences in alcoholism involving DNA methylation, gene (exon) and microRNA expression profiles which play a significant role in cell differentiation and function in human brain tissue (Miranda et al., 2010).
1.2. Whole genome approach to study DNA methylation
Increased interest in DNA methylation profiling has resulted in the development of techniques for the identification of methylated DNA regions and quantification of methylation levels in a sensitive and site-specific manner. Methylation loci may be targeted for isolation using methylation-specific restriction enzyme digestion, immunoprecipitation of methylated DNA regions, or bisulfite conversion of non-methylated cytosine to uracil. These methods have now been adapted for high throughput whole genome microarrays capable of detecting global differences at very low levels of methylation [summarized by Bibikova and Fan (2010) and Laird (2010)]. However, each approach carries with it a specific set of limitations. For example, restriction enzyme-based analytical approaches require enzyme recognition sites within the interrogated region which limits the applicability to high throughput whole-genome investigation. Restriction enzyme-based approaches also require more DNA than immunoprecipitation or bisulfite techniques. Whereby, bisulfite conversion requires exposure to harsh conditions that may fragment or degrade DNA in an uncontrolled fashion. Bisulfite sequencing techniques have been used to map methylation loci in humans including the brain (Ladd-Acosta et al., 2007; Lister et al., 2009; Xin et al., 2011).
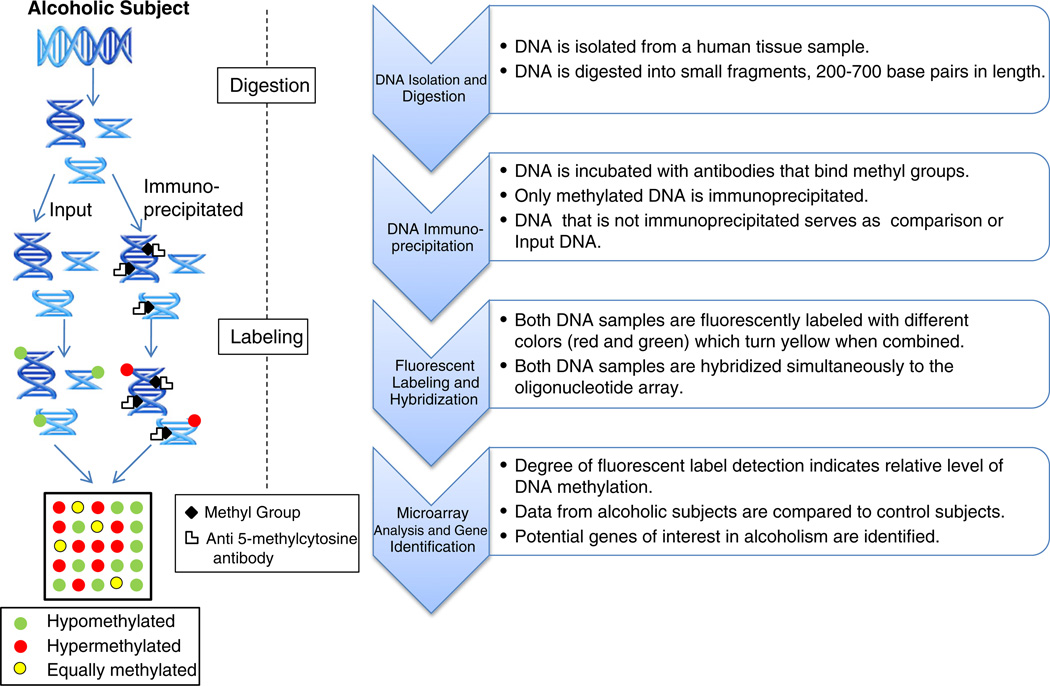
For our study, we have chosen the Roche NimbleGen platform (Madison, WI) which employs methylation-specific immunoprecipitation in combination with hybridization of input DNA on an oligonucleotide tiling array to interrogate the entire human genome (see Fig. 1). Tiling arrays are composed of non-overlapping or partially overlapping probes which are “tiled” or spaced at regular intervals across the entire genome (Gregory and Belostotsky, 2009). The relative comparison of methylated (immunoprecipitated DNA) vs unmethylated (input DNA) fractions has been shown to substantially improve sensitivity of observed differences (Schumacher et al., 2006). The Kolmogorov–Smirnov (KS) test was used to identify statistically significant differences between immunoprecipitated and input DNA samples (Scacheri et al., 2006). The two-sample KS test is a general non-parametric method of comparing differences in both location and shape of the empirical cumulative distribution functions or the underlying probability distributions in the two samples (Shorak and Wellner, 1986). Immunoprecipitation may preferentially enrich methylated CpG-rich sequences over methylated CpG-poor sequences (Bibikova and Fan, 2010); however, oligonucleotide tiling arrays partially compensate for this bias through signal averaging across neighboring genomic regions thereby improving both sensitivity and specificity (Irizarry et al., 2008). This approach should diminish the potential influences of high sequence similarity amongst gene families (e.g., histones) and their promoters which could confound peak mapping assignments.
Fig. 1.
Roche NimbleGen Human DNA Methylation 2.1M Deluxe Promoter Array Analysis Protocol.
The NimbleGen arrays permit site-specific identification and quantification of DNA methylated loci broadly across the genome at relatively high resolution and reliability. The approach is useful for examining global DNA methylation profiles as well as generation of hypothetical gene targets located in neighboring regions of methylated DNA sequences. The present study uses the Roche NimbleGen Human DNA Methylation 2.1 M Deluxe Promoter Array to interrogate methylation loci in the brain of alcoholics relative to matched controls in order to identify global methylation disturbances and affected candidate genes for alcoholism.
2. Methods
2.1. Samples
Methylation patterns were obtained for DNA isolated from frozen, intact frontal cortex tissue (Brodman Area 9) of 10 alcoholics [7 males, 3 females; mean (± SD) age = 49.5 (± 5.8) yrs, range 41–57] and 10 age and gender-matched control subjects [7 males, 3 females; mean (±SD) age = 49.6 (± 6.0) yrs, range 37–56] to account for differences that age may contribute to methylation patterns (Fraga et al., 2002). The preponderance of males in our study reflects the sex ratio distribution found in the general population of alcoholics. Samples were procured from the New South Wales Tissue Resource Centre (Sydney, Australia) according to a standardized protocol (Sheedy et al., 2008) and in compliance with ethical guidelines established by the Sydney South West Area Health Service Human Ethics Committee (X03-0074). Informed written consent was obtained from the nearest living relative. Tissues were intact with no evidence of necrosis. The mean (± SD) post-mortem interval (PMI) for tissue acquisition was 28.0 (± 11.8) hours with a range of 13 to 60 h. The mean (± SD) sample pH was 6.6 (± 0.21). All samples tested negative for viral hepatitis and human immunodeficiency virus.
All subjects were of European descent and alcohol dependent subjects met criteria for Diagnostic and Statistical Manual for Mental Disorders, Fourth Edition and National Health and Medical Research Council/World Health Organization criteria. Controls were social drinkers (non-abstainers for alcohol use) who did not meet criteria for alcohol abuse or dependence. Case subjects had an average estimated duration of alcohol dependence of 21.5 (±9.8) years (range 10–39 years). The individual causes of death varied across participants, but the most common causes were cardiovascular or respiratory problems or infection. Direct alcohol toxicity or overdose was indicated in two deaths of alcoholic subjects. Family history of alcohol problems was either negative or unknown for all subjects.
2.2. Microarray
The Roche NimbleGen Human DNA Methylation 2.1M Deluxe Promoter Array (Roche NimbleGen, Inc.; Madison, WI, USA) was used to examine DNA methylation disturbances of all annotated gene and non-coding microRNA promoters and their CpG islands in Alcohol Dependent subjects relative to age and gender-matched controls. The array specifications included the following information: Array Type— NimbleGen 100205_HG18_Promoter_MeDIP_HX1; Format— 2.1M; Source— University of California, Santa Cruz; Build— all probe locations used the human genome reference GRCh36/hg18 assembly (http://genome.ucsc.edu/cgi-bin/hgGateway?db=hg18); Probe Length— 50 to 75mer; and Storage— arrays stored at room temperature in desiccated conditions. This Deluxe Promoter Array consists of probes for 28,226 CpG islands and 475 microRNA promoters. Probe tiling for the Deluxe Promoter Array extended from 7250 bp upstream to 3250 bp downstream for each CpG island. Each probe was distributed approximately 135 bp apart with about 75 probes per CpG island for determination of methylation status. The NimbleGen platform maps methylated DNA regions using a combination of affinity-based enrichment, Methylated DNA Immunoprecipitation (MeDIP) and Methylated CpG Island Recovery Assay, followed by microarray analysis (Fig. 1). The arrays use long oligonucleotides combined with high-stringency hybridization protocols to increase sensitivity and specificity.
2.2.1. Immunoprecipitation procedure
DNA was isolated from brain tissue and fragmented via digestion with MseI enzyme following manufacturer's protocols. Immunoprecipitation of methylated DNA was then performed using anti-5-methylcytosine antibodies. A portion of the fragmented DNA that was not immunoprecipitated from each subject was used as reference or input DNA. Input and immunoprecipitated DNA were amplified then differentially labeled and co-hybridized to microarrays following standardized protocols (see Fig. 1).
2.2.2. Microarray data processing and bioinformatics (performed by Roche NimbleGen)
An algorithm derived from a modified Kolmogorov–Smirnov test was used to predict enriched regions representing methylated CpG islands across multiple adjacent probes in sliding-windows 100 base pairs in length (Scacheri et al., 2006). Differentially enriched regions of immunoprecipitated vs input DNA were identified based on number and coverage of bound probes to methylated fragments. The mean log-ratio of immunoprecipitated to input samples was integrated across the enriched regions. Regions showing enrichment at 4 or more consecutive loci were integrated together to form a single “peak”. Clusters of enriched regions separated by more than 500 base pairs were integrated as separate peaks. The output included the number of identified peaks, the total number of peak and gene combinations, and peak log2-ratios (peak scores) which reflected the probability of methylation for the designated peak and/or gene at a p-value of less than 0.01.
2.2.3. Microarray post processing: identification of hypothetical gene targets
Statistical analysis of output microarray data was conducted using SAS statistical analytical software version 9.2 (SAS Inc., Cary, NC). Array data for all study subjects were formatted, uploaded and concatenated into a single large SAS data file. Initial data processing included a descriptive analysis of all peak and gene target combinations (overall and by chromosome) for alcoholic and control subjects. Secondary data processing was conducted to isolate and characterize all unique peaks and their associated gene targets. This analysis selected peaks associated with known annotated gene targets and deleted “tiled regions” not linked to known genes. Subsequent processing eliminated duplicate feature/peak combinations to provide a list of unique genes representing hypothetical methylation targets. Peak scores were used to quantify the probability of methylation for individual gene targets based upon peak linkage. Unique methylated promoter regions for genes were filtered based upon the maximum peak score of all gene associated peaks.
2.3. Statistical analysis
Descriptive analyses of gross differences used simple one-way analysis of variance to compare the mean number of peak/gene combinations and the mean number of peaks and genes by group. Mean peak scores of individual gene targets were also used as a measure of methylation to examine disturbances by subject group. The Chi Square test was used to examine group differences in the frequency of promoter methylation for individual target genes.
3. Results
3.1. Summary statistics
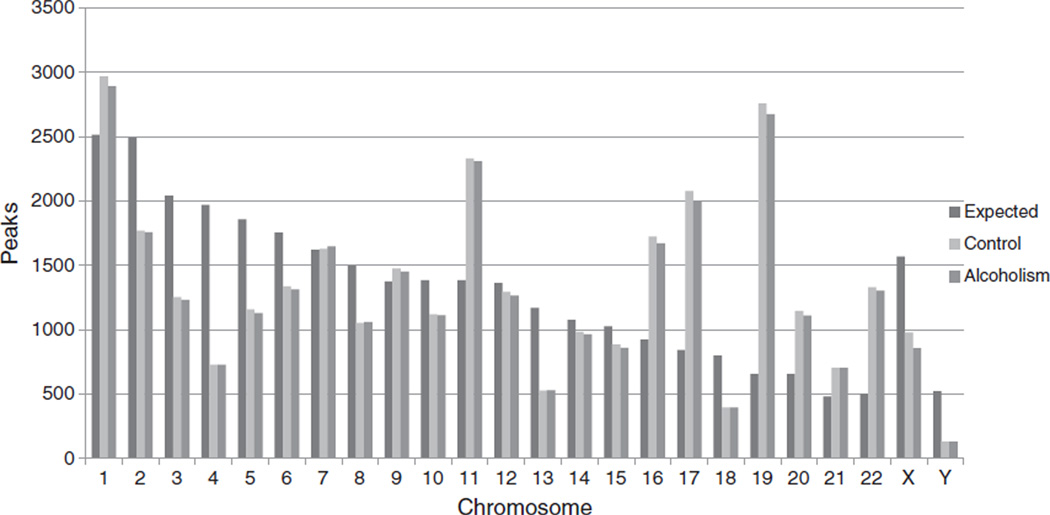
Our high resolution DNA methylation microarray analysis of human frontal cortex samples taken from alcoholics and controls identified more than 180,000 unique feature and peak combinations per subject (x̄ = 180,001; SD = 9,793; range 154,189 to 188,131 for alcoholics; x̄ = 183,875; SD = 6,707; range 176,195 to 200,285 for controls). As noted in the methods, peak/gene combinations were not limited to a one to one ratio. Any individual peak may be associated with multiple gene targets; similarly, any individual gene may be associated with multiple peaks. Gene/peak combinations were correlated with over 30,000 distinct, integrated peaks distributed across the 28,266 target CpG islands (x̄ = 31,217; SD = 1,553; range 27,110 to 32,460 for alcoholics; x̄ = 31,780; SD = 1,067; range 30,244 to 34,059 for controls). These peaks were linked to over 17,000 separate annotated and named genes (16,902; x̄ = 13,252; SD = 109; range 13,080 to 13,469 for alcoholics; 17,193; x̄ = 13,330; SD = 397; range 12,936 to 14,091 for controls) per subject. No global differences were observed between alcoholics and controls in either the number of peak/feature combinations (F = 1.07; p = 0.3157); the mean number of identified peaks (F = 0.89; p = 0.357), or genes (F = 0.35; p = 0.5593) per subject. It should also be noted that no methylation was observed at any of the 475 target miRNA promoter loci for either alcoholic or control subjects. An examination of the distribution of probe binding across chromosomes (Fig. 2) found the highest density of coverage in relationship to chromosome size (number of base pairs) for chromosome 19 and the smallest for chromosome 12 suggesting that the degree of probe binding is not necessarily proportional to the length of the chromosome.
Fig. 2.
Observed vs expected number of peaks by diagnostic category. Expected values are based upon the proportion of total peaks standardized by the relative length of the chromosome in base pairs. Simple Analysis of Variance found no significant differences between alcoholic and control subjects for any chromosome (p<0.05). A 2-sided t-test found a significant difference between observed and expected values (p<0.05) for all chromosomes in both groups except for chromosome 7.
3.2. Peak score analysis
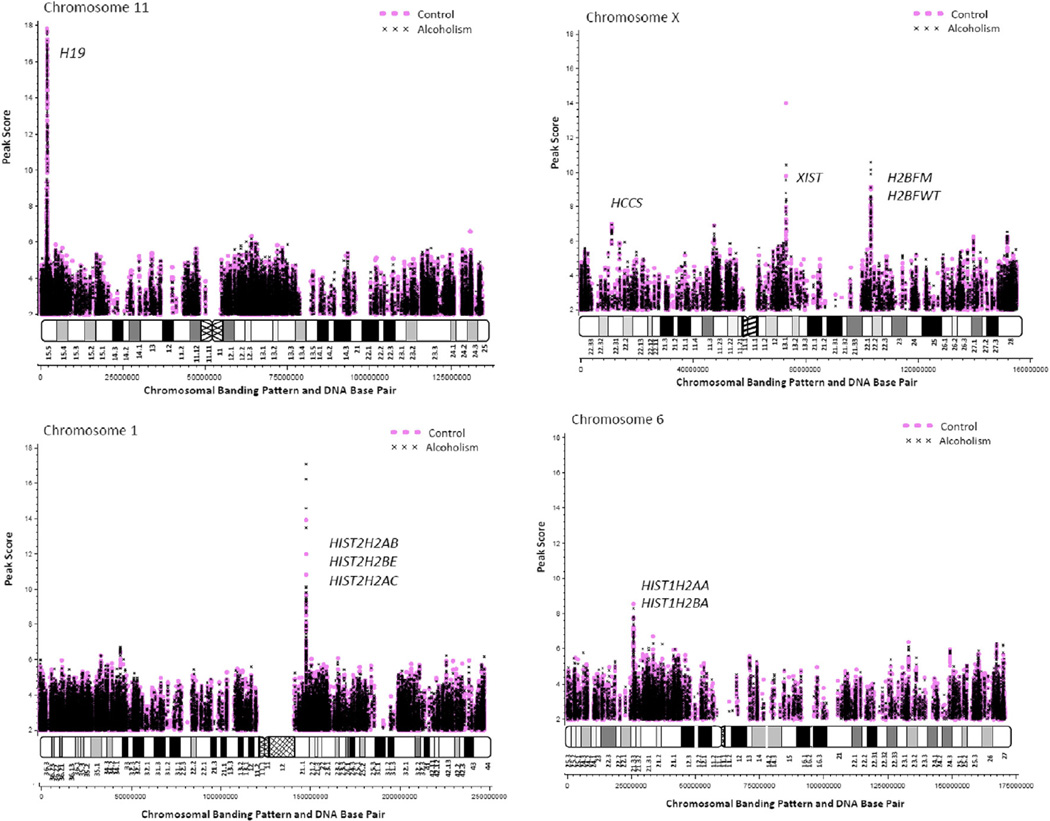
Peak scores followed an approximately normal distribution with a mean (SD) = 2.93 (0.68) and range from 2 to 17.8. Mean scores per subject did not differ significantly between alcoholic [2.92 (0.072), range 2.8–3.0] and control groups [2.93 (0.048), range 2.9–3.0; F = 0.09, p = 0.765]. Profile plots of peak scores according to peak loci (peak start site per chromosome) illustrate areas of high density methylation which can be visualized as banding along the length of the chromosome. Plots of selected chromosomes were generated from raw data incorporating all integrated (but not duplicated) peaks and differentially labeled for alcoholic and control samples (Fig. 3). As shown in Fig. 3, the methylation density patterns representing enriched CpG islands, corresponding to gene locations, visually correlate with recognized chromosome banding ideograms with pale (negative) G-banding corresponding to CG base pair enriched chromosome regions and dark (positive) bands corresponding to AT enriched regions. Evidence of extreme methylation at specific gene loci can be visualized as large spikes in the graph. These spikes correspond with loci near to knownmethylatedmammalian genes (e.g., H19 on chromosome 11) supporting internal confirmation of probe specificity and array methodology. Spikes on chromosomes 1, 6, 11, 20 and X correspond with extreme methylation gene targets as listed in Table 1. There was no statistically significant difference in the mean peak scores between alcoholic and control subjects for 10 of 11 identified “extreme methylation” gene targets. However, alcoholic subjects did show a significantly greater probability of methylation than controls at the promoter region near the histone HIST2H2AC gene.
Fig. 3.
Selected plots of methylated DNA loci with chromosome banding patterns and DNA base pair assignments.
Table 1.
Mean peak scores for highly methylated genes in all alcohol dependent and control subjects.
| Gene | Chromosome | Alcoholic mean (SD) | Range | Control mean (SD) | Range | F | p-value |
|---|---|---|---|---|---|---|---|
| GNAS | 20 | 7.65 (0.99) | 6.25–9.57 | 8.36 (1.50) | 5.61–10.47 | 1.0 | 0.388 |
| H19 | 11 | 7.75 (1.34) | 5.84–9.50 | 7.96 (1.35) | 5.89–10.61 | 0.69 | 0.513 |
| HIST2H2AB | 1 | 9.66 (5.09) | 4.28–17.08 | 7.86 (3.23) | 3.98–13.93 | −0.66 | 0.521 |
| HIST2H2BE | 1 | 9.66 (5.09) | 4.28–17.08 | 7.86 (3.23) | 3.98–13.93 | −0.66 | 0.521 |
| H2BFM | X | 7.90 (1.57) | 5.83–10.56 | 7.72 (0.85) | 6.45–9.15 | 0.01 | 0.990 |
| H2BFWT | X | 7.89 (1.58) | 5.83–10.56 | 7.66 (0.85) | 6.41–9.15 | −0.04 | 0.967 |
| BOLA1 | 1 | 6.81 (2.02) | 4.76–10.07 | 7.25 (1.71) | 4.40–9.71 | 0.71 | 0.487 |
| HIST1H2AA | 6 | 7.11 (0.78) | 5.64–8.32 | 7.06 (0.78) | 6.07–8.56 | 0.73 | 0.476 |
| HCCS | X | 6.46 (0.31) | 5.84–6.96 | 6.63 (0.39) | 5.85–7.02 | 1.15 | 0.265 |
| HIST1H2BA | 6 | 6.66 (0.80) | 5.25–7.81 | 6.63 (0.89) | 4.73–7.79 | 0.81 | 0.429 |
| HIST2H2AC | 1 | 7.18 (2.93) | 2.69–10.16 | 4.52 (1.56) | 3.04–7.86 | −3.02 | 0.008a |
Genes with mean peak scores ≥6.0 in either alcoholic or control groups. Peak scores reflect group means with N = 10 per group. Statistical comparisons used Two-Way Analysis of Variance adjusting for gender using a Tukey test to adjust for multiple comparisons.
Significant main effect of diagnosis.
3.3. Gene target analysis
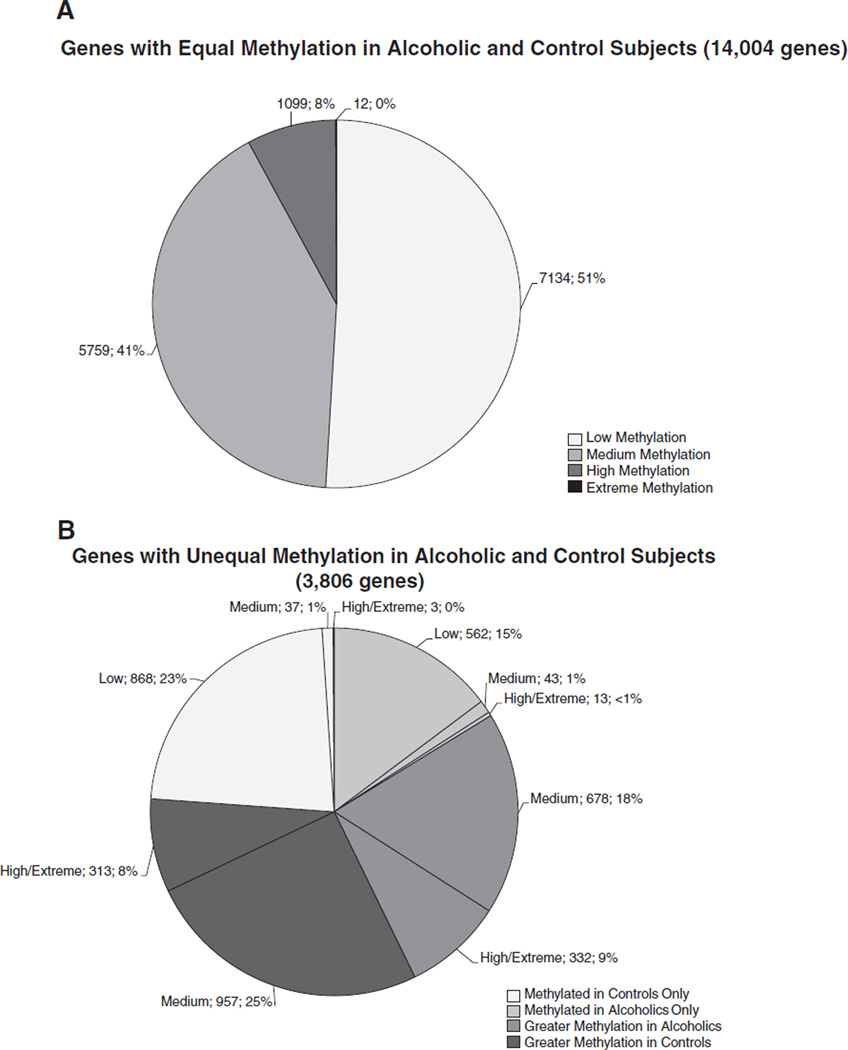
Hypothetical gene targets were subdivided into four methylation levels (low, medium, high and extreme) based on the approximate standard deviation of the overall distribution of peak scores. Peak scores from 2 to ≤3 were categorized as “low”; from > 3 to <4 were “medium”; ≥ 4 to <6 were “high” and ≥6 were “extreme” (Fig 4). A high level of overlap in methylated gene targets was observed between alcoholic and control subjects with about 80% of genes in the low (7134 genes, 40%), medium (5759 genes, 32%), high (1099 genes, 6%) and extreme (12 genes, <1%) peak score ranges common to both alcoholic and control subjects (Fig. 4A). Fig. 4B illustrates the distribution of the remaining 3806 identified hypothetical gene targets in the different methylation categories grouped by alcoholism or control. Alcoholics exhibited higher mean peak scores than control subjects for a subset of 1010 genes with 678 genes in the medium category, and 331 genes in the high category only. These same genes were in the low or medium categories in control subjects. Control subjects exhibited higher mean peak scores than alcoholic subjects for a subset of 1270 genes with 957 genes grouped in the medium category and 313 in the high category. Two other subsets of genes were exclusively methylated in alcoholic subjects (618 genes) only or in control subjects (908 genes) only.
Fig. 4.
Distribution and assortment of hypothetical gene targets based upon methylation level. Human DNA methylation data obtained using the Roche NimbleGen Human DNA Methylation 2.1M Deluxe Promoter Array grouped by diagnosis and level of methylation [low (2 to ≤3); medium (>3 to <4); high (≥4 to <6); extreme (≥6)] based upon mean peak score per gene. Labels indicate the level of gene methylation; number of genes per category; and percentage of genes. A total of 17,810 genes are reported for N = 10 subjects per group. Extreme methylation accounts for <1% of all methylated genes.
4. Discussion
The present study provides an extensive and detailed examination of DNA methylation profiles in the brain of severely Alcohol Dependent subjects. Preliminary descriptive analyses identified approximately 180,000 unique peak/gene combinations which corresponded to ~30,000 distinct integrated peaks or methylated DNA regions and 17,810 annotated/named genes. The distribution of DNA methylation across the genome was not correlated with chromosome length, and appeared to be systematic (i.e., did not appear to result from random influences). Array output data indicated high methylation at promoter regions near to known methylated mammalian genes, and plots of raw data revealed high methylation density in banding patterns analogous to chromosomal ideograms. Replication of this characteristic chromosomal banding pattern supports the general reliability of the technique to detect meaningful effects broadly across the genome. A high level of overlap (~80%) was observed in hypothetical methylation targets between alcoholic and control subjects, although the frequency and level of methylation varied. Study data were processed according to the standard recommended parameters for Roche NimbleGen microarray analysis using a sliding window 100 bp in length and a peak separation of 500 bp. The absolute number of peaks and the integrated peak scores were dictated by these fixed parameters. Peak score data consisted of the integrated log2 ratio of immunoprecipitated to input DNA of bound probes within the designated peak area. The approximate number and location of identified peaks showed good internal consistency across study subjects and between subject groups with little to no difference in the number of peaks per gene observed across subjects.
Microarray output showed good internal consistency across study subjects and subject groups with generally low variability in peak score measures. Standard deviation in mean peak score values were small enough to identify large effect sizes (>0.8) with a relatively small subject number, even after multiple linear regression modeling was adjusted for confounding factors such as sex, age and PMI (data not shown). We did not find global differences in DNA methylation in the same frontal cortex region of alcoholic compared to control samples. While methylation levels between differing brain regions can vary in non-alcoholic controls (Ladd-Acosta et al., 2007), significant methylation differences within any given brain region per subject may not be compatible with life. Observed differences were bidirectional and appeared to be systematic. Confirmatory functional analyses will be necessary to draw firm conclusions regarding the observed effects and are pending but were not the focus of the present report.
Zilberman and others in 2007 indicated that the use of the long oligonucleotide probes in the NimbleGen platform provides “a good balance between sensitivity, specificity and noise” in the absence of enhanced statistical manipulation (Penterman et al., 2007; Zilberman and Henikoff, 2007; Zilberman et al., 2007), and reported that raw data from NimbleGen arrays are published routinely with good results (Mito et al., 2005, 2007; Penterman et al., 2007). Further, the ability to detect small differences even after controlled regression analysis increases confidence in the identified differences found in the study. Hypothetical gene targets are mapped to the region of methylation based upon their sequence and known genetic loci, and represent possible candidate genes for future studies on alcoholism. However, other modifications of cytosine which could impact on gene expression include hydroxylation, formylation, carboxymethylation were not analyzed separately in our report.
Extreme methylation was observed in promoter regions near several known methylated mammalian genes including H19, GNAS, HCCS, XIST and BOLA1 supporting the biological relevance of epigenetic regulation of gene expression at these loci. The identification of extreme methylation of promoters near to several specific histone genes was unexpected. The list of 17,000 unique annotated hypothetical gene targets of promoter methylation included 38 different histone genes distributed across the 3 histone binding domains (HIST1, HIST2, and HIST3). In hepatocytes, ethanol promotes histone H3 acetylation on lysine 9 which is associated with transcriptional activation (Choudhury and Shukla, 2008). Alcohol dependent subjects showed a significantly greater degree of methylation than control subjects at the promoter region near HIST2H2AC suggesting the possibility of epigenetic disturbances in histone expression in alcoholism. Chromatin remodeling in the brain through acetylation, phosphorylation and methylation of histone proteins are believed to be major mediators of epigenetic regulation of gene expression. Histone modification has been observed in response to cocaine exposure in animal models of addiction (Maze and Nestler, 2011). These effects were associated with functional changes in cellular signaling and gene expression that may be associated with addictive behaviors. Zhou et al., 2011 found that the greatest disturbance in mRNA expression, common to both alcohol dependent and cocaine dependent subjects, was a 1–2 fold increase in the expression of HIST1H4E. This result is consistent with the present findings of possible functional disturbances in histone expression in alcoholism. However, as with most new genetic tests and protocols confirmation of study findings is required. Thus, the whole-genome changes in methylation would need to be individually cross-checked with expression studies as well as detailed examination of individual CpG islands in order to make valid assumptions concerning the nature of individual deregulation caused by alcohol abuse.
5. Conclusions
The present extensive and detailed examination of DNA methylation has provided insight into global epigenetic regulation in the human brain and global differences in brain epigenetic regulation between alcohol dependent and control subjects. Alcohol dependent subjects did not differ from controls in global methylation parameters, most gene targets or their level of methylation in most cases. Group differences were bi-directional and appeared to be systematic. As expected, high levels of methylation were observed in both alcohol dependent and control subjects near known methylated mammalian genes such as H19 and GNAS. However, several variants of histone proteins showed high levels of DNA methylation, particularly those with gene loci in the HIST2 domain of chromosome 1 supporting the possibility of epigenetic disturbances in histone expression in alcoholism. More research is needed to elucidate the functional role of these competing influences on gene expression, the mechanisms underlying them and their potential impact on substance abuse behaviors.
Acknowledgments
We would like to thank Brandon Hidaka for his assistance in sample preparation. Tissues were received from the New South Wales Tissue Resource Centre at the University of Sydney supported by the National Health and Medical Research Council of Australia, Schizophrenia Research Institute and the National Institute of Alcohol Abuse and Alcoholism [NIH (NIAAA) R24AA012725]. This investigation was supported by a grant from the Hubert & Richard Hanlon Trust, NICHD HD02528 and NIAAA K01-AA015935.
Abbreviations
- Bp
Base pair
- DNA
Deoxyribonucleic acid
- DNMT
DNA methyltransferase
- KS
Kolmogorov-Smirnov
- MeDIP
Methylated DNA immunoprecipitation
- PMI
Post mortem interval
- RNA
Ribonucleic acid
- miRNA
Micro ribonucleic acid
- SAM
S-adenosyl methionine
- SNPs
Single nucleotide polymorphisms
- SD
Standard deviation
Footnotes
This investigation was supported by a grant from the Hubert & Richard Hanlon Trust, NICHD HD02528 and NIAAA K01-AA015935.
References
- Abdolmaleky HM, et al. Hypermethylation of the reelin (RELN) promoter in the brain of schizophrenic patients: a preliminary report. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2005;134:60–66. doi: 10.1002/ajmg.b.30140. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Lynskey MT. Are there genetic influences on addiction: evidence from family, adoption and twin studies. Addiction. 2008;103:1069–1081. doi: 10.1111/j.1360-0443.2008.02213.x. [DOI] [PubMed] [Google Scholar]
- Awofala AA. Genetic approaches to alcohol addiction: gene expression studies and recent candidates from Drosophila. Invert. Neurosci. 2011;11:1–7. doi: 10.1007/s10158-010-0113-y. Review. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Fan J. Genome-wide DNA methylation profiling. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010;16:210–223. doi: 10.1002/wsbm.35. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bleich S, et al. Epigenetic DNA hypermethylation of the HERP gene promoter induces down-regulation of its mRNA expression in patients with alcohol dependence. Alcohol. Clin. Exp. Res. 2006;30:587–591. doi: 10.1111/j.1530-0277.2006.00068.x. [DOI] [PubMed] [Google Scholar]
- Butler MG. Genomic imprinting disorders in humans: a mini-review. J. Assist. Reprod. Genet. 2009;26:477–486. doi: 10.1007/s10815-009-9353-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury M, Shukla SD. Surrogate alcohols and their metabolites modify histone H3 acetylation: involvement of histone acetyl transferase and histone deacetylase. Alcohol. Clin. Exp. Res. 2008;32:829–839. doi: 10.1111/j.1530-0277.2008.00630.x. [DOI] [PubMed] [Google Scholar]
- Ducci F, Goldman D. Genetic approaches to addiction: genes and alcohol. Addiction. 2008;103:1414–1428. doi: 10.1111/j.1360-0443.2008.02203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatscher-Bader T, et al. Alcohol-responsive genes in the frontal cortex and nucleus accumbens of human alcoholics. J. Neurochem. 2005;93:359–370. doi: 10.1111/j.1471-4159.2004.03021.x. [DOI] [PubMed] [Google Scholar]
- Fraga MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl. Acad. Sci. U. S. A. 2002;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin DW. Alcoholism and heredity: a review and hypothesis. Arch. Gen. Psychiatry. 1979;36:57–61. doi: 10.1001/archpsyc.1979.01780010063006. [DOI] [PubMed] [Google Scholar]
- Goodwin DW, Schulsinger F, Moller N, Hermansen L, Winokur G, Guze SB. Drinking problems in adopted and nonadopted sons of alcoholics. Arch. Gen. Psychiatry. 1974;31:164–169. doi: 10.1001/archpsyc.1974.01760140022003. [DOI] [PubMed] [Google Scholar]
- Grayson DR, et al. Reelin promoter hypermethylation in schizophrenia. Proc. Natl. Acad. Sci. 2005;102:9341–9346. doi: 10.1073/pnas.0503736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory BD, Belostotsky DA. Whole-genome microarrays: applications and technical issues. Methods Mol. Biol. 2009;553:39–56. doi: 10.1007/978-1-60327-563-7_3. [DOI] [PubMed] [Google Scholar]
- Hamid A, Wani NA, Kaur J. New perspectives on folate transport in relation to alcoholism-induced folate malabsorption—association with epigenome stability and cancer development. FEBS. J. 2009;276:2175–2191. doi: 10.1111/j.1742-4658.2009.06959.x. [DOI] [PubMed] [Google Scholar]
- Heberlein A, et al. Epigenetic down regulation of nerve growth factor during alcohol withdrawal. Addict. Biol. 2011 Mar 11; doi: 10.1111/j.1369-1600.2010.00307.x. (Electronic publication ahead of print). [DOI] [PubMed] [Google Scholar]
- Hillemacher T, Frieling H, Hartl T, Wilhelm J, Kornhuber J, Bleich S. Promoter specific methylation of the dopamine transporter gene is altered in alcohol dependence and associated with craving. J. Psychiatr. Res. 2009;43:388–392. doi: 10.1016/j.jpsychires.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, et al. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd-Acosta C, et al. DNA methylation signatures within the human brain. Am. J. Hum. Genet. 2007;81:1304–1315. doi: 10.1086/524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird PW. Principles and challenges of genome wide DNA methylation analysis. Nat. Rev. Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield RD, Harris RA, Schuckit MA. Genetic factors influencing alcohol dependence. Br. J. Pharmacol. 2008;154:275–287. doi: 10.1038/bjp.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Nestler EJ. The epigenetic landscape of addiction. Ann. N. Y. Acad. Sci. 2011;1216:99–113. doi: 10.1111/j.1749-6632.2010.05893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda RC, et al. MicroRNAs: master regulators of ethanol abuse and toxicity? Alcohol. Clin. Exp. Res. 2010;34:575–587. doi: 10.1111/j.1530-0277.2009.01126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S. Histone replacement marks the boundaries of cis-regulatory domains. Science. 2007;315:1408–1411. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- Muschler MA, Hillemacher T, Kraus C, Kornhuber J, Bleich S, Frieling H. DNA methylation of the POMC gene promoter is associated with craving in alcohol dependence. J. Neural Transm. 2010;117:513–519. doi: 10.1007/s00702-010-0378-7. [DOI] [PubMed] [Google Scholar]
- Ouko LA, Shantikumar K, Knezovich J, Haycock P, Schnugh DJ, Ramsay M. Effect of alcohol consumption on CpG methylation in the differentially methylated regions of H19 and IG-DMR in male gametes: implications for fetal alcohol spectrum disorders. Alcohol. Clin. Exp. Res. 2009;33:1615–1627. doi: 10.1111/j.1530-0277.2009.00993.x. [DOI] [PubMed] [Google Scholar]
- Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL. DNA demethylation in the Arabidopsis genome. Proc. Natl. Acad. Sci. U. S. A. 2007;104:6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesskaya OO, Aston C, Sokolov BP. Allele C-specific methylation of the 5-HT2A receptor gene: evidence for correlation with its expression and expression of DNA methylase DNMT1. J. Neurosci. Res. 2006;83:362–373. doi: 10.1002/jnr.20732. [DOI] [PubMed] [Google Scholar]
- Portela A, Esteller M. Epigenetic modifications and human disease. Nat. Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- Prescott CA, Kendler KS. Genetic and environmental contributions to alcohol abuse and dependence in a population-based sample of male twins. Am. J. Psychiatry. 1999;156:34–40. doi: 10.1176/ajp.156.1.34. [DOI] [PubMed] [Google Scholar]
- Prescott CA, Aggen SH, Kendler KS. Sex differences in the sources of genetic liability to alcohol abuse and dependence in a population-based sample of U.S. twins. Alcohol. Clin. Exp. Res. 1999;23:1136–1144. doi: 10.1111/j.1530-0277.1999.tb04270.x. [DOI] [PubMed] [Google Scholar]
- Scacheri PC, Crawford GE, Davis S. Statistics for ChIP-chip and DNase hypersensitivity experiments on NimbleGen arrays. Methods Enzymol. 2006;411:270–282. doi: 10.1016/S0076-6879(06)11014-9. Review. [DOI] [PubMed] [Google Scholar]
- Schumacher A, et al. Microarray-based DNA methylation profiling: technology and applications. Nucleic Acids Res. 2006;34:528–542. doi: 10.1093/nar/gkj461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheedy D, et al. An Australian brain bank: a critical investment with a high return! Cell Tissue Bank. 2008;9:205–216. doi: 10.1007/s10561-008-9076-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorak GG, Wellner JA. Empicical Processes with Applications to Statistics. New Jersey: Wiley; 1986. [Google Scholar]
- Taqi MM, et al. Prodynorphin CpG-SNPs associated with alcohol dependence: elevated methylation in the brain of human alcoholics. Addict. Biol. 2011;16:499–509. doi: 10.1111/j.1369-1600.2011.00323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant GE. Natural history of male psychological health. III. Empirical dimensions of mental health. Arch. Gen. Psychiatry. 1975;32:420–426. doi: 10.1001/archpsyc.1975.01760220032003. [DOI] [PubMed] [Google Scholar]
- Vaillant GE. Natural history of male psychological health. V. The relation of choice of ego mechanisms of defense to adult adjustment. Arch. Gen. Psychiatry. 1976;33:535–545. doi: 10.1001/archpsyc.1976.01770050003001. [DOI] [PubMed] [Google Scholar]
- Wilson AG. Epigenetic regulation of gene expression in the inflammatory response and relevance to common diseases. J. Periodontol. 2008;79:1514–1519. doi: 10.1902/jop.2008.080172. [DOI] [PubMed] [Google Scholar]
- Xin Y, et al. MethylomeDB: a database of DNA methylation profiles of the brain. Nucleic Acids Res. 2011;40:D1245–D1249. doi: 10.1093/nar/gkr1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Yuan Q, Mash DC, Goldman D. Substance-specific and shared transcription and epigenetic changes in the human hippocampus chronically exposed to cocaine and alcohol. Proc. Natl. Acad. Sci. U. S. A. 2011;108:6626–6631. doi: 10.1073/pnas.1018514108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Henikoff S. Genome-wide analysis of DNA methylation patterns. Development. 2007;134:3959–3965. doi: 10.1242/dev.001131. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]