Summary
The importance of neuronal morphology in brain function has been recognized for over a century. The broad applicability of “digital reconstructions” of neuron morphology across neuroscience sub-disciplines has stimulated the rapid development of numerous synergistic tools for data acquisition, anatomical analysis, three-dimensional rendering, electrophysiological simulation, growth models, and data sharing. Here we discuss the processes of histological labeling, microscopic imaging, and semi-automated tracing. Moreover, we provide an annotated compilation of currently available resources in this rich research “ecosystem” as a central reference for experimental and computational neuroscience.
1. Introduction
Neurons communicate via axons and dendrites, functionally and morphologically specialized tree-like processes. The importance of these branching structures is underscored by their broad morphological diversity across and within brain regions (Figure 1). In the central nervous system (CNS), the shape of the dendritic arbor is related to the cell-type specificity and large number of synaptic inputs. Furthermore, the extent of dendritic arbors, at least in peripheral nervous system sensory neurons, physically defines their receptive fields (Hall and Treinin, 2011), and axonal topology is known to affect synaptic output (Sasaki et al., 2012).
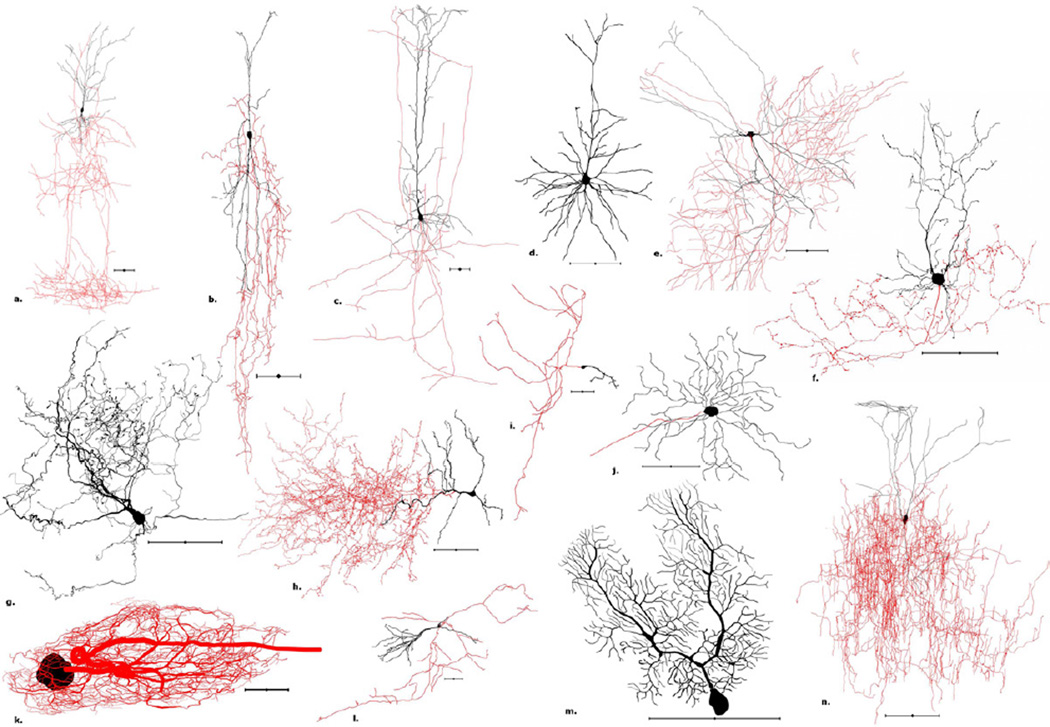
Figure 1. Morphological diversity: A representative sample of reconstructed neurons from NeuroMorpho.Org.
a. Rat neocortex Martinotti cell (NMO_00351). b. Rat neocortex bipolar cell (NMO_06144). c. Rat neocortex pyramidal cell (NMO_05729). d. Mouse neocortex pyramidal cell (NMO_05538). e. Mouse hippocampus Schaffer collateral associated neuron (NMO_07893). f. Mouse cerebellum Golgi cell (NMO_06902). g. Cat brainstem vertical cell (NMO_06171). h. Rat olfactory bulb deep short-axon cell (NMO_06222). i. Mouse neocortex Cajal-Retzius cell (NMO_07521) j. Mouse retina ganglion cell (NMO_06379). k. Spiny lobster stomatogastric ganglion motoneuron (NMO_06635). l. Rat hippocampus granule cell (NMO_06778). m. Mouse cerebellum Purkinje cell (NMO_00865). n. Rat neocortex layer 2/3 interneuron (NMO_04548). Scale bar: 100µm; somas and dendrites: black; axons: red.
The structure-function relation is central to many questions in neuroscience across all scales of investigation, from whole brain to macromolecules. Thus, the study of axonal and dendritic morphology plays a prominent role in the continuous investigation of neuronal activity and function. Yet, even some basic questions remain outstanding. For example, one of the most studied neuron types, cortical pyramidal cells, are characterized by morphologically distinct basal and apical dendrites, which receive distinctly organized synaptic inputs from different afferents and brain regions, but the functional implication of such a design is still not fully understood (Spruston, 2008). Computational models have shown that dendritic geometry can be responsible for producing the entire spectrum of firing patterns displayed across different cortical neuron types (Mainen and Sejnowski, 1996) and within a single class of electrophysiologically heterogeneous hippocampal neurons (Krichmar et al., 2002). The morphological development of these arbors influences synaptic organization and neural activity, which leaves a critical open question about the relationship between structure and function during growth.
Here, we briefly review the earlier history of the scientific characterization of axonal and dendritic morphology, leading to the current digital era (for a more thorough account, see Senft, 2011). We then outline how the establishment of a standard digital format for reconstructions of neuronal arbors catalyzed the emergence of a thriving research community that spans sub-disciplines, techniques, and scientific questions.
A brief historical overview of neuronal tracing
In the late 19th and early 20th centuries, Ramon y Cajal adopted Golgi’s staining technique to produce a revolutionary series of drawings of dendritic and (unmyelinated) axonal morphology that remain to this day absolutely remarkable for both their sheer amount and level of detail. This collection provided the foundation to approach the investigation of the structure-function relationship in nervous systems. The fundamental principles recognized by Cajal included the directional flow of impulses between neurons, the diversity of microcircuit motifs, and the specificity of network connectivity. Cajal’s work also established the intertwined relationship of three key processes in the characterization of neuronal morphology: histological preparation, light microscopic visualization, and accurate tracing. The spectacular morphological exuberance of axons and dendrites revealed by the Golgi stain could only be properly captured by faithful tracing of the arbors and their circuits. It also became apparent that neuronal trees, due to their enormous span relative to the caliber of individual branches, could not simply be reproduced (e.g. photographically), but needed to be reconstructed from numerous focal depths and fields of view.
Subsequently, interest in cellular neuroanatomy has seen its ups and downs, reflecting stages of advances and stagnation. In the 1960s and 1970s the development of easier, more sensitive, and more specific staining techniques led to a flourishing of descriptive neuromorphological studies across species, brain regions, and developmental stages. This progress spurred parallel strides in reconstruction technology. Glaser and Van der Loos (1965) used a “computing light microscope” to trace dendrites from 100 µm sections of the cerebral cortex while recording the location of the stage (x- and y-coordinates) and fine focus (z-coordinate). The system reproduced a two dimensional representation of Golgi-stained neurons and generated accurate measurements of dendritic length. Subsequently, similar reconstructions were obtained from micrographs (Macagno et al., 1979) or film strips (Levinthal and Ware, 1972) of serially sectioned tissue at the electron microscopy (EM) level. Ensuing advancements in computer hardware and software progressively shifted tracing and analysis from analogic media to a digital interface with the light microscope. Computerized microscopy systems recorded not just the position of the soma and dendrites, but also the tree origin, bifurcation, and terminal points (Wann et al., 1973). A system developed by Capowski (1977) additionally recorded process thickness, assigned an order to the traced points, and allowed differentiating natural terminations from cut ends due to tissue sectioning. The resulting Eutectic Neuron Tracing System could display reconstructed neurons graphically in three dimensions, becoming the first broadly adopted commercial product.
Further advancements in digital tracing for the past 35 years have focused mainly on ergonomic improvement, as it became increasingly clear that neuronal reconstruction was the most labor intensive and time consuming step of the process to extract axonal and dendritic morphology data from the brain. At present, the majority of neuromorphological tracing involves a human operator (Donohue and Ascoli, 2011), but promising attempts to develop completely automatic digital reconstruction of neuronal morphology will be discussed below.
An “ecological niche” of biomedical research
The increasing user-friendliness of digital reconstruction systems from light microscopy led to the widespread adoption of a standard vector-style representation of neuronal morphology as a branching sequence of interconnected tubules (Cannon et al., 1998; Ascoli et al., 2001). This simple format is compatible with diverse techniques and experimental approaches, from intracellular label injection and bright field visualization in vitro to genetic marker expression and confocal microscopy in vivo. Digital reconstruction constitutes a research hub bridging a host of neuroscientific topics. Interactions across sub-disciplines fostered the synergistic development of many tools for data acquisition, anatomical analysis, three-dimensional (3D) visualization, electrophysiological simulation, developmental modeling, and connectivity estimation. Open sharing of available digital reconstructions catalyzed the emergence of a continuously growing collection of interoperable resources. Such an “ecosystem” of three-dimensional reconstructions of neuron morphology is similar to that seen in other mature domains like gene expression patterns and non-invasive brain imaging.
2. From neuronal morphology to digital reconstructions
Three-dimensional reconstruction of neuronal morphology has been an established and widespread laboratory technique for three decades but recent progress in neurobiology, microscopy, and information technology has recently expanded both the breadth and the depth of these studies. We can now selectively label various neuron types, confirming their stunning phenotypic diversity and allowing identification of their distinguishing properties (PING, 2008). Advancements in light microscopy are increasing the resolution, contrast, speed, and applicability of neuronal imaging, revealing more refined and previously inaccessible morphological details. Continuous increase of computational power and algorithmic sophistication are constantly adding to the available applications of data processing.
Labeling Neurons
Cell labeling and tract tracing have long been pursued to elucidate the complex neuronal network architecture. Different staining methods developed over the years have yielded a rich histological toolbox (Figure 2a). Certain techniques are better suited for specific experiments and preparations and selecting the appropriate method is crucial. Basic criteria include clear contrast between the neurite and background tissue and maximum labeling extent of the neuronal arbor. Here, we overview a selection of labeling approaches (for more comprehensive coverage of these topics, please see Kobbert et al., 2000; Lanciego and Wouterlood, 2011).
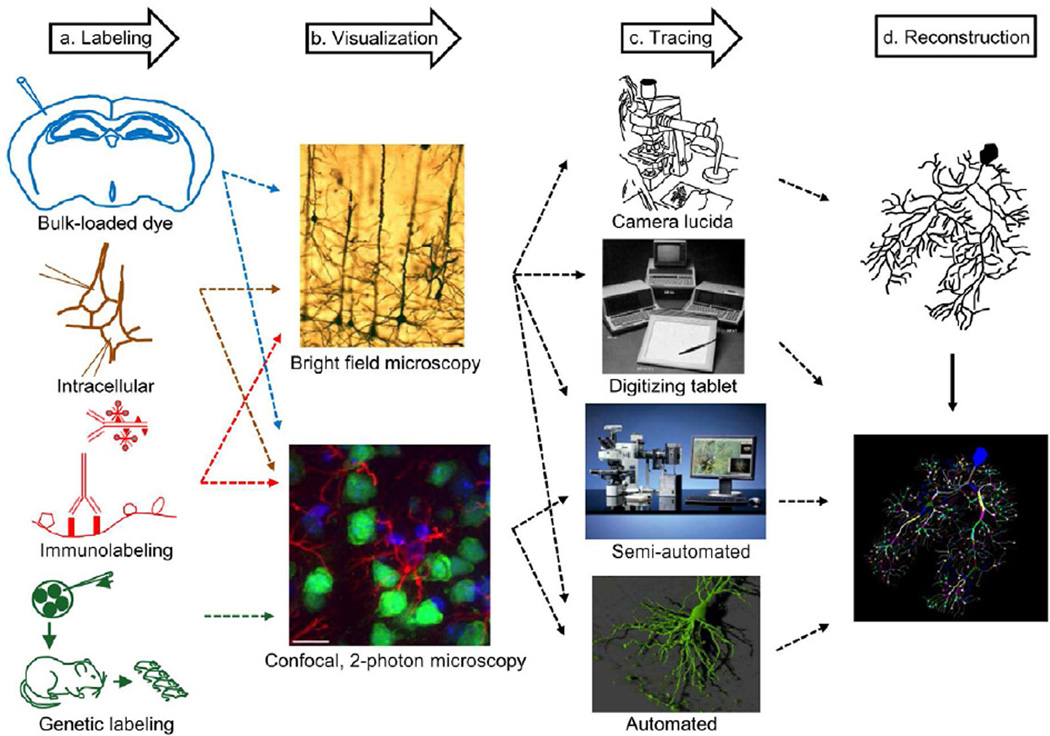
Figure 2. From nervous systems to digital reconstructions of neuronal morphology.
a. Staining techniques that label individual neurons include bulk extracellular loads, intracellular tracer injection, immunolabeling of cellular proteins, and genetic labeling that marks neurons intrinsically, b. Various optical microscopy visualization techniques (also depending on the labeling method) can be used to acquire images, which are then used to trace individual neurons, c. Tracing techniques have evolved over the years starting with pencil drawings using camera lucida, to a digitizing tablet that logs the tracing coordinates, followed by semi-automated methods with a computer interface and automatic algorithm-generated reconstructions that minimize manual intervention, d. Digital reconstructions files are produced by all systems interfaced with a computer. Analog camera lucida tracings can be scanned but require substantial post-processing for conversion to ‘vector’-style representation.
Immonulabeling (adapted from oncoprof.net/Generale2000/g04_Diagnostic/Histologie/Technique-texte/dg_gb_ap_tech06.html with permission from Prof. J.F.Heron) Bright field microscopy (from Lanciego and Wouterlood, 2011) Confocal, 2-photon microscopy (from Lemmens et al., 2010) Camera lucida (adapted from e-book.lib.sjtu.edu.cn/iupsys/Proc/mont2/mpv2ch05.html with permission from Dr. Nick Hammond) Digitizing tablet (This image is included under the fair use exemption and is restricted from further use) Semi-automated (with permission from MBF Bioscience, Inc.) Automated (with permission from Dr. A. McKinney, McGill University)
Bulk dye loading is used to visualize the gross morphology and connectivity patterns of neurons which can then be traced individually or as networks. The following is a selection of common dyes employed in morphological studies.
- Horseradish peroxidase (HRP)
HRP is visualized by histochemical analysis and its sensitivity is enhanced by conjugation with a non-toxic fragment of cholera toxin or with wheat germ agglutinin (Trojanowski et al., 1982), which slows removal from the loaded neurons and allows for visualization of the full structure.
- Biotinylated dextran amines (BDA)
The dextran amine is conjugated to a fluorescent dye and is detected by peroxidase and 3, 3’-diaminobenzidine tetrahydrochloride (DAB) reaction. The reaction product is distributed homogenously and fills the entire neuronal structure (Reiner et al., 2000).
- Phaseolus vulgaris Leucoagglutinin (PHA-L)
PHA-L is an anterograde tracer with unknown receptor-based uptake mechanism. Using antibodies against the lectin, PHA-L staining can be detected in the entire neuronal structure, including axon collaterals and terminals.
- Fluoro-Gold
The bleach-resistant properties of Fluoro-Gold (hydroxystilbamidine), an unconjugated fluorescent dye, make it a ‘gold standard’ in labeling. As a retrograde tracer it has been combined with up to two anterograde tracers like BDA and PHA-L where all three are visualized simultaneously in the same section (Lanciego and Wouterlood, 2011).
- Tritiated amino acids
[3H]-L-leucine locally injected in vivo is taken up by cell bodies, incorporated into proteins, and transported to the axons. The radioactive label is detected by autoradiography, which can be followed by standard histological staining to display the underlying cytoarchitecture.
- Golgi method
Fixed tissue immersion in solutions of potassium dichromate and silver nitrate fills the neurons with brown precipitate of silver chromate against a translucent yellow background. The Golgi stain impregnates only a fraction of neurons in the tissue by a yet unknown mechanism, highlighting fine details such as dendritic spines, but not myelinated axons. This characteristic is desirable yet constitutes at the same time a limitation. On the one hand, staining only a fraction of neurons makes it easier to identify the extent of individual dendritic arbors. On the other, this method fails to reveal the whole neural circuitry since it does not stain the full axonal network. Much of today’s knowledge about neuroanatomy and connectivity is owed to the Golgi staining technique.
- Lipophilic fluorescent carbocynanine dyes
Lipophilic fluorescent carbocynanine dyes like Dil and DiO are versatile as they can stain neurons in cultures as well as in living and fixed tissue. Dye diffusion in fixed tissue is limited to the labeled neuron, whereas in living tissue certain dyes like Dil can diffuse trans-neuronally. Dil, DiO and other carbocyanines such as DiAsp and DiA can withstand intense illumination and their strong fluorescence remains stable for up to one year (Kobbert et al., 2000). Particle-mediated ballistic delivery of these same dyes has shown to be successful in labeling individual neurons in both living and fixed tissue (Gan et al., 2000).
- Viral vectors
Viral vectors make excellent trans-neuronal retrograde markers, labeling the entire neuronal structures including small spines and distal dendrites of up to third-order neurons. Thus, they are particularly useful for studies of connectivity. The two main classes of tracers are derived from alpha-herpes viruses (Herpes Simplex virus Type 1 and Pseudorabies) and rhabdovirus (Rabies virus). The latter is better suited for studies of neuronal morphology because it is transported unidirectionally, is entirely specific to the neurons it propagates through, and does not induce neurodegeneration (Ugolini, 2010). Post-injection and incubation, immunohistochemistry reveals Golgi-like staining of neurons with fine details of thin dendrites and no background staining. Culture preparations also provide ideal conditions for viral gene transfer. Organotypic slices are simply incubated in the viral vector suspension for viral transduction to take place. Fluorescently labeled cells can be visualized as early as 24 hours after transfection and can be maintained for as long as three weeks (Teschemacher et al., 2005).
Intracellular injections of dyes directly into the neuron during electrophysiological recording allows for specific staining of single neurons, hence providing a straight link between neuronal structure and activity.
- Biocytin and Biotinamide (Neurobiotin)
Biocytin is a small conjugate of biotin and lysine naturally found in eukaryotic organisms. Due to its low molecular weight and biocompatibility, it constitutes a valuable tool in whole-cell or juxtacellular recordings, as it is incorporated into living neurons without perturbing ionic balance or membrane properties. Biocytin stains both axons and dendrites and is not transported trans-neuronally. Biotinamide (trademarked by Vector Laboratories as Neurobiotin) is the chloride salt of biotin with labeling capabilities very similar to biocytin. Both are soluble in electrolyte solutions for intracellular recordings. However, biocytin can be electrophoresed into neurons by either positive or negative currents, whereas biotinamide is selectively electrophoresed with positive currents (Kita and Armstrong, 1991). Since hyperpolarization is necessary to stabilize neurons after patching, the selectivity of biotinamide prevents spurious labeling of neurons before their viability is determined. Both biocytin and biotinamide have high affinity for avidin, and the tissue is processed post-fixation using an avidin-biotin-peroxidase complex followed by DAB reaction.
- Lucifer Yellow (LY)
LY, an intensely fluorescent non-toxic dye, is a popular intracellular label for both living and fixed tissue, though inexplicably it does not fill axons post-fixation. LY can be injected into the cell body by pressure or iontophoresis, and can also be used as a retrograde tracer after backfilling axonal terminals. Its low molecular weight allows for greater mobility between neurons and hence results in “dye-coupling”, where neurons morphologically connected to the filled neuron are also labeled. At times, these cells are found to be also electrically coupled, thus uncovering functional connectivity by morphological means. LY is compatible with other tracers (like HRP), but its unstable fluorescence requires photoconversion in presence of DAB to create a permanent record of labeled neurons for later reconstruction. Though both HRP and LY can label living neurons, these tracers make it more challenging to obtain accurate electrophysiological recordings. LY increases electrode resistance thus affecting the patch stability and modifying the intrinsic properties of the neuron.
Immunolabeling (immunohistochemistry and immunocytochemistry) tags and localizes proteins in intact tissue or isolated cells. Specific epitopes of proteins are targeted using antibodies, which when bound can be detected via conjugated secondary antibody binding and standard staining protocols. Multi-color double- or triple-immunostaining may reveal simultaneous presence of various proteins in the same neurons. Techniques like array tomography increase immunolabeling efficiency by allowing multiple staining cycles for the same tissue sample (Micheva and Smith, 2007). This method uses physical instead of optical sectioning to improve z-axis resolution in addition to depth-independent immunostaining. That is accomplished with arrays of ultrathin (50 nm) serial section ribbons of tissue on a single slide, which can be stained, imaged, eluted, and re-stained with different combinations of antibodies. The majority of antigen distribution is conserved during several staining cycles, without fluorescent intensity reduction or tissue damage.
Genetic labeling combines cytochemistry with molecular manipulations to color live biological systems intrinsically with genetically encoded fluorescent proteins (Lavis, 2011). Transgenic lines with exclusively labeled populations of cells, such as parvalbumin-expressing interneurons (Meyer et al., 2002) and astroglia (Nolte et al., 2001) are now the norm. The Brainbow technique incorporates genetic recombination to impart several dozen distinct colors in individual neurons and glia in the mouse nervous system (Livet et al., 2007). Similar techniques have been successfully applied in Drosophila (Hadjieconomou et al., 2011; Hampel et al., 2011).
Visualizing Neurons
In imaging the neuronal architecture of the brain, two main aspects should be considered: resolution and field of view. Visualizing large volumes of the brain, sufficient to include the entire territory invaded by a single axonal arborization, sacrifices resolution at the individual neuron level. Higher resolution imaging, useful to capture the finer details of spines, boutons, and synaptic contacts, is typically restricted to smaller regions. The future of imaging is a combination of both high resolution and large field of view without sacrificing either. Here we briefly discuss the types of light microscopy (Figure 2b) most relevant to neuromorphological reconstructions. In all these cases, resolution in the plane of illumination is generally greater than in the depth of the tissue.
Bright field microscopy
The majority of dendritic and axonal morphology reconstructions to date are based on bright field microscopy (Halavi et al., 2012), due to its broad compatibility with histological staining methods. In conventional bright field microscopy, as the name suggests, the tissue background is illuminated by transmitted light, whereas the stained neuron absorbs the light and is visible in dark contrast against the bright background. However, for certain applications or depending on user preference, simple image processing can be employed to invert this contrast (Myatt et al., 2012). Thus this modality should be more precisely referred to as trans-illumination or transmitted light microscopy. Unlike confocal microscopy, which requires fluorescent labels, bright field microscopy can visualize Golgi stain preparations and intracellular labels like biocytin. Even neurons labeled with fluorescent markers can be permanently labeled by DAB reaction and imaged with bright field microscopy. Moreover, the ability to enhance the signal intensity by counterstaining renders bright field microscopy largely unsurpassed for reconstructions of whole axonal arbors up to the very thin (and typically faint) terminals.
Confocal, 2-photon and super-resolution microscopy
Both confocal and 2-photon microscopy use point illumination, which narrows planar focus. In confocal microscopy, the emitted signal is spatially filtered via a pinhole aperture. The light emitted from a single plane creates an image, and a progression of images is captured through the thickness of the tissue, resulting in optical sectioning of the specimen (Wilson, 1990). Thus confocal microscopy eliminates the need for re-sectioning thick slices typically employed in electrophysiological recording preparations. Furthermore, confocal microscopy provides high spatial resolution, which is particularly important for 3D reconstructions. The temporal stability of the sample is especially relevant to neuronal reconstructions. Since fluorescently labeled specimens have a limited viability period, it is often necessary to collect image stacks for later offline tracing of the arbor structure.
In 2-photon microscopy, fluorophores are excited by the simultaneous absorption of two photons (Denk et al., 1990). The two photons converge simultaneously only at the focal point, yielding sharper images with less background noise. Such a specific illumination removes the need for spatial filtering. Additionally, since the fluorophores outside the focal point are not excited, the specimen undergoes less photobleaching and photodamage (Denk and Svoboda, 1997). Multiphoton microscopy, an extension of 2-photon, uses more than two photons to excite the fluorophores, resulting in a narrower emission region and even less out-of-focus noise. Since the stimulation energy is split between two (or more) photons, a drawback of 2-photon (or multiphoton) microscopy is its lower spatial resolution relative to confocal, due to the longer excitation wavelength. Moreover, the necessity to scan the specimen one point at a time greatly increases the time necessary to capture the same field of view (Lemmens et al., 2010).
Even with the higher resolution afforded by confocal and 2-photon microscopy, subcellular details such as synaptic contacts remain elusive and until recently, neuronal ultrastructure remained the purview of electron microscopy. However, the advent of super-resolution fluorescence microscopy addresses this limit. Two main approaches exist to enable super-resolution: one involves the sequential and stochastic on and off switching of fluorophores; the other uses patterned illumination to modulate fluorophore emission. The former includes Stochastic Optical Reconstruction Microscopy (STORM; Rust et al., 2006), and (Fluorescence) Photo Activated Localization Microscopy (PALM; Betzig et al., 2006; or FPALM; Hess et al., 2006). In this class of techniques, only a subset of fluorophores are illuminated in each imaging cycle and localized to be imaged and reconstructed, and the process is repeated to capture the full distribution of fluorophores. The second category includes Stimulated Emission Depletion (STED; Klar and Hell, 1999) and Saturated Structured Illumination Microscopy (SSIM; Gustafsson, 2005). STED increases lateral resolution by modulating the excited fluorescent molecule on the outer ring of the focal spot and preventing light emission via negative patterning. SSIM achieves the same effect by positive patterning with two interfering light beams. Super-resolution techniques allow inspection of neuronal morphology at the scale of tens of nanometers, and are thus suitable to identify location, architecture, dynamics, and molecular content of synapses (Huang et al., 2010). A recent study (Lakadamyali et al., 2012) tested the feasibility of tracing axons of cultured hippocampal neurons using multicolor 3D STORM and found the subsequent reconstructions to be more accurate than those with confocal imaging. With certain improvements in labeling density and their optical properties for better resolution in volume imaging of brain tissue, STORM may thus become a useful tool in mapping neural connectivity.
Tracing
There are multiple ways to digitize neuronal morphology once it has been visualized by optical microscopy. The structure of interest may be represented volumetrically by identifying all the voxels it occupies or as a surface contour delineating its spatial boundaries. A more effective alternative is to describe the tree-like branching of axons and dendrites as a sequence of interconnected cylinders (as in the widely used, non-proprietary SWC file format). In this “vector” representation, each uniform segment in the arbor can be parsimoniously characterized by only five values, corresponding to the three Euclidean coordinates and diameter of its ending location, plus the identity of the “parent” segment from which it originates. Tracing techniques have evolved over the years from the basic camera lucida to automated algorithms (Figure 2c) that generate digital reconstructions of neuron morphologies. While more modern reconstruction approaches are facilitated by increasingly automated computational algorithms, human intervention is still required in all cases at least to ensure error-checking and quality control.
Although the majority of existing reconstructions have been so far acquired with the commercial reconstruction software Neurolucida (Halavi et al., 2012), several alternatives exist. The availability of numerous options helps accommodate the wide variety of user preferences and data set characteristics. However, all reconstruction systems ultimately implement the same general process. Digital tracing of neuronal morphology converts large amounts of imaging information into a simple and compact representation (Figure 2d) that is easy to visualize, quantify, archive, and share (Meijering, 2010), thus maximizing the opportunity to exploit the full potential of collected experimental data through secondary discovery and meta-analysis (Ascoli, 2006). While only limited morphometric measures can be manually extracted from images and camera lucida tracings, digital reconstructions allow virtually unrestricted morphometric analyses (Costa et al., 2010), biophysically realistic computational modeling, and potential connectivity mapping. By reproducing branch topology and meandering, digital reconstructions faithfully capture both global properties and local features of neurons. Thus, digital reconstructions recapitulate the functional essence of neuronal morphology (Figure 3).
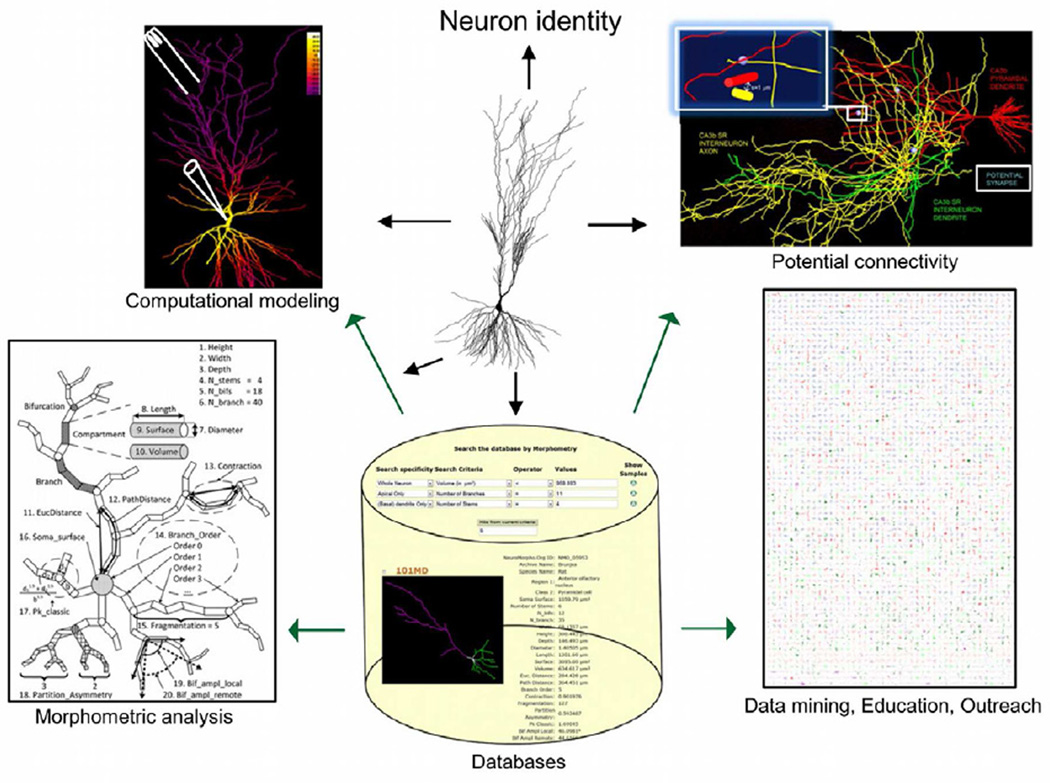
Figure 3. Scientific applications of digital reconstructions.
Three-dimensional tracing of axonal and dendritic morphology are typically acquired for one of several purposes, such as establishing neuronal identity, implementing anatomically and biophysically realistic simulations of neuronal electrophysiology, performing morphometric and stereological analyses, and determining potential connectivity. Data deposition in central repositories makes the reconstructions easily accessible for reuse in any of these applications, as well as data mining, education, and outreach.
Morphometric analysis (from Costa et al., 2010)
Potential connectivity (from Ropireddy and Ascoli, 2011)
Distillation of morphological data by digital reconstruction
Results obtained in cellular anatomy with the aid of digital reconstructions include comparative morphological characterizations of neurons, quantification of changes during development and pathology, determination of the genetic underpinning of neuronal structure, and establishment of general principles underlying neural circuitry. Moreover, three-dimensional tracing is now routinely employed to implement detailed computational simulations of biophysical mechanisms underlying growth and electrophysiological activity.
Early neuronal digital reconstructions were primarily used for quantitative morphological description of axons and dendrites in a range of species (Halavi et al., 2012). Neuronal reconstructions have been employed in direct comparative studies across species (Chmykhova et al., 2005), cell types (Bui et al., 2003; Andjelic et al., 2009), and hemispheres (Hayes and Lewis, 1996). Morphological investigations have also led to the discovery of new neuron types (e.g. Le Magueresse et al., 2011). Additionally, digital reconstructions can quantify morphological aberrations in pathological conditions, experience-dependent morphological changes, and morphological changes during development. Finally, the ever-increasing use of transgenic mice has vastly expanded research on the genetic factors in axonal and dendritic morphology, including protein regulation in the maturation and specification of neuron identity (Franco et al., 2012; Sulkowski et al., 2011; Michaelsen et al., 2010).
Statistical distributions of geometrical features extracted from digital reconstructions have aided the search for general principles underlying dendritic and axonal branching (Cuntz et al., 2008; Wen and Chklovskii, 2008; Snider et al., 2010; Teeter and Stevens, 2011) and computation (Seidl et al., 2010). Virtually embedding three-dimensional tracings in a template atlas of the brain enables analysis of system stereology, such as space occupancy (Oberlaender et al., 2011; Ropireddy et al., 2012). In recent years, whole brain 3D atlases have been acquired along with internally registered neuronal reconstructions in several insect models, constituting important progress towards the generation of comprehensive connectivity maps in these species (Kvello et al., 2009; Wei et al., 2010; Rybak et al., 2010; Chiang et al., 2011).
Even the morphological reconstructions of a handful of individual neurons can allow derivation of potential connectivity patterns by computational analysis of the spatial overlap between axons and dendrites (Stepanyants et al., 2002). Passage of an axon within a spine length of a dendrite is assumed to constitute a potential synapse (Stepanyants and Chklovskii, 2005). This “neurogeometry” theoretical framework has functional implications in neural circuits. For example, the maximum number of synaptic connectivity patterns resulting from spine remodeling, related to the network information storage capacity, can be quantitatively estimated based on the number of existing synapses and the shape and distribution of axons and dendrites (Escobar et al., 2008). Digital tracing of axons and dendrites in the cat visual cortex in vivo revealed distinct potential connectivity organizations in excitatory and inhibitory neurons relative to columnar domains (Stepanyants et al., 2008). Similar application of 3D reconstructions have investigated potential connectivity patterns in several other systems, from drosophila olfactory centers (Jefferis et al., 2007) to rat hippocampus (Ropireddy and Ascoli, 2011).
The relationship between neuronal morphology and network connectivity has always constituted a major motivation of digital reconstructions. Intracellular labeling was used to reconstruct connections in macaque visual cortex (Yabuta et al., 1998) and between excitatory neurons in rat barrel cortex (Feldmeyer et al., 1999), paired with dual whole cell patch recording to establish functional connectivity. With the neuronal reconstruction boom in the new millennium, connectivity patterns were rapidly characterized among other regions in the subiculum (Harris and Stewart, 2001), spinal cord (Dityatev et al., 2001), somatosensory cortex (Feldmeyer et al., 2005; Frick et al., 2008), and main olfactory bulb (Eyre et al., 2008), suggesting specific rules for the microcircuit architecture (Packer and Yuste, 2011). As with dendritic morphology, reconstructions were also heavily involved in determining the changes of connectivity patterns in response to environmental conditions such as stress (Vyas et al., 2006), hibernation (Magariños et al., 2006) or during neural circuit development (Peng et al., 2009).
One of the most important applications of digital reconstructions is in the implementation of biophysical simulations of electrophysiology. The neuronal arborization is represented as interconnected compartments, each sufficiently small to adequately reflect significant local variations of the distribution of membrane potential and membrane current along the length of each neurite. The compartment longitudinal and transverse resistances are set to reproduce the neuronal axial and membrane resistances, respectively. Additional terms (varying among compartments) describe the spatially distributed gradients of voltage-gated and synaptic properties. Such a framework enables simulation of fundamental aspects of neuronal function at the sub-cellular, cellular, and circuit levels.
The contributions of digital reconstructions to results of electrophysiological modeling are far too numerous to be overviewed comprehensively, ranging from network (Lytton and Sejnowski, 1991; McIntyre and Grill, 2002; Poirazi et al., 2003a; Ferrante et al., 2009) to cellular signaling (Schiller et al., 2000; Bartos et al., 2002; Destexhe et al., 1998; Gasparini et al., 2004). Notable discoveries about synaptic functioning involve signal integration (Stuart and Hausser, 2001; Spruston et al., 1994; Softky and Koch, 1993; Cauller and Connors, 1994), learning (Sah and Bekkers, 1996; Buonomano, 2000; Watanabe et al., 2002), and scaling (Liu et al., 2011) or lack thereof (Perez-Rosello et al., 2011). Mechanisms elucidated with this approach include action potential initiation and propagation (Hoffman et al., 1997; Hausser et al., 2001; Alle et al., 2009; Kole et al., 2008), information encoding (Cutsuridis et al 2010), neuron communication (Solinas et al., 2006; Traynelis et al., 1993; DiGregorio et al., 2002; Gulledge and Stuart, 2003; Silberberg and Markram, 2007), and oscillation (Atunes et al., 2003; Fransen et al., 2004; Margrie and Schaefer, 2003). Morphologically and biophysically realistic model of electrophysiology have also shed light on the neuronal structure-function relationship (Kim and Connors, 1993; Markram et al., 1997; Magee and Cook, 2000; Brecht et al., 2003), including effects of pathology (Chan et al., 2007; Chen et al., 2001; McIntyre et al., 2004) and drugs on neuronal activity (Poolos et al., 2002; Ferrante et al., 2008).
Another key application of digital reconstruction in computational neuroscience is to the modeling of neuronal morphology itself (Ascoli, 2002). Virtual generation of axonal and dendritic arbors is useful to explore mechanisms of growth (Eberhard et al., 2006; van Ooyen, 2011) and to construct biologically realistic neural networks (Koene et a., 2009). Moreover, reproducing in silico relevant geometrical features of experimental reconstruction data identifies the necessary and sufficient metrics to describe neuronal morphology, eliminating redundant descriptors. In these simulations, model parameters are randomly sampled from the statistical distributions of metrics extracted from real neurons. The stochastic nature of this process can generate an infinite number of non-identical neurons from a finite sample within each morphological class. Thus, neuromorphological models achieve both data compression and amplification.
3. Tools and resources for digital reconstructions
A defining feature of the ecosystem of neuronal reconstructions is the breadth and depth of the electronic toolbox currently available to the research community. This Section describes each of these digital resources from the user’s perspective, starting from the suitability for specific application domains and particularly noteworthy features. We comment on usability, including documentation, available support, user-friendliness, and whether the resource is actively maintained or under continuous development. We also consider accessibility in terms of compatible hardware and software platforms, noting whether the resource is commercial, free or open-source. Lastly, we discuss evidence of impact: published results, user base, course works, symposia, and books. Several of these details and additional pointers, such as literature references, contact information, and internet addresses, are summarized in Table 1. This “user’s digest” is organized in four Sections: a) digital tracing of morphologies from microscopic imaging; b) analysis and visualization, including post-process editing and morphometric extraction; c) simulation environments for single neuron and network modeling; and d) databases providing curation and free public access to reconstructions. Figure 4 illustrates representative user interface examples from the four categories. A brief compilation of relevant complementary tools is also included at the end of each Section.
Table 1.
Resources and tools of 3D reconstructions of neuron morphology: Unabridged collection of essential pointers and literature references
Resource:  ;
;  ; Free;
; Free;
morphological modeling (as opposed to electrophysiological)
Platform: W: Windows; M: Mac; L: Linux; (C: C/C++; J: Java; I: ImageJ plugin; O: Others, i.e. Matlab, Python, Lisp or Fortran)
Support: E: E-mail; F: Forum; G: Guide (Manual); L: mailing List; N: Newsletter; T: Tutorial; D: ongoing Development; A: Actively maintained
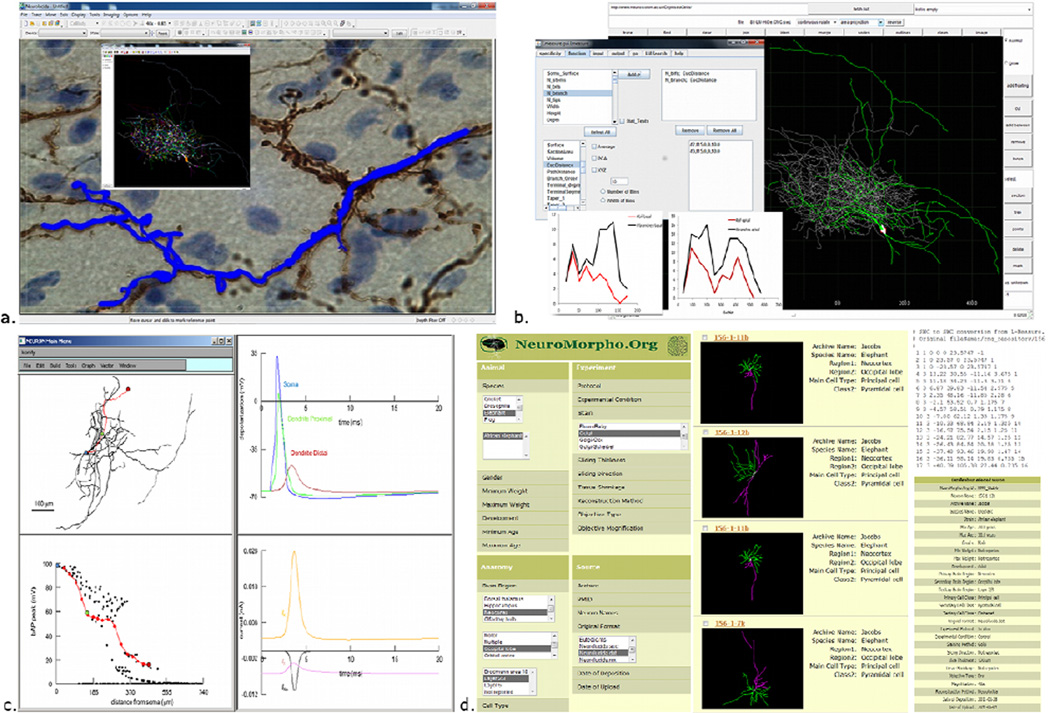
Figure 4. The ecosystem of digital reconstructions: representative resources and tools.
a. Neurolucida allows live or offline tracing and its companion module NeuroExplorer (inset) performs quantitative analyses, b. Digital reconstructions can be viewed and edited using Cvapp, and extensive morphometric analysis can be performed using L-Measure (inset. c. The NEURON simulation environment can distribute biophysical properties on imported neuronal reconstructions (in this example, a hippocampal interneuron; top left) for electrophysiological simulations. Here, the membrane depolarization is recorded (top right) at the soma (blue), proximal dendrite (green) and distal dendrite (red). The peak of back-propagating action potential along the dendrite decreases in amplitude with distance from the soma (bottom left), due to distinct current contributions (bottom right). d. NeuroMorpho.Org hosts digital reconstruction of neuronal morphologies that are published in peer-reviewed journals. Searching across different metadata categories (left) returns a summary result list (middle), which can be individually browsed to download the reconstruction as a standard SWC file (top right) or for additional metadata details (bottom right).
Digital tracing software
As described above, computer-aided reconstruction of neuronal morphology creates vector-format compartmental representations of dendritic and axonal arbors visualized by light microscopy. All existing tracing software requires a certain amount of user intervention, varying from manually drawing neurites to selecting parameters for automated or semi-automated reconstructions. Most tracing programs allow visualization of the reconstructed structure and offer some basic post-reconstruction editing and analysis functions, as well as file conversion utilities. Several reconstruction and visualization tools were created as plugins of the broad umbrella program ImageJ (rsb.info.nih.gov/ij). Initially introduced as a low-cost image-analysis software for the bench scientist (Schneider et al., 2012), this popular software has grown to include over 500 plugins performing functions from image acquisition, editing, and analysis to reconstruction and quantification. We made an effort to include all publicly available tracing programs. Other software for digital reconstruction may be in use in individual labs that was custom produced or is no longer distributed (e.g. Wolf et al., 1995).
Neurolucida (MBF Bioscience, Williston, VT, USA) is a comprehensive commercial package for three-dimensional neuronal reconstruction and brain mapping. Semi-manual tracing can be performed live from the microscope feed through specialized companion hardware or offline on collected image stacks (Figure 4a). The user clicks along the center line of the neurite, assigns the diameter with a circular cursor, and the software connects each point with the previous one. The AutoNeuron extension module (mbfbioscience.com/autoneuron) automatically reconstructs neurons from image stacks of sufficient quality and moderate complexity after adequate parameter setting. Neuron reconstructions can be viewed and edited in Neurolucida or exported into ASCII or binary files. Complementary programs include Neurolucida Explorer (see Visualization and analysis below) and Stereolnvestigator, which performs stereological analyses such as unbiased cell count and volumetric measurements. Extensive and detailed documentation is available in published books and user guides in addition to an online FAQs section. Webinars and web tutorials, training workshops, a user forum, and live remote assistance provide active and thorough support to users. Presentations at scientific meetings indicate a broad user base. A list of publications using Neurolucida is also maintained on their website. Neurolucida and its modules only run on Windows.
Filament Tracer is a module of the commercial Imaris package (Bitplane, Zurich, Switzerland) for detection of neurons, microtubules, and filaments in 2D, 3D, and 4D. Three-dimensional tracing is overlaid on imaging data in real time and a progression of reconstruction strategies is available, ranging from manual to fully automatic detection and segmentation methods. The software includes visualization and editing utilities and performs morphometric calculations as well as dendrite and spine tracking from time lapse datasets. Reconstructions are saved in Imaris ASCII files that can be converted into standard SWC format with L-Measure or NeuronLand (see Visualization and analysis below). Online video instructions cover all offered functionalities and an extensive FAQs section provides detailed answers and step-by-step troubleshooting. A list of publications using FilamentTracer is also available on their webpage along with a “Knowledge Base” illustrating case studies of specific features used in various experiments. Imaris hosts an annual User Group Meeting and offers education webinars. Filament Tracer runs on Windows and Mac.
Amira (Visualization Sciences Group) is a commercial software package with numerous extensions for different data sets. The Skeletonization extension supports reconstruction and analysis of vascular and dendritic networks. An image processing tool performs semi-automatic neuronal reconstruction from confocal images and stacks. Additional features include options for advanced visualization, simulation, and quantitative analysis. Several features in the Skeletonization extension are also present in the Filament Editor, part of a separate Microscopy extension. Reconstructions can be post-processed for high quality presentation and exported as movie files or in custom ASCII format that can be converted to SWC with L-Measure or NeuronLand. A list of publications utilizing Amira is available on their website. Customer support is provided and documentation includes release notes and FAQs. Amira Skeletonization runs on Windows, Mac, and Linux.
3DMA-Neuron was developed at the State University of New York at Stony Brook and is commercially distributed by Nihon Visual Science (Tokyo, Japan). This software performs automated reconstruction from high resolution images of single neurons along with various morphological measurements and integrated analysis of spine morphology. Output file formats include standard SWC and NeuroML. The program runs on Linux and other Unix operating systems including SGI Irix and SUN.
Eutectic NTS (Neuron Tracing System) was produced commercially, but the executable is now freely available from the developer. Eutectic allows tracing of neurons directly from a single image, and was employed in the majority of the early publications using digital reconstructions. The user can edit, merge, filter, and view the morphology in 3D, though this display is not fully interactive. The program performs numerous mathematical manipulations and maintains statistical summaries of morphology. Eutectic runs on Windows and user guide documentation is available online.
NeuronJ is a Java plugin of ImageJ that allows semi-automatic tracing and quantification of elongated structures from 8-bit gray-scale or indexed color images. The user indicates the tracing start point and moves the mouse along the neurite. The algorithm computes the optimal path between the initial point and the current position of the mouse and displays the path. The user can accept the suggested path with a click and continue. For more accurate representation of the neurite, users may try moving in shorter segments to avoid straight-line shortcuts that mis-represent neuritic curvature. Although it only enables 2D reconstruction, NeuronJ enjoys fairly broad usage, as it was the first freeware product to facilitate neurite tracing and analysis across computer platforms (Windows, Mac, Unix). Tracings can be saved in the native .ndf (NeuronJ data file) format or in various options of text files. The software is under active maintenance, but is no longer developed and the source code is not released.
NeuRa (NEUron Reconstruction Algorithm) automatically converts image stacks obtained by 2-photon microscopy into vector-style digital tracings. Image pre-processing is required to mark and delete unnecessary structures. After reconstruction, the file can be manually edited and saved in NEURON format (see Computational modeling below). The program is no longer developed nor actively maintained, but a concise manual is available on the website. NeuRa runs only on Mac with a simple graphical user interface. SpineLab, based on NeuRA, is a software tool that reconstructs dendritic trees as well as changes in spine density over time (Jungblut et al., 2012).
NeuronStudio is a self-contained free program for manual, semi-manual or automatic tracing of neuronal structures from confocal and multiphoton images. Reconstructions can be inspected and edited in 2D or 3D with the companion NeuroGL software (Rodriguez et al., 2003) and saved in standard SWC or NeuroZoom (parkerdigital.com/neurome) ASCII file formats. The software also performs simple measurements and image processing, including dendritic spine detection and classification. A detailed online manual provides step-by-step instruction for all functionality. Active user support is available via email and regular updates are posted by the developer. NeuronStudio runs on Windows and an OpenGL-capable video card is required for the 3D NeuroGL viewer only.
ORION (Online Reconstruction and Imaging Of Neurons) automatically reconstructs neuronal morphology from tiled image stacks acquired by confocal or multiphoton microscopy. The software accounts for experimental variables like background noise, visual artifacts of patch pipettes, and image alignment. Full dendritic arbors can be traced in several minutes without human intervention and saved in standard SWC or NEURON formats. Written in C++ and Matlab, ORION runs on Windows and Linux. An online blog serves as a discussion forum for users and developers.
Neuron_Morpho is an open source ImageJ plugin for semi-manual digital tracing of neurons from image stacks. The user outlines the surface of the neurite or soma by drawing straight lines across the width of the section. Thus an ellipsoid soma can be represented as a series of straight lines perpendicular to the length of the ellipsoid. Information about the line’s order, type, connectivity, x, y, z coordinates, and radius is recorded in a table. The contents of the table itself can be edited further. Though very easy to use, Neuron_Morpho may be cumbersome for complex neurons, yet remains extremely useful for simpler morphologies. Reconstructions can be saved in standard SWC and NEURON formats. Neuron_Morpho is no longer actively supported, but feedback is available from the developer. The code, written in Java, is open source and runs on Windows and Mac.
Farsight (Fluorescence Association Rules for multi-dimensional inSight) is an image analysis toolkit collaboratively developed by biological and computational investigators. The Open-Snake Tracing System of this toolkit allows automated reconstruction from 3D image stacks (Wang et al., 2011). The algorithm is self-contained including various reconstruction aspects from image pre-processing to tracing, editing, and generating different output formats. The Tracing System is currently under construction though the code for basic 3-D operations is available online. The Trace Editor feature of the Farsight toolkit (Luisi et al., 2011; www.farsight-toolkit.org/wiki/Trace_Editor) allows closer inspection of the reconstruction along with rapid identification and correction of errors using a graphical interface. Though these are presently independent modules, current development is focused on integrating the Trace Editor with the Tracing System. Farsight, written in C++, is open source and runs on Windows, Mac, and Linux.
Trees Toolbox allows manual and automatic neuronal reconstruction from image stacks as well as editing, visualization, quantitative branch measurements, and electrotonic analysis. Furthermore, the package contains a neuron construction module that uses a growth algorithm to build virtual dendritic arbors. The user accesses the toolbox in Matlab and can call a graphical interface to trace neuronal morphologies as cylinder-based OpenGL renderings. Morphologies can be exported in formats compatible with NEURON and neuroConstruct (see Computational modeling below) or as simple movies. Video tutorials as well as a detailed, clear online manual are available along with a user forum and developer mailing list. The open source Matlab code runs on Windows, Mac, and Linux. Trees Toolbox is actively maintained and under continuous development.
Vaa3D (3D Visualization Assisted Analysis) is a multi-functional system for bioimage and surface object analyses. The Vaa3D-Neuron module traces morphology from large image stacks of numerous file formats, visualizes and measures the resulting 3D structure, and facilitates comparative analyses. Video tutorials are available on the website along with a list of publications citing Vaa3D and test datasets that allow hands-on tracing experience with the graphical interface. An online discussion forum and mailing list provide user support, bug reports and miscellaneous announcements. Vaa3D is under active maintenance and continuous development. Developers can write their own plugins for the platform and community participation is encouraged via “hackathon” events. The C code is open source and runs on Windows, Mac, and Linux.
Simple Neurite Tracer is an ImageJ plugin true to its name: it is easy to install and its intuitive graphical tracing interface makes creating 3D reconstructions simple. The exploratory tracing system efficiently finds the best path between user-selected points in large image stacks. For a novice user, the option to visually select parameters to increase contrast from a preview palette of the preprocessed image is a very intuitive process. The program offers interactive 3D visualization, performs basic morphometric analysis, and interacts with other ImageJ plugins. Simple Neurite Tracer supports numerous imaging formats and exports reconstructions as raw comma separated values, standard SWC files or as its native XML-based format. A number of online video tutorials provide step-by-step instructions to the functionalities, and additionally a user guide and FAQs offer detailed documentation. A user mailing list, online bug tracking and reporting, and e-mail support are also available. The open source Java program runs on Windows, Mac, and Linux.
Neuromantic is another reconstruction software with a simple interface that allows manual and semi-automatic neuromorphological reconstruction from single images or image stacks. In the semiautomatic tracing system, the user indicates the starting point and then moves the mouse along the centerline of the neurite, while the program automatically estimates the radii of the process. Neuromantic allows the user to view and interact with the tracing within the same application as it is being created. Its intuitive editing features make it a powerful resource for 3D reconstructions. The reconstructions can be saved in SWC format and conversion with MorphML is also available. Video tutorials are offered online along with a blog of updates and a list of publications utilizing Neuromantic. Neuromantic is open source and runs on Windows.
COMPLEMENTARY TOOLS
Two other types of software programs are particularly relevant to digital tracing of neuronal morphology. The first consists of algorithms for fully automating the reconstruction process. At present, automated systems are not sufficiently general and robust to replace manual reconstructions in most cases. In part, this is due to the broad variation in tissue preparation, staining methods, and imaging techniques described above. Nevertheless, automated tracing of neuronal morphology holds the promise of high throughput reconstruction, changing the type of scientific questions that can be asked (Svoboda, 2011; Donohue and Ascoli, 2011). The DIADEM Challenge (Digital reconstructions of Axonal and DEndritic Morphology) recently screened a number of software entries remotely developed for automatically tracing representative neuroscience data sets (Brown et al., 2011) with a novel custom-designed metric for quantitative comparison against the manual benchmark (Gillette et al., 2011). The five finalist algorithms are freely available for download at diademchallenge.org. Other automated tracing algorithms are being developed in individual laboratories (e.g. Chiang et al., 2011; Peng et al., 2011). In particular, the software that enabled the first (and so far only) high-throughput reconstruction study, 16,000 drosophila neurons (Lee et al., 2012), can be downloaded as executable for different operating systems (flycircuit.tw/NT/Win32.zip; flycircuit.tw/NT/Win64.zip; flycircuit.tw/NT/Linux_x86.zip), but lacks user-friendly documentation.
The second type of electronic tool related to three-dimensional reconstruction of neuronal morphology consists of software to trace neurons in a format other than vector-style. The most common alternative is the ‘surface’ representation typically adopted to reconstruct neurons from high resolution imaging such as EM. A popular tool for this style of neuronal tracing is Reconstruct (synapses.clm.utexas.edu/tools/reconstruct/reconstruct.stm), a free editor that facilitates montaging, alignment, analysis, and visualization of serial sections. Reconstruct enables tracing of different structures over large number of sections and images. A semi-automated tracing utility determines the boundary of the region surrounding a location selected by the operator according to user-defined parameters. The 3D surface rendering is generated from the z-traces drawn over multiple serial sections. Reconstruct has a detailed online manual, is open source and runs on Windows.
Other reconstruction plugins for ImageJ include IJ-MorphDig (retina.anatomy.upenn.edu/~rob/ncman3) that allows morphological tracing from confocal image stacks to be used specifically with “Retsim”, a retinal simulation package included with NeuronC (see Computational modeling below); Skeletonize 3D (fiji.sc/Skeletonize3D), which is based on the implementation of a previous 3D thinning algorithm (Lee et al., 1994); Neurite Tracer (Pool et al., 2008; fournierlab.mcgill.ca/neuritetracer.html); and the more recent NeuronPersistentJ (imagejdocu.tudor.lu/doku.php?id=plugin:utilities:neuronpersistentj:start). These latter three only produce ‘volumetric’ reconstructions without generating segment-based arbor connectivity. Thus, they are suitable for visualization and limited analysis, but not for broader application such as compartmental modeling and extensive morphometric characterizations.
Visualization and analysis
Increasing adoption of digital reconstruction software created the demand for powerful and user-friendly tools for visualization and analysis. As mentioned above, these functionalities are often included within the same software environments that allow for morphological tracing. However, a few additional stand-alone resources are also available, which we describe here.
Neurolucida Explorer is a 3D visualization and morphometric analysis program (Figure 4a inset) that accompanies Neurolucida. Automatic morphometric analysis can be performed on an entire data set or on selected objects within a data set collected with Neurolucida. Reconstructions and analysis tables can be exported into other graphics programs and MS-Excel, respectively. User support and system requirements are the same as described for Neurolucida.
NeuronLand offers two freely available utilities. The Morphology Converter supports format conversions of 3D reconstruction files between more than 20 different formats. This command line program also performs limited statistical morphological analysis and multiple other functionalities to manipulate the reconstructed branching structure, such as cutting, splicing, flipping, and stitching subtrees. The second application, NeuronLand Morphology Viewer, provides 3D interactive display importing and exporting data in the same formats supported by the Converter. These programs are regularly updated and actively maintained. Online tutorial and instruction manual are available as well as a user support mailing list. NeuronLand Morphology Converter runs on Windows and Mac, while the Viewer only runs on Windows.
Cvapp is a visualization, editing, and file conversion tool with a user-friendly interface to zoom, pan, and rotate Neurolucida and SWC reconstructions in 3D (Figure 4b). Cvapp can export reconstruction files into NEURON and GENESIS formats (see Computational modeling). Online documentation is available, but the software is no longer developed or maintained. The free and open source Java code runs on Windows, Mac, and Unix, and can be customized to meet specific requirements.
L-Measure is a free, open source program that extracts over 30 quantitative morphological parameters from neuronal reconstructions in a variety of file formats. Morphological measurements can be computed from the entire neuron or from a user-specified portion of the neuronal structure (Figure 4b inset). The C/C++ program can be executed via command line or through a user-friendly graphical interface written in Java. A help section within L-Measure provides detailed instructions for running the programs and in-depth explanations for each parameter. Additional user support is available via mailing list and discussion group. The software is available as an online applet or downloadable stand-alone application, and the distributed source code has been incorporated in other applications such as Farsight and Vaa3D. L-Measure is actively maintained, under continuous development, and runs on Windows, Mac, and Linux.
COMPLEMENTARY TOOLS
Quantitative analysis is not restricted to the morphometry of vector-style digital reconstructions. Stereological parameters such as cell counts or volume and surface measures can be extracted from optical microscopy images with StereoInvestigator (mbfbioscience.com/stereo-investigator), Neuron Image Quantitator (NeuronIQ: cbi-tmhs.org/Neuroniq), a Matlab program with code available upon request, NEuron MOrphological Analysis Tool (NEMO: Billeci et al., 2013; centropiaggio.unipi.it/content/nemo-neuron-morphological-analysis-tool) that performs dynamic morphometric analysis on images and the ImageJ plugin NeuronMetrics (Narro et al., 2007; ibridgenetwork.org/arizona/ua07-56-neuronmetrics). Huygens software (svi.nl/HuygensSoftware) is another image processing and analysis package used to quantify light microscopy data sets in neuroscience that runs on Windows, Mac, and Linux. Similar applications are offered by several leading commercial microscopic imaging systems. An additional related development is MorphML (Crook et al., 2007), a formal mark-up language to describe the compartmental representation of reconstructed neuronal morphology like SWC files (neuroml.org).
Computational modeling: virtual growth and electrophysiological simulations
Use of digital reconstructions in morphologically and biophysically realistic simulations and network models allows direct investigation of the neuronal structure-activity-function relation. The following is an overview of currently available modeling software, all of which is free and typically open source. The first four programs are for morphological modeling (marked with an asterisk in Table 1). The remaining are for biophysical simulations of neuronal electrophysiology.
L-Neuron is a computational tool to generate anatomically accurate virtual neurons of various morphological classes. L-Neuron resamples statistical distributions extracted from experimental data to generate virtual neurons according to algorithms that implement established anatomical rules. The 3D morphological models can be visualized with the companion L-Viewer program and exported into classic graphic formats (bitmap, VRML, DXF, POV, Blob) or standard SWC format for morphometric analysis and electrophysiological simulations. Online documentation and e-mail user support are available. Actively maintained but no longer developed, L-Neuron is written in C/C++ and runs on Windows and Linux. The source code is available upon request bundled within L-Measure.
NeuGen utilizes experimental data to extract relevant parameters, based on which it generates synaptically connected somatosensory barrel cortical neurons of different morphological classes (pyramidal, star pyramidal, and stellate cells). Morphologies are created in a format compatible with NEURON. The online manual is detailed with screen shots providing step-by-step guidance. NeuGen is free, open source, and under ongoing development. The program is written in Java and runs on Windows, Mac, and Linux.
NetMorph simulates neuronal morphogenesis to generate large-scale 3D neuronal networks. Axonal and dendritic morphology is modeled according to realistic growth cone dynamics with synaptic formation along the way. Animated output files allow visualization of the developing structure over time. Full anatomy or abstracted circuit connectivity can be exported as text files. NetMorph is under ongoing development and online documentation is available. The program is written in C++, open source, and runs on Windows, Mac, and Linux.
CX3D simulates the realistic development of large 3D neural circuits. Mimicking biology, cells are generated by replication, then migrate into the layered cortex and extend their neurites to form synaptic connectivity. CX3D models branching patterns and growth based on intracellular protein concentrations and diffusion of extracellular signaling molecules. The connected network can be exported in the XML format. A manual and tutorial are available online in addition to a discussion forum. CX3D is written in Java, open source, and runs on Windows and Mac.
NEURON is the leading simulation environment for compartmental modeling of neuronal biophysics and electrophysiology. The user-friendly graphical interface allows for file input, format conversion, and output, new cell model specification, implementation of small circuits including gap junctions and extracellular electric fields, model analysis and optimization (Figure 4c). Extensive documentation includes “The NEURON Book” (Carnevale and Hines, 2006), online tutorials, articles, and additional material. NEURON has a large user base and support is available via mailing list, blog, and discussion forum. Workshops and summer courses are also regularly held. As of January 2012, this software was used in 1176 publications (www.neuron.yale.edu/neuron/static/bib/usednrn.html). Written in C++, NEURON is actively maintained and under continuous development. The program runs on Windows, Mac, Linux, and on parallel architectures including Beowulf clusters, IBM Blue Gene, and Cray XT3. The popularity of NEURON also encouraged the implementation of ancillary tools such as Neuronvisio, an enhanced interface allowing interactive 3D model representations and saving/reloading of simulation results (Mattioni et al., 2012; michelemattioni.me/neuronvisio).
GENESIS (GEneral NEural Slmulation System) supports implementation of biologically realistic models of neuronal activity ranging from biochemical reactions at the subcellular level to large networks at the systems level. The environment is intended to allow users to focus on simulation design, execution, and analysis (with a graphical user interface), while enabling developers to continuously expand available functionality for education, collaboration, and research. To resolve recent difficulties with community contribution to the simulator implementation, GENESIS is currently undergoing a software reconfiguration based on a federated architecture under the Computational Biology Initiative. Extensive documentation is available in the form of tutorials, articles, and the free internet edition of “The Book of Genesis: Exploring Realistic Neural Models” (genesis-sim.org/GENESIS/bog/bog.html), which is also popular in print (Bower and Beeman, 1998). An online forum and a mailing list provide support and allow interactions between users and developers. GENESIS runs on Windows with Cygwin, Mac, and Linux, and a parallel version is available for cluster architectures.
NeuronC allows construction, 3D visualization, and biophysically realistic physiological simulation of up to 10,000 neurons in the retina circuit. NeuronC is actively maintained and regularly updated. A detailed manual is available online along with a discussion mailing list. NeuronC runs on Unix, is written in C/C++, and is open-source.
Surf-Hippo is a simulation system to investigate morphologically and biophysically detailed compartmental models of single neurons and networks. Cells and circuits can be constructed using built-in functions and various anatomy file formats. Surf-Hippo is similar to NEURON and GENESIS, but is written in Lisp. A new release of Surf-Hippo, which will support NeuroML, is under development. Online manual and a mailing list are available. Surf-Hippo is open source and runs on Linux.
Catacomb is a modeling system for defining, building, and exploring biological models with a focus on biologically plausible neurons, synapses, and networks in the context of whole animal behavior. Online documentation includes screen shots and accompanying explanations. Written in Java, Catacomb is not open source and runs on Windows, Mac, and Linux.
neuroConstruct is designed to simplify development, visualization, and analysis of complex networks of biologically realistic neurons in 3D by generating script files for other simulation platforms like NEURON, GENESIS, Moose, PSICS, and PyNN (see below). Neuronal morphologies can be imported directly into neuroConstruct. Simulator-independent, conductance-based cell models can be generated and spatially arranged to create synaptically connected networks. Network simulations are then scripted out to e.g. NEURON or GENESIS, and the results stored in text files. Simulations can be loaded back into neuroConstruct for visualization and analysis. neuroConstruct is under active maintenance and ongoing development. A mailing list and online tutorials are available. The program is written in Java, open source, and runs on Windows, Mac, and Linux.
Moose (Multiscale Object-Oriented Simulation Environment) allows large, detailed, numerical simulations across the scales of single molecules, subcellular networks, individual cells, neuronal networks, and larger systems. Moose is backward compatible with GENESIS and forward compatible with Python and XML-based model definition standards. A graphical toolkit runs the Python user interface to Moose for simulations. Python allows cross-simulator interoperability, such as combining a NEURON electrophysiological model with a GENESIS biochemical signaling model. Detailed documentation is available online along with a bug tracking and reporting system. Moose is open source and currently in beta release.
PSICS (Parallel Stochastic Ion Channel Simulator) enables simulation of neuronal activity in complex morphologies with arbitrary localization of ion channels. The Interactive Channel INsertion Graphical user interface (ICING) allows definition and editing of cell properties and distribution of membrane conductances over the neuron structure. Morphologies can be imported from NEURON, neuroConstruct, and NeuroMorpho.Org (see Database availability) in NeuroML. Several built-in models, an online user guide, and links to external useful resources are also provided. PSICS is written in Fortran and Java, open source, and runs on Windows, Mac, and Linux.
COMPLEMENTARY TOOLS
The above list does not include the numerous published computational models of neuronal morphology (e.g. Samsonovich and Ascoli, 2005; Lopez-Cruz et al., 2011), as this compilation focuses on research tools available to the scientific community as opposed to individual custom solutions restricted to the domain of a single laboratory. The software programs listed for computational modeling of neuron electrophysiology include resources for circuit simulation, but always with the ability of representing dendritic and axonal morphology. Various other neural network simulators largely or exclusively consider point neurons as the elementary unit of computation, sacrificing neuron-level realism for larger-scale modeling. For example, PCSIM (Parallel neural Circuit SIMulator; lsm.tugraz.at/pcsim) allows parallel simulation of large-scale heterogeneous spiking and analog neural networks with up to millions of different point neurons and billions of synapses. Emergent (grey.colorado.edu/emergent) models neural dynamics at the level of activity rates. Topographica (http://topographica.org) focuses on modeling activity in cortical maps. CNS (Cortical Network Simulator cbcl.mit.edu/jmutch/cns) is designed specifically for graphical processing units (GPUs).
Other neural network simulators include CNRun (johnhommer.com/academic/code/cnrun), LENS (tedlab.mit.edu/~dr/Lens), Nodus (www.tnb.ua.ac.be/software/nodus/nodus_info.shtml), Simbrain (simbrain.net), and NEST (Neural Simulation Technology; nest-initiative.org). PyNN (neuralensemble.org/trac/PyNN) is a Python-based, simulator-independent language for building neuronal network models. The same unmodified PyNN code runs with several supported programs, including NEURON, PCSIM, NEST, and Brian (briansimulator.org), which itself similarly allows implementation seamlessly compatible on GPUs and traditional hardware (Goodman, 2010). NeuroTools (neuralensemble.org/trac/NeuroTools) provides supporting tools for tasks associated with a simulator such as setup, parameterization, data management, analysis, and visualization. NeurAnim (sourceforge.net/projects/neuranim) animates neural network simulations in 3D. These latter two resources may be valuable for use with circuit simulations implemented in the environments described above and in Table 1.
Database availability
The explosive and continuing acceleration in collection and application of digital reconstruction of neuronal morphology has created a pressing need for organized efforts towards data curation, annotation, storage, and distribution. Meta-analysis of existing primary data pooled from many studies and reanalysis of selected datasets can lead to remarkable secondary discoveries (Ascoli, 2007). In turn, data sharing and re-use by the community increases the visibility and impact of the original studies for which the data were collected. Therefore, several labs are freely posting their reconstructions online after result publication, either on their own web site, or in public resources such as NeuroMorpho.Org. This section describes existing free databases of neuronal reconstructions, all of which are actively maintained and regularly updated. Since NeuroMorpho.Org mirrors data from other existing collections in addition to allowing direct deposition from researchers, individual laboratory databases are not described separately (though they are listed at the end of the Section), unless they contain relevant complementary data not included elsewhere.
-
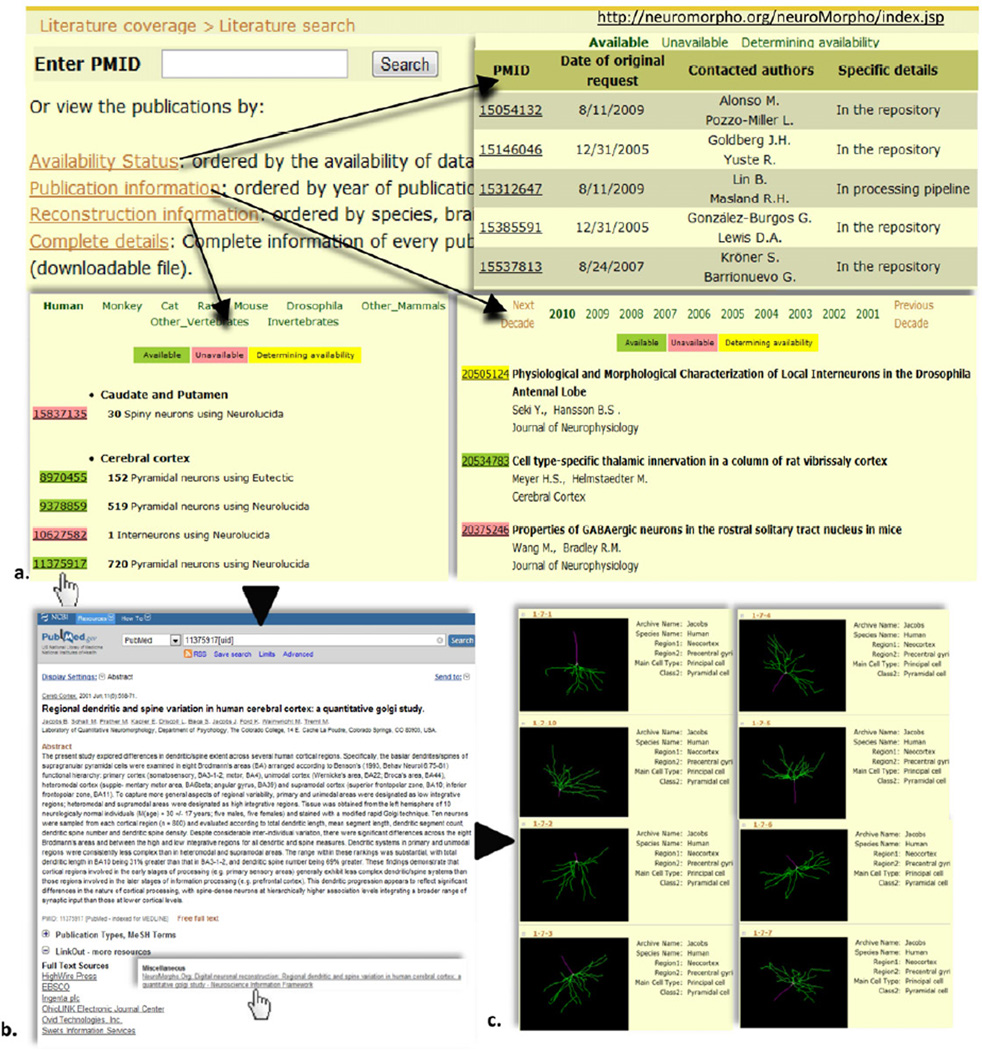
NeuroMorpho.Org is a public, NIH-sponsored, central repository of digital reconstructions of neuronal morphologies. Version 5.5 (Winter 2013) had 8858 reconstructions contributed by 108 laboratories from 19 species and 20 brain regions, representing the broad diversity of dendritic and axonal morphology (Figure 1). Morphologies can be browsed and searched with drop-down menus (Figure 4d) or with a Google-like keyword bar by any metadata category (e.g. neuron type, experimental conditions, and file format), as well as by morphometry (number of branches, volume, etc.). Neurons are returned as summary lists or organized by animal species, brain region, neuron type or lab of origin, and can be downloaded or inspected in the browser with an intuitive interactive display. All available rodent neurons are also accessible through a voxel density map visualized in a 3D mouse brain atlas. A quick-start guide, FAQs, and numerous links to many other relevant resources described in this review are also available online. NeuroMorpho.Org is recognized as a major enabling resource in the research ecosystem of digital neuromorphology.
New reconstructions are regularly added to the database. Original digital tracing files received from contributors are processed to generate a standardized SWC format, 2D image, and 3D animation of the morphology. The original, standardized, and rendered files are all freely downloadable along with log files reporting changes enacted in the conversion process and detailed notes. Each reconstruction in NeuroMorpho.Org is further annotated with rich information including animal strain, age, gender, weight, histological protocol, staining method, and microscopy technique. Moreover, all morphologies are associated with their corresponding PubMed references. In turn, PubMed abstracts of publications whose morphologies are deposited in NeuroMorpho.Org enable direct “linkout” access to the digital reconstructions from the database. Clear terms of use ensure that contributors are appropriately cited when their data are downloaded and used in published studies. Reconstructions posted on NeuroMorpho.Org have been utilized in over 120 peer-reviewed publications. More than 2.4 million files have been downloaded in the past six years in over 100,000 visits from 125 countries. NeuroMorpho.Org also maintains extensive literature coverage of publications containing neuromorphological tracings since the inception of digital reconstruction technology (Halavi et al., 2012). Publications can be perused by entering the PubMed identifier and browsed by reconstruction information, year of publication or availability status of the described data (Figure 5).
CercalDB (Cricket cercal system database) contains over 100 freely downloadable reconstructions of sensory neurons and interneurons of the cricket cercal system, categorized by type, length, and directional tuning. The concise and specific nature of this database facilitates seamless browsing and searching. Selected reconstructions can be viewed together along with an on-line comparison of their neuronal properties.
CCDB (Cell Centered Database) provides free access to digital reconstructions from various systems in addition to underlying raw microscopy image sets. CCDB contains high resolution 2D, 3D, and 4D data from light and electron microscopy as well as image files of protein localization, brain mosaics, tomographic registrations, and analyses. The database also includes a Subcellular Anatomy Ontology linked to the Neuroscience Information Framework (see Complementary Tools).
Fly Circuit is a database of Drosophila neurons containing approximately 16,000 reconstructions from the nine major neurotransmitter systems in the optic lobe and central brain of the fruit fly. Genetically labeled local and projection neurons in 29 neuropil regions from each hemisphere were individually reconstructed with a custom automated algorithm after manual preprocessing of the confocal images. Standard brain models of 6-day-old male and female adult flies allow users to register neurons in a common anatomical framework. Reconstructions can be visualized in 3D in different modes such as volume renderings, skeleton mode, and by soma location within the atlas. Each reconstruction is accompanied by metadata including the animal gender, age, and GFP expression driver, as well as the putative neurotransmitter and spatial distribution of axon terminals. This information is coded to allow rapid, full-scale similarity searchers. Reconstructions can be downloaded as Amira files.
Figure 5. Literature database of references reporting digital reconstructions of neuronal morphology.
a. Articles describing neuromorphological tracings are categorized in NeuroMorpho.Org based on availability of the reconstructions, year of publication, and species from which the morphologies are traced. b. Each publication is listed with its PubMed identifier (PMID) linking to the online abstract. c. If the reconstructions from the publication are available in the repository, they can be directly retrieved from PubMed via linkout mechanism.
COMPLEMENTARY TOOLS
Several laboratories maintain publicly available databases of reconstructions from their own studies. These include the collections of Drs. Alexander Borst (www.neuro.mpg.de/30330/borst_modelfly_downloads), Brenda Claiborne (utsa.edu/claibornelab), Alain Destexhe (http://cns.iaf.cnrs-gif.fr/alain_geometries.html), Attila Gulyas (www.koki.hu/~gulyas/ca1cells), Patrick Hof (research.mssm.edu/cnic/repository), Gregory Jefferis (flybrain.stanford.edu), William Kath (dendrites.esam.northwestern.edu), Dennis Turner (www.compneuro.org/CDROM/nmorph), and Rafael Yuste (columbia.edu/cu/biology/faculty/yuste/databases). These databases are mirrored into NeuroMorpho.Org for centralized access to all reconstructions and associated metadata. The Virtual Neuromorphology Electronic Database (krasnow1.gmu.edu/cn3/L-Neuron/database) contains virtual models of neuronal morphology generated with the L-Neuron program (see Computational modeling), which can also be re-analyzed or employed for biophysical simulations of electrophysiology.
The Invertebrate Brain Platform (invbrain.neuroinf.jp; Ikeno et al., 2008) is a repository of confocal images and electrical responses of neurons in systems including honeybee, silkworm, cockroach, and crayfish. Experimental data are integrated with mathematical models and research tools relevant to the study of invertebrate brain, neurons, and behavior. The DIADEM Challenge web site also provides access to image stacks useful as training material and test beds for reconstruction systems. Another complementary resource is the Neuroscience Information Framework (neuinfo.org), a continuously updated inventory of neuroscience tools, data, and materials accessible online. In addition to NeuroMorpho.Org and CCDB, NIF-federated initiatives include ModelDB (senselab.med.yale.edu/modeldb) and associated resources, which serve to store and retrieve neuron and network electrophysiological models and related biophysical properties. A common usage pipeline in computational neuroscience consists of integrating a morphological reconstruction from NeuroMorpho.Org with a NEURON model from ModelDB to run new simulation experiments.
4. Future directions, opportunities, and needs
Neuronal morphology is a core neuroscience interest and digital reconstruction is a mature, popular, and powerful method. Nevertheless, the field continues to evolve. Scientific developments in molecular genetics, paired with technological advances in light microscopy, are radically changing the scope and prospects of neuromorphological data acquisition and analysis. It is now plausible to envision the complete structural characterization of an entire mammalian nervous system at the cellular level. The sheer complexity of the problem, in terms of diversity of neuronal types and number of individual neurons, dictates the employment of computer technology to acquire, handle, and analyze the data. Advances in automation in every aspect of neuronal reconstruction, from tissue preparation and microscopic imaging to post-processing techniques and tracing systems, will yield an unprecedented wealth of data. The resulting knowledge will be invaluable in addressing the brain structure-function relationship at systems level cellular scale.
An active area of ongoing research development parallels the shifting focus from single neurons to connected sub-circuits, and eventually to dense networks. Relative to the prevalence of individual neuron digital tracing, reconstructing neural circuits remains elusive. The key obstacle is that neurons are tightly packed and their arbors span macroscopic volumes. Thus, the region spanned by a single projection axon is invaded by branches extending from millions of other neurons. To reconstruct a circuit, multiple neurons sharing the same space must all be visualized, yet distinguished from each other. Several lines of progress described in this review are particularly relevant to solving this issue. Advanced genetic methods allow multicolor labeling of neurons and axon tracts (Lichtman et al., 2008), and Brainbow was especially devised to map circuitry (Livet et al., 2007). An alternative approach is to immunolabel distinct neural populations sequentially with multiple staining cycles of the same tissue preparation as enabled by array tomography (Micheva and Smith, 2007). In principle, voltage-sensitive dyes could afford conceptually similar temporal discrimination leveraging the sparse asynchronous firing of cortical networks (Mukamel et al., 2009), possibly aided by causal analysis (Gerhard et al., 2011). In practice, to enable visualization of neural sub-circuits, all these approaches must still overcome non-trivial limitations, such as insufficient spectral separation (Brainbow), antibody depth penetration (array tomography), and inadequate signal intensity in the distal branches (voltage-sensitive dyes). Synaptic connectivity may be revealed by other means (Kim et al., 2011; Zador et al., 2012), but the problem of reconstructing the neuronal arbors of the connected network remains.
Another crucial aspect of future emphasis is the temporal dimension. Neuronal reconstruction time lapse series reflect morphological changes in development, neurodegeneration or other observable time courses, such as physiological cycles, response to environmental conditions, and learning. During development, dendrites and axons undergo periods of dramatic branch addition, outgrowth, pruning, or elimination. More subtle, but equally important, structural plasticity continues in many mature networks. Advanced imaging techniques allow routine acquisition of in vivo and in vitro time lapse data. However, digital reconstruction of the captured 4D data is still rare. Outstanding challenges include alignment and correspondence identification of the changing morphological components (He and Cline, 2011). New tools for the comparative analysis of (temporally) serial reconstruction of axonal and dendritic arbors in normal and pathological states will glean valuable insight into the mechanisms and implications of change over time (Lee et al., 2013). Whole circuit reconstructions and temporal series will both necessitate new standardized formats and curation procedure to facilitate data accessibility as well as integration with the continuously evolving analysis, modeling, and database resources of the digital neuromorphology ecosystem.
Elucidating the complex organization of the brain will require synthesis of information about neuron types, about the spatial patterns of their dendritic and axonal arborization, cell counts and densities, synapse number and location (DeFelipe, 2010). Large-scale simulation projects are leveraging the state of the art of data and tools reviewed here in morphometry, biophysics, and stereology, to build realistic network models. Ongoing efforts focus on the organization, connectivity, and function of rat barrel cortex (NeuroDUNE; www.neurodune.org), hippocampus (Ropireddy and Ascoli, 2011), and neocortical columns (Blue Brain Project; Markram, 2006; bluebrain.epfl.ch), with plans aiming at the whole human brain (Abbott and Schiermeier, 2013). The overarching goal to generate virtual functioning nervous systems in silico (Roysam et al., 2009) emphasizes the need for even greater interaction between experimental and computational research.
Most models involving digital reconstructions are constrained and validated by measurements from experiments. For this purpose, the goal of real-scale simulations shifts the demand to massive experimental datasets, not only to ensure sufficient statistical power for adequate estimation of all model parameters, but also to capture the natural diversity of neuron types (Hill et al., 2012). The amount of necessary experimental data requires fully automated digital tracing. Yet a century after Cajal’s drawings, the majority of publicly available morphological data is still being reconstructed manually (Halavi et al., 2012), because the extensive heuristic expertise of humans has not yet been matched by computer algorithms (Donohue and Ascoli, 2011). As recent developments pull within reach of full automation (e.g. Chiang et al., 2011), the emphasis will move to generalization of high quality results to all routine laboratory preparations. An important lesson taught by the DIADEM Challenge is that success hinges not only on independent advancements in imaging technology and algorithm design, but also on specifically tailoring the experimental details to the computational goal. As large volumes of reconstructions become attainable by high-throughput pipelines, quality control will still require human validation, which will likely become the ultimate bottleneck.
In this review, we highlighted the research designs and digital resources that fuel the thriving scientific progress of neuromorphology reconstruction in so many areas of neuroscience. Applications abound in morphometric and stereological analyses, biophysically realistic simulations of neuronal activity, computational models of developmental growth and migration, and stochastic generation of synaptically connected networks. Real scale, four-dimensional reconstructions of entire plastic circuits at the single neuron level promise to make the next decade the most exciting yet.
Acknowledgements
This work was supported in part by grants R01-NS39600 from the National Institutes of Health and MURI-N00014-10-1-0198 from the Office of Naval research. We are grateful to Dr. Michele Ferrante for Figure 4c. We thank Mr. Todd Gillette, Dr. Kerry Brown, and Dr. Michele Ferrante for feedback on an earlier version of this manuscript. We express gratitude to Dr. Meredith LeMasurier, Deputy Editor of Neuron, for her outstanding help and vision throughout the editing process.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbott A, Schiermeier Q. Research prize boost for Europe. Nature. 2013;493:585–586. doi: 10.1038/493585a. [DOI] [PubMed] [Google Scholar]
- Alle H, Roth A, Geiger JR. Energy-efficient action potentials in hippocampal mossy fibers. Science. 2009;325:1405–1408. doi: 10.1126/science.1174331. [DOI] [PubMed] [Google Scholar]
- Andjelic S, Gallopin T, Cauli B, Hill EL, Roux L, Badr S, Hu E, Tamas G, Lambolez B. Glutamatergic nonpyramidal neurons from neocortical layer VI and their comparison with pyramidal and spiny stellate neurons. J. Neurophysiol. 2009;101:641–654. doi: 10.1152/jn.91094.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascoli GA. Computational Neuroanatomy: Principles and methods. New York: Humana Press; 2002. [Google Scholar]
- Ascoli GA. Mobilizing the base of neuroscience data: the case of neuronal morphologies. Nat. Rev. Neurosci. 2006;7:318–324. doi: 10.1038/nrn1885. [DOI] [PubMed] [Google Scholar]
- Ascoli GA. Successes and rewards in sharing digital reconstructions of neuronal morphology. Neuroinform. 2007;5:154–160. doi: 10.1007/s12021-007-0010-7. [DOI] [PubMed] [Google Scholar]
- Ascoli GA, Krichmar J, Scorcioni R, Nasuto SJ, Senft SL. Computer generation and quantitative morphometric analysis of virtual neurons. Anat. Embryol. 2001;204:283–301. doi: 10.1007/s004290100201. [DOI] [PubMed] [Google Scholar]
- Atunes G, Simones-de-Souza FM, Roque AC. Sensitivity of AMPA receptor channel to calcium oscillations: a computational study. Neurocomputing. 2003;52–54:341–346. [Google Scholar]
- Bartos M, Vida I, Frotscher M, Meyer A, Monyer H, Geiger JR, Jonas P. Fast synaptic inhibition promotes synchronized gamma oscillations in hippocampal interneuron networks. PNAS. 2002;99:13222–13227. doi: 10.1073/pnas.192233099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Borg-Graham LJ. Additional efficient computation of branched nerve equations: adaptive time step and ideal voltage clamp. J. Comput. Neurosci. 2000;8:209–226. doi: 10.1023/a:1008945925865. [DOI] [PubMed] [Google Scholar]
- Bower JM, Beeman D. The Book of GENESIS: Exploring Realistic Neural Models with the GEneral NEural Simulation System. New York: Springer-Verlag; 1998. [Google Scholar]
- Brecht M, Roth A, Sakmann B. Dynamic receptive fields of reconstructed pyramidal cells in layers 3 and 2 of rat somatosensory barrel cortex. J. Physiol. 2003;553:243–265. doi: 10.1113/jphysiol.2003.044222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broser PJ, Schulte R, Lang S, Roth A, Helmchen F, Waters J, Sakmann B, Wittum G. Nonlinear anisotropic diffusion filtering of three-dimensional image data from two-photon microscopy. J. Biomed. Opt. 2004;9:1253–1264. doi: 10.1117/1.1806832. [DOI] [PubMed] [Google Scholar]
- Brown KM, Barrionuevo G, Canty AJ, De Paola V, Hirsch JA, Jefferis GSXE, Lu J, Snippe M, Sugihara I, Ascoli GA. The DIADEM data sets: Representative light microscopy images of neuronal morphology to advance automation of digital reconstructions. Neuroinform. 2011;9:143–157. doi: 10.1007/s12021-010-9095-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KM, Donohue DE, D’Alessandro G, Ascoli GA. A cross-platform freeware tool for digital reconstruction of neuronal arborizations from image stacks. Neuroinform. 2005;3:343–360. doi: 10.1385/NI:3:4:343. [DOI] [PubMed] [Google Scholar]
- Bui TV, Cushing S, Dewey D, Fyffe RE, Rose PK. Comparison of the morphological and electrotonic properties of Renshaw cells, la inhibitory interneurons, and motoneurons in the cat. J. Neurophysiol. 2003;90:2900–2918. doi: 10.1152/jn.00533.2003. [DOI] [PubMed] [Google Scholar]
- Buonomano DV. Decoding temporal information: A model based on short-term synaptic plasticity. J. Neurosci. 2000;20:1129–1141. doi: 10.1523/JNEUROSCI.20-03-01129.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon RC, Hasselmo ME, Koene RA. From biophysics to behavior: Catacomb2 and the design of biologically-plausible models for spatial navigation. Neuroinform. 2003;1:3–42. doi: 10.1385/NI:1:1:003. [DOI] [PubMed] [Google Scholar]
- Cannon RC, O’Donnell C, Nolan MF. Stochastic ion channel gating in dendritic neurons: morphology dependence and probabilistic synaptic activation of dendritic spikes. PLoS Comp. Biol. 2010;6(8) doi: 10.1371/journal.pcbi.1000886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon RC, Turner DA, Pyapali GK, Wheal HV. An on-line archive of reconstructed hippocampal neurons. J. Neurosci. Methods. 1998;84:49–54. doi: 10.1016/s0165-0270(98)00091-0. [DOI] [PubMed] [Google Scholar]
- Capowski JJ. Computer-aided reconstruction of neuron trees from several serial sections. Comput. Biomed. Res. 1977;10:617–629. doi: 10.1016/0010-4809(77)90017-9. [DOI] [PubMed] [Google Scholar]
- Cauller LJ, Connors BW. Synaptic physiology of horizontal afferents to layer I in slices of rat SI neocortex. J. Neurosci. 1994;14:751–762. doi: 10.1523/JNEUROSCI.14-02-00751.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CS, GUzman JN, llijic C, Mercer JN, Rick C, Tkatch T, Meredith GE, Surmeier DJ. “Rejuvenation” projects neurons in mouse models of Parkinson’s disease. Nature. 2007;447:1081–1086. doi: 10.1038/nature05865. [DOI] [PubMed] [Google Scholar]
- Chen K, Aradi I, Thon N, Eqbhal-Ahmadi M, Baram TZ, Soltesz I. Persistently modified h-channels after complex febrile seizures convert the seizure-induced enhancement of inhibition to hyperexcitability. Nat. Med. 2001;7:331–337. doi: 10.1038/85480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang AS, Lin CY, Chuang CC, Chang HM, Hsieh CH, Yeh CW, Shih CT, Wu JJ, Wang GT, Chen YC, et al. Three-dimensional reconstruction of brain-wide wiring networks in Drosophila at single-cell resolution. Curr. Biol. 2011;21:1–11. doi: 10.1016/j.cub.2010.11.056. [DOI] [PubMed] [Google Scholar]
- Chmykhova NM, Adanina VO, Karamian OA, Kozhanov VM, Vesselkin NP, Clamann HP. Comparative study of spinal motoneuron axon collaterals. Brain Res. Bull. 2005;66:381–386. doi: 10.1016/j.brainresbull.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Costa Lda. F, Zawadzki K, Miazaki M, Viana MP, Taraskin SN. Unveiling the neuromorphological space. Front. Comput. Neurosci. 2010;4:150. doi: 10.3389/fncom.2010.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuntz H, Forstner F, Borst A, Hausser M. The TREES toolbox-probing the basis of axonal and dendritic branching. Neuroinform. 2011;9(1):91–6. doi: 10.1007/s12021-010-9093-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuntz H, Forstner F, Haag J, Borst A. The morphological identity of insect dendrites. PLoS Comp.Biol. 2008;4(12) doi: 10.1371/journal.pcbi.1000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutsuridis V, Cobb S, Graham BP. Encoding and retrieval in a model of the hippocampal CA1 microcircuit. Hippocampus. 2010;20:423–446. doi: 10.1002/hipo.20661. [DOI] [PubMed] [Google Scholar]
- DeFelipe J. From the connectome to the synaptome: an epic love story. Science. 2010;330:1198–1201. doi: 10.1126/science.1193378. [DOI] [PubMed] [Google Scholar]
- Denk W, Svoboda K. Photo upmanship: Why multiphoton imaging is more than a gimmick. Neuron. 1997;18:351–357. doi: 10.1016/s0896-6273(00)81237-4. [DOI] [PubMed] [Google Scholar]
- Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- Destexhe A, Neubig M, Ulrich D, Huguenard J. Dendritic low-threshold calcium currents in thalamic relay cells. J. Neurosci. 1998;18:3574–3588. doi: 10.1523/JNEUROSCI.18-10-03574.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGregorio DA, Nusser Z, Silver RA. Spillover of glutamate onto synaptic AMPA receptors enhances fast transmission at a cerebellar synapse. Neuron. 2002;35:521–533. doi: 10.1016/s0896-6273(02)00787-0. [DOI] [PubMed] [Google Scholar]
- Dityatev AE, Chmykhova NM, Dityateva GV, Babalian AL, Kleinle J, Clamann HP. Structural and physiological properties of connections between individual reticulospinal axons and lumbar motoneurons of the frog. J. Comp. Neurol. 2001;430:433–447. doi: 10.1002/1096-9861(20010219)430:4<433::aid-cne1041>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Donohue DE, Ascoli GA. Automated reconstruction of neuronal morphology: an overview. Brain Res. 2011;24:94–102. doi: 10.1016/j.brainresrev.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard JP, Wanner A, Wittum G. NeuGen: A tool for the generation of realistic morphology of cortical neurons and neural networks in 3D. Neurocomputing. 2006;70:327–342. [Google Scholar]
- Escobar G, Fares T, Stepanyants A. Structural plasticity of circuits in cortical neuropil. J. Neurosci. 2008;28:8477–8488. doi: 10.1523/JNEUROSCI.2046-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre MD, Antal M, Nusser Z. Distinct deep short-axon cell subtypes of the main olfactory bulb provide novel intrabulbar and extrabulbar GABAergic connections. J. Neurosci. 2008;28:8217–8229. doi: 10.1523/JNEUROSCI.2490-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmeyer D, Egger V, Lubke J, Sakmann B. Reliable synaptic connections between pairs of excitatory layer 4 neurones within a single “barrel” of developing rat somatosensory cortex. J. Physiol. 1999;521:169–190. doi: 10.1111/j.1469-7793.1999.00169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmeyer D, Roth A, Sakmann B. Monosynaptic connections between pairs of spiny stellate cells in layer 4 and pyramidal cells in layer 5a indicate lemniscal and paralemniscal afferent pathways converge in the infragranular somatosensory cortex. J. Neurosci. 2005;25:3423–3431. doi: 10.1523/JNEUROSCI.5227-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante M, Blackwell KT, Migliore M, Ascoli GA. Computational models of neuronal biophysics and the characterization of potential neuropharmacological targets. Curr. Med. Chem. 2008;15:2456–2471. doi: 10.2174/092986708785909094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante M, Migliore M, Ascoli GA. Feed-forward inhibition as a buffer of the neuronal input-output relation. PNAS. 2009;106:18004–18009. doi: 10.1073/pnas.0904784106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco A, Knafo S, Banon-Rodriguez I, Merino-Serrais P, Fernaud-Espinosa I, Nieto M, Garrido JJ, Esteban JA, Wandosell F, Anton IM. WIP is a negative regulator of neuronal maturation and synaptic activity. Cereb. Cortex. 2012;22:1191–1202. doi: 10.1093/cercor/bhr199. [DOI] [PubMed] [Google Scholar]
- Fransen E, Alonso AA, Dickson CT, Magistretti J, Hasselmo ME. Ionic mechanisms in the generation of subthreshold oscillations and action potentials clustering in entorhinal layer II stellate neurons. Hippocampus. 2004;14:368–384. doi: 10.1002/hipo.10198. [DOI] [PubMed] [Google Scholar]
- Frick A, Feldmeyer D, Helmstaedter M, Sakmann B. Monosynaptic connections between pairs of L5a pyramidal neurons in column of juvenile rat somatosensory cortex. Cereb. Cortex. 2008;18:397–406. doi: 10.1093/cercor/bhm074. [DOI] [PubMed] [Google Scholar]
- Gan W-B, Grutzendler J, Wong WT, Wong ROL, Lichtman JW. Multicolor “DiOlistic” labeling of the nervous system using lipophilic dye combinations. Neuron. 2000;27:219–225. doi: 10.1016/s0896-6273(00)00031-3. [DOI] [PubMed] [Google Scholar]
- Gasparini S, Migliore M, Magee JC. On the initiation and propagation of dendritic spikes in CA1 pyramidal neurons. J. Neurosci. 2004;24:11046–11056. doi: 10.1523/JNEUROSCI.2520-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhard F, Pipa G, Lima B, Neuenschwander S, Gerstner W. Extraction of network topology from multi-electrode recordings: is there a small-world effect? Front. Comput. Neurosci. 2011;5(4) doi: 10.3389/fncom.2011.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillette TA, Brown KM, Ascoli GA. The DIADEM metric: comparing multiple reconstructions of the same neuron. Neuroinform. 2011;9:233–245. doi: 10.1007/s12021-011-9117-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser JR, Glaser EM. Neuron imaging with neurolucida – A PC-based system for image combining microscopy. Comput. Med. Imaging Graph. 1990;14(5):307–317. doi: 10.1016/0895-6111(90)90105-k. [DOI] [PubMed] [Google Scholar]
- Glaser EM, Van der Loos H. A semi-automatic computer-microscope for the analysis of neuronal morphology. IEEE Trans. Biomed. Eng. 1965;12:22–31. [PubMed] [Google Scholar]
- Gleeson P, Steuber V, Silver RA. neuroConstruct: A tool for modeling networks of neurons in 3D space. Neuron. 2007;54:219–235. doi: 10.1016/j.neuron.2007.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman DF. Code generation: a strategy for neural network simulators. Neuroinform. 2010;8:183–196. doi: 10.1007/s12021-010-9082-x. [DOI] [PubMed] [Google Scholar]
- Gulledge AT, Stuart GJ. Excitatory actions of GABA in the cortex. Neuron. 2003;37:299–309. doi: 10.1016/s0896-6273(02)01146-7. [DOI] [PubMed] [Google Scholar]
- Gustafsson MGL. Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution. PNAS. 2005;102:13081–13086. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjieconomou D, Rotkopf S, Alexandre C, Bell DM, Dickson BJ, Salecker I. Flybow: genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster. Nature Methods. 2011;8:260–268. doi: 10.1038/nmeth.1567. [DOI] [PubMed] [Google Scholar]
- Halavi M, Hamilton KA, Parekh R, Ascoli GA. Digital reconstructions of neuronal morphology: three decades of research trends. Front. Neurosci. 2012;6(49) doi: 10.3389/fnins.2012.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DH, Treinin M. How does morphology relate to function in sensory arbors? Trends Neurosci. 2011;34:443–451. doi: 10.1016/j.tins.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampel S, Chung P, McKellar CE, Hall D, Looger LL, Simpson JH. Drosophila Brainbow: a recombinase-based fluorescence labeling technique to subdivide neural expression patterns. Nature Methods. 2011;8:253–259. doi: 10.1038/nmeth.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E, Stewart M. Propagation of synchronous epileptiform events from subiculum backward into area CA1 of rat brain slices. Brain Res. 2001;895:41–49. doi: 10.1016/s0006-8993(01)02023-6. [DOI] [PubMed] [Google Scholar]
- Hayes TL, Lewis DA. Magnopyramidal neurons in the anterior motor speech region. Dendritic features and interhemispheric comparisons. Arch Neurol. 1996;53:1277–1283. doi: 10.1001/archneur.1996.00550120089021. [DOI] [PubMed] [Google Scholar]
- He HY, Cline HT. Diadem X: Automated 4 dimensional analysis of morphological data. Neuroinform. 2011;9:107–112. doi: 10.1007/s12021-011-9098-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess ST, Girirajan TPK, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SL, Wang Y, Riachi I, Schurmann F, Markram H. Statistical connectivity provides a sufficient foundation for specific functional connectivity in neocortical circuits. PNAS. 2012;109:E2885–E2894. doi: 10.1073/pnas.1202128109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines ML, Carnevale NT. The NEURON simulation environment. Neural Comput. 1997;9:1179–1209. doi: 10.1162/neco.1997.9.6.1179. [DOI] [PubMed] [Google Scholar]
- Hoffman DA, Magee JC, Colbert CM, Johnston D. K+ channel regulation of signal propagation in dendrites of hippocampal pyramidal neurons. Nature. 1997;387:869–75. doi: 10.1038/43119. [DOI] [PubMed] [Google Scholar]
- Huang B, Babcock H, Zhuang X. Breaking the diffraction barrier: super-resolution imaging of cells. Cell. 2010;143:1047–1058. doi: 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferis GS, Potter CJ, Chan AM, Marin EC, Rohlfing T, Maurer CR, Jr., Luo L. Comprehensive maps of Drosophila higher olfactory centers: spatially segregated fruit and pheromone representation. Cell. 2007;128:1187–1203. doi: 10.1016/j.cell.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungblut D, Vlachos A, Schuldt G, Zahn N, Deller T, Wittum G. SpineLab: tool for three-dimensional reconstruction of neuronal morphology. J. Biomed. Opt. 2012;17 doi: 10.1117/1.JBO.17.7.076007. [DOI] [PubMed] [Google Scholar]
- Kim J, Zhao T, Petralia RS, Yu Y, Peng H, Magee JC. mGrasp enables mapping mammalian synaptic connectivity with ligh microscopy. Nat. Methods. 2011;9:96–102. doi: 10.1038/nmeth.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita H, Armstrong W. A biotin-containing compound N-(2-aminoethyl) biotinamide for intracellular labeling and neuronal tracing studies: a comparison with biocytin. J. Neurosci. Methods. 1991;37:141–150. doi: 10.1016/0165-0270(91)90124-i. [DOI] [PubMed] [Google Scholar]
- Klar TA, Hell SW. Subdiffraction resolution in far-field fluorescence microscopy. Opt. Lett. 1999;24:954–956. doi: 10.1364/ol.24.000954. [DOI] [PubMed] [Google Scholar]
- Kobbert C, Apps R, Bechmann I, Lanciego JL, Mey J, Thanos S. Current topics in neuroanatomical tracing. Prog. Neurobiol. 2000;62:327–351. doi: 10.1016/s0301-0082(00)00019-8. [DOI] [PubMed] [Google Scholar]
- Koene RA, Tijms B, van Hees P, Postma F, de Ridder A, Ramakers GJA, van Pelt J, van Ooyen A. NETMORPH: A framework for the stochastic generation of large scale neuronal networks with realistic neuron morphologies. Neuroinform. 2009;7:195–210. doi: 10.1007/s12021-009-9052-3. [DOI] [PubMed] [Google Scholar]
- Koh IY, Lindquist WB, Zito K, Nimchinsky EA, Svoboda K. An image analysis algorithm for dendritic spines. Neural Comput. 2002;14(6):1283–310. doi: 10.1162/089976602753712945. [DOI] [PubMed] [Google Scholar]
- Kole MH, llschner SU, Kampa BM, Williams SR, Ruben PC, Stuart GJ. Action potential generation requires a high sodium channel density in the axon initial segment. Nat. Neurosci. 2008;11:178–186. doi: 10.1038/nn2040. [DOI] [PubMed] [Google Scholar]
- Krichmar JL, Nasuto SJ, Scorcioni R, Washington SD, Ascoli GA. Effects of dendritic morphology on CA3 pyramidal cell electrophysiology: a simulation study. Brain Res. 2002;941:11–28. doi: 10.1016/s0006-8993(02)02488-5. [DOI] [PubMed] [Google Scholar]
- Kvello P, Lofaldi BB, Rybak J, Menzel R, Mustaparta H. Digital, three-dimensional average shaped atlas of the heliothis virescens brain with integrated gustatory and olfactory neurons. Front. Syst. Neurosci. 2009;3(14) doi: 10.3389/neuro.06.014.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakadamyali M, Babcock H, Bates M, Zhuang X, Lichtman J. 3D multicolor super-resolution imaging offers improved accuracy in neuron tracing. PLoS ONE. 2012;7(1) doi: 10.1371/journal.pone.0030826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciego JL, Wouterlood FG. A half century of experimental neuroanatomical tracing. J. Chem. Neuroanat. 2011;42:157–183. doi: 10.1016/j.jchemneu.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Lavis LD. Histochemistry: Live and in color. J. Histochem. Cytochem. 2011;59:139–145. doi: 10.1369/0022155410395760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Magueresse C, Alfonso J, Khodosevich K, Arroyo Martin AA, Bark C, Monyer H. “Small axonless neurons”: postnatally generated neocortical interneurons with delayed functional maturation. J. Neurosci. 2011;31:16731–16747. doi: 10.1523/JNEUROSCI.4273-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PC, Chuang CC, Chiang AS, Ching YT. High-throughput computer method for 3D neuronal structure reconstruction from the image stack of the Drosophila brain and its applications. PLoS Comput. Biol. 2012;8(9) doi: 10.1371/journal.pcbi.1002658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PC, He H, Lin CY, Ching YT, Cline HT. Computer aided alignment and quantitative 4D structural plasticity analysis of neurons. Neuroinformatics. 2013 doi: 10.1007/s12021-013-9179-0. [Epub ahead of print. Feb 14] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T-C, Rangasami KL, Chong-Nam C. Building skeleton models via 3-D medial surface/axis thinning algorithms. Computer Vision, Graphics and Image Processing. 1994;56:462–478. [Google Scholar]
- Lemmens MAM, Steinbusch HWM, Rutten BPF, Schmitz C. Advanced microscopy techniques for quantitative analysis in neuromorphology and neuropathology research: current status and requirements for the future. J. Chem. Neuroanat. 2010;40:199–209. doi: 10.1016/j.jchemneu.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Levinthal C, Ware R. Three dimensional reconstruction from serial sections. Nature. 1972;238:207–210. [Google Scholar]
- Lichtman JW, Livet J, Sanes JR. A technicolor approach to the connectome. Nat. Rev. Neurosci. 2008;9:417–422. doi: 10.1038/nrn2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livet J, Weissman TA, Kang H, Draft RW, Bennis RA, Sanes JR, Lichtman JW. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- Longair MH, Baker DA, Armstrong JD. Simple Neurite Tracer: Open Source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics. 2011;27:2453–2454. doi: 10.1093/bioinformatics/btr390. [DOI] [PubMed] [Google Scholar]
- Lopez-Cruz PL, Bielza C, Larranaga P, Benavides-Piccione R, DeFelipe J. Models and simulation of 3D neuronal dendritic trees using Bayesian networks. Neuroinform. 2011;9:347–369. doi: 10.1007/s12021-011-9103-4. [DOI] [PubMed] [Google Scholar]
- Losavia BE, Liang Y, Santamaria-Pang A, Kakadiaris IA, Colbert CM, Saggu P. Live neuron morphology automatically reconstructed from multiphoton and confocal imaging data. J. Neurophysiol. 2008;100:2422–2429. doi: 10.1152/jn.90627.2008. [DOI] [PubMed] [Google Scholar]
- Luisi J, Narayanaswamy A, Galbreath Z, Roysam B. The FARSIGHT trace editor: an open source tool for 3-D inspection and efficient pattern analysis aided editing of automated neuronal reconstructions. Neuroinform. 2011;9(2–3):305–15. doi: 10.1007/s12021-011-9115-0. [DOI] [PubMed] [Google Scholar]
- Lytton WW, Sejnowski TJ. Simulations of cortical pyramidal neurons synchronized by inhibitory interneurons. J. Neurophysiol. 1991;66:1059–1079. doi: 10.1152/jn.1991.66.3.1059. [DOI] [PubMed] [Google Scholar]
- Macagno ER, Levinthal C, Sobel I. Three-dimensional computer reconstruction of neurons and neuronal assemblies. Ann. Rev. Biophys. Bioeng. 1979;8:323–351. doi: 10.1146/annurev.bb.09.060180.002023. [DOI] [PubMed] [Google Scholar]
- Magee JC, Cook EP. Somatic EPSP amplitude is independent of synapse location in hippocampal pyramidal neurons. Nat. Neurosci. 2000;3:895–903. doi: 10.1038/78800. [DOI] [PubMed] [Google Scholar]
- Mainen ZF, Sejnowski TJ. Influence of dendrite structure on firing pattern in model neocortical neurons. Nature. 1996;382:363–366. doi: 10.1038/382363a0. [DOI] [PubMed] [Google Scholar]
- Margrie TW, Schaefer AT. Theta oscillation coupled spike latencies yield computational vigour in a mammalian sensory system. J. Physiol. 2003;546:363–374. doi: 10.1113/jphysiol.2002.031245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markram H. The blue brain project. Nat. Rev. Neurosci. 2006;7:153–160. doi: 10.1038/nrn1848. [DOI] [PubMed] [Google Scholar]
- Markram H, Lubke J, Frotscher M, Roth A, Sakmann B. Physiology and anatomy of synaptic connections between thick tufted pyramidal neurones in the developing rat neocortex. J. Physiol. 1997;500:409–440. doi: 10.1113/jphysiol.1997.sp022031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martone ME, Tran J, Wong WW, Sargis J, Fong L, Larson S, Lamont SP, Gupta A, Ellisman MH. The cell centered database project: an update on building community resources for managing and sharing 3D imaging data. J. Struct. Biol. 2008;161:220–231. doi: 10.1016/j.jsb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattioni M, Cohen U, Le Novere N. Neuronvisio : a graphical user interface with 3D capabilities for NEURON. Front. Neuroinform. 2012;6(20) doi: 10.3389/fninf.2012.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre CC, Grill WM. Extracellular stimulation of central neurons: influence of stimulus waveform and frequency on neuronal output. J. Neurophysiol. 2002;88:1592–1604. doi: 10.1152/jn.2002.88.4.1592. [DOI] [PubMed] [Google Scholar]
- McIntyre CC, Grill WM, Sherman DL, Thakor NV. Cellular effects of deep brain stimulation: model-based analysis of activation and inhibition. J. Neurophysiol. 2004;91:1457–1469. doi: 10.1152/jn.00989.2003. [DOI] [PubMed] [Google Scholar]
- Meijering E. Neuron tracing in perspective. Cytometry A. 2010;77:693–704. doi: 10.1002/cyto.a.20895. [DOI] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria J-CF, Steiner P, Hirling H, Unser M. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A. 2004;58A:167–176. doi: 10.1002/cyto.a.20022. [DOI] [PubMed] [Google Scholar]
- Meyer AH, Katona I, Blatow M, Rozov A, Monyer H. In vivo labeling of parvalbumin-positive interneurons and analysis of electrical coupling in identified neurons. J. Neurosci. 2002;22:7055–7064. doi: 10.1523/JNEUROSCI.22-16-07055.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelsen K, Murk K, Zagrebelsky M, Dreznjak A, Jockusch BM, Rothkegel M, Korte M. Fine-tuning of neuronal architecture requires two profilin isoforms. PNAS. 2010;107:15780–15785. doi: 10.1073/pnas.1004406107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheva KD, Smith SJ. Array tomography: A new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron. 2007;55:25–36. doi: 10.1016/j.neuron.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukamel EA, Nimmerjahn A, Schnitzer MJ. Automated analysis of cellular signals from large-scale calcium imaging data. Neuron. 2009;63:747–760. doi: 10.1016/j.neuron.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myatt DR, Hadlington T, Ascoli GA, Nasuto SJ. Neuromantic-from semi-manual to semi-automatic reconstruction of neuron morphology. Front. Neuroinform. 2012;6(4) doi: 10.3389/fninf.2012.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narro ML, Yang F, Kraft R, Wenk C, Efrat A, Restifo LL. NeuronMetrics: Software for Semi-Automated Processing of Cultured-Neuron Images. Brain Res. 2007;1138:57–75. doi: 10.1016/j.brainres.2006.10.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte C, Matyash M, Pivneva T, Schipke CG, Ohlemeyer C, Hanisch U-K, Kirchhoff F, Kettenmann H. GFAP promoter-controlled EGFP expressing transgenic mice: A tool to visualize astrocytes and astrogliosis in living brain tissue. Glia. 2001;33:72–86. [PubMed] [Google Scholar]
- Oberlaender M, de Kock CP, Bruno RM, Ramirez A, Meyer HS, Dercksen VJ, Helmstaedter M, Sakmann B. Cell-type specific three-dimensional structure of thalamocortical circuits in a column of rat vibrissal cortex. Cereb. Cortex. 2011 doi: 10.1093/cercor/bhr317. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AM, Yuste R. Dense, unspecific connectivity of neocortical parvalbumin-positive interneurons: a canonical microcircuit for inhibition? J. Neurosci. 2011;31:13260–13271. doi: 10.1523/JNEUROSCI.3131-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Long F, Myers G. Automatic 3D neuron tracing using all-path pruning. Bioinformatics. 2011;27:1239–1247. doi: 10.1093/bioinformatics/btr237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Ruan Z, Long F, Simpson JH, Myers EW. V3D enables real-time 3D visualization and quantitative analysis of large-scale biological image data sets. Nat. Biotechnol. 2010;28:348–353. doi: 10.1038/nbt.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YR, He S, Marie H, Zeng SY, Ma J, Tan ZJ, Lee SY, Malenka RC, Yu X. Coordinated changes in dendritic arborization and synaptic strength during neural circuit development. Neuron. 2009;61:71–84. doi: 10.1016/j.neuron.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Rosello T, Baker JL, Ferrante M, Iyengar S, Ascoli GA, Barrionuevo G. Passive and active shaping of unitary responses from associational/commissural and perforant path synapses in hippocampal CA3 pyramidal cells. J. Comput. Neurosci. 2011;31:159–182. doi: 10.1007/s10827-010-0303-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PING (Petilla Interneuron Nomenclature Group) Ascoli GA, Alonso-Nanclares L, Anderson SA, Barrionuevo G, Benavides-Piccione R, Burkhalter A. coauthors Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat. Rev. Neurosci. 2008;9:557–568. doi: 10.1038/nrn2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirazi P, Brannon T, Mel BW. Arithmetic of subthreshold synaptic summation in a model CA1 pyramidal cell. Neuron. 2003a;37:977–987. doi: 10.1016/s0896-6273(03)00148-x. [DOI] [PubMed] [Google Scholar]
- Pool M, Thiemann J, Bar-Or A, Fournier AE. NeuriteTracer: A novel ImageJ plugin for automated quantification of neurite outgrowth. J. Neurosci. Methods. 2008;168:134–139. doi: 10.1016/j.jneumeth.2007.08.029. [DOI] [PubMed] [Google Scholar]
- Poolos NP, Migliore M, Johnston D. Pharmacological upregulation of h-channels reduces the excitability of pyramidal neuron dendrites. Nat. Neurosci. 2002;5:767–774. doi: 10.1038/nn891. [DOI] [PubMed] [Google Scholar]
- Ray S, Bhalla US. PyMOOSE: Interoperable scripting in Python for MOOSE. Front. Neuroinform. 2008;2:6. doi: 10.3389/neuro.11.006.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A, Leonardus Veenman C, Medina L, Jiao Y, Del Mar N, Honig MG. Pathway tracing using biotinylated dextran amines. J. Neurosci. Methods. 2000;103:23–27. doi: 10.1016/s0165-0270(00)00293-4. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Ehlenberger DB, Kelliher KT, Einstein M, Henderson SC, Morrison JH, Hof PR, Wearne SL. Automated reconstruction of three-dimensional neuronal morphology from laser scanning microscopy images. Methods. 2003;30:94–105. doi: 10.1016/s1046-2023(03)00011-2. [DOI] [PubMed] [Google Scholar]
- Ropireddy D, Ascoli GA. Potential synaptic connectivity of different neurons onto pyramidal cells in a 3D reconstruction of the rat hippocampus. Front. Neuroinform. 2011;5(5) doi: 10.3389/fninf.2011.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropireddy D, Bachus SE, Ascoli GA. Non-homogeneous stereological properties of the rat hippocampus from high-resolution 3D serial reconstructions of thin histological sections. Neuroscience. 2012;205:91–111. doi: 10.1016/j.neuroscience.2011.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roysam B, Shain W, Ascoli GA. The central role of neuroinformatics in the National Academy of Engineering’s grandest challenge: reverse engineer the brain. Neuroinform. 2009;7:1–5. doi: 10.1007/s12021-008-9043-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nature Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak J, Kuss A, Lamecker H, Zachow S, Hege HC, Lienhard M, Singer J, Neubert K, Menzel R. The digital bee brain: integrating and managing neurons in a common 3D reference system. Front. Syst. Neurosci. 2010;4:30. doi: 10.3389/fnsys.2010.00030. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah P, Bekkers JM. Apical dendritic location of slow afterhyperpolarization current in hippocampal pyramidal neurons: implications for the integration of long-term potentiation. J. Neurosci. 1996;16:4537–4542. doi: 10.1523/JNEUROSCI.16-15-04537.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsonovich AV, Ascoli GA. Statistical determinants of dendritic morphology in hippocampal pyramidal neurons: a hidden Markov model. Hippocampus. 2005;15:166–183. doi: 10.1002/hipo.20041. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Matsuki N, Ikegaya Y. Effects of axonal topology on the somatic modulation of synaptic outputs. J. Neurosci. 2012;32:2868–2876. doi: 10.1523/JNEUROSCI.5365-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller J, Major G, Koester HJ, Schiller Y. NMDA spikes in basal dendrites of cortical pyramidal neurons. Nature. 2000;404:285–289. doi: 10.1038/35005094. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorcioni R, Polavaram S, Ascoli GA. L-Measure: a web-accessible tool for the analysis, comparison and search of digital reconstructions of neuronal morphologies. Nat. Protoc. 2008;3:866–876. doi: 10.1038/nprot.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl AH, Rubel EW, Harris DM. Mechanisms for adjusting interaural time differences to achieve binaural coincidence detection. J. Neurosci. 2010;30:70–80. doi: 10.1523/JNEUROSCI.3464-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senft SL. A brief history of neuronal reconstruction. Neuroinform. 2011;9:119–128. doi: 10.1007/s12021-011-9107-0. [DOI] [PubMed] [Google Scholar]
- Silberberg G, Markram H. Disynaptic inhibition between neocortical pyramidal cells mediated by Martinotti cells. Neuron. 2007;53:735–746. doi: 10.1016/j.neuron.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Smith RG. NeuronC: A computational language for investigating functional architecture of neural circuits. J. Neurosci. Methods. 1992;43:83–108. doi: 10.1016/0165-0270(92)90019-a. [DOI] [PubMed] [Google Scholar]
- Snider J, Pillai A, Stevens CF. A universal property of axonal and dendritic arbors. Neuron. 2010;66:45–56. doi: 10.1016/j.neuron.2010.02.013. [DOI] [PubMed] [Google Scholar]
- Softky WR, Koch C. The highly irregular firing of cortical cells in inconsistent with temporal integration of random EPSPs. J. Neurosci. 1993;13:334–350. doi: 10.1523/JNEUROSCI.13-01-00334.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinas SM, Maex R, De Schutter E. Dendritic amplification of inhibitory postsynaptic potentials in a model Purkinje cell. Eur. J. Neurosci. 2006;23:1207–1218. doi: 10.1111/j.1460-9568.2005.04564.x. [DOI] [PubMed] [Google Scholar]
- Spruston N. Pyramidal neurons: dendritic structure and synaptic integration. Nat. Rev. Neurosci. 2008;9:206–221. doi: 10.1038/nrn2286. [DOI] [PubMed] [Google Scholar]
- Spruston N, Jaffe DB, Johnston D. Dendritic attenuation of synaptic potentials and currents: the role of passive membrane properties. Trends Neurosci. 1994;17:161–166. doi: 10.1016/0166-2236(94)90094-9. [DOI] [PubMed] [Google Scholar]
- Stepanyants A, Chklovskii DB. Neurogeometry and potential synaptic connectivity. Trends Neurosci. 2005;28:387–394. doi: 10.1016/j.tins.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Stepanyants A, Hirsch JA, Martinez LM, Kisvarday ZF, Ferecsko AS, Chklovskii DB. Local potential connectivity in cat primary visual cortex. Cereb. Cortex. 2008;18:13–28. doi: 10.1093/cercor/bhm027. [DOI] [PubMed] [Google Scholar]
- Stepanyants A, Hof PR, Chklovskii DB. Geometry and structural plasticity of synaptic connectivity. Neuron. 2002;34:275–288. doi: 10.1016/s0896-6273(02)00652-9. [DOI] [PubMed] [Google Scholar]
- Stuart GJ, Hausser M. Dendritic coincidence detection of EPSPs and action potentials. Nat. Neurosci. 2001;4:63–71. doi: 10.1038/82910. [DOI] [PubMed] [Google Scholar]
- Sulkowski MJ, Iyer SC, Kurosawa MS, Iyer EP, Cox DN. Turtle functions downstream of Cut in differentially regulating class specific dendrite morphogenesis in Drosophila. PLoS One. 2011;6(7) doi: 10.1371/journal.pone.0022611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda K. The past, present and future of single neuron reconstructions. Neuroinform. 2011;9:97–98. doi: 10.1007/s12021-011-9097-y. [DOI] [PubMed] [Google Scholar]
- Teeter CM, Stevens CF. A general principle of neural arbor branch density. Curr. Biol. 2011;21:2105–2108. doi: 10.1016/j.cub.2011.11.013. [DOI] [PubMed] [Google Scholar]
- Teschemacher AG, Paton JFR, Kasparov S. Imaging living central neurones using viral gene transfer. Adv. Drug Deliv. Rev. 2005;57:79–93. doi: 10.1016/j.addr.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Silver RA, Cull-Candy SG. Estimated conductance of glutamate receptor channels activated during EPSCs at the cerebellar mossy fiber-granule cell synapse. Neuron. 1993;11:279–289. doi: 10.1016/0896-6273(93)90184-s. [DOI] [PubMed] [Google Scholar]
- Troyer TW, Levin JE, Jacobs GA. Construction and analysis of a database representing a neural map. Microsc. Res. Tech. 1994;29:329–343. doi: 10.1002/jemt.1070290502. [DOI] [PubMed] [Google Scholar]
- Ugolini G. Advances in viral transneuronal tracing. J. Neurosci. Methods. 2010;194:2–20. doi: 10.1016/j.jneumeth.2009.12.001. [DOI] [PubMed] [Google Scholar]
- van Ooyen A. Using theoretical models to analyse neural development. Nat. Rev. Neurosci. 2011;12:311–326. doi: 10.1038/nrn3031. [DOI] [PubMed] [Google Scholar]
- Vyas A, Jadhav S, Chattarji S. Prolonged behavioral stress enhances synaptic connectivity in the basolateral amygdala. Neuroscience. 2006;143:387–393. doi: 10.1016/j.neuroscience.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Wang Y, Narayanaswamy A, Tsai CL, Roysam B. A broadly applicable 3-D neuron tracing method based on open-curve snake. Neuroinform. 2011;9(2–3):193–217. doi: 10.1007/s12021-011-9110-5. [DOI] [PubMed] [Google Scholar]
- Wann DF, Woolsey TA, Dierker ML, Cowan WM. An on-line digital-computer system for the semiautomatic analysis of golgi-impregnated neurons. IEEE Trans. Biomed. Eng. BME. 1973;20(4) doi: 10.1109/TBME.1973.324187. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Hoffman DA, Migliore M, Johnston D. Dendritic K+ channels distribution to spike-timing dependent long-term potentiation in hippocampal pyramidal neurons. PNAS. 2002;99:8366–8371. doi: 10.1073/pnas.122210599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wearne SL, Rodriguez A, Ehlenberger DB, Rocher AB, Henderson SC, Hof PR. New techniques for imaging, digitization and analysis of three-dimensional neuron morphology on multiple scales. Neuroscience. 2005;136:661–680. doi: 10.1016/j.neuroscience.2005.05.053. [DOI] [PubMed] [Google Scholar]
- Wei H, el Jundi B, Homberg U, Stengl M. Implementation of pigment-dispersing factor-immunoreactive neurons in a standardized atlas of the brain of the cockroach Leucophaea maderae. J. Comp. Neurol. 2010;518:4113–4133. doi: 10.1002/cne.22471. [DOI] [PubMed] [Google Scholar]
- Wen Q, Chklovskii DB. A cost-benefit analysis of neuronal morphology. J. Neurophysiol. 2008;99:2320–2328. doi: 10.1152/jn.00280.2007. [DOI] [PubMed] [Google Scholar]
- Wolf E, Birinyi A, Pomahazi S. A fast 3-dimensional neuronal tree reconstruction system that uses cubic polynomials to estimate dendritic curvature. J. Neurosci. Methods. 1995;63:137–145. doi: 10.1016/0165-0270(95)00102-6. [DOI] [PubMed] [Google Scholar]
- Zador AM, Dubnau J, Oyibo HK, Zhan H, Cao G, Peikon ID. Sequencing the connectome. PLoS Biol. 2012;10(10) doi: 10.1371/journal.pbio.1001411. el001411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubler F, Douglas R. A framework for modeling the growth and development of neurons and networks. Front. Comput. Neurosci. 2009;3(25) doi: 10.3389/neuro.10.025.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]