Abstract
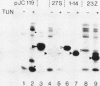
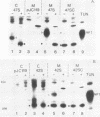
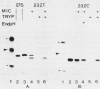
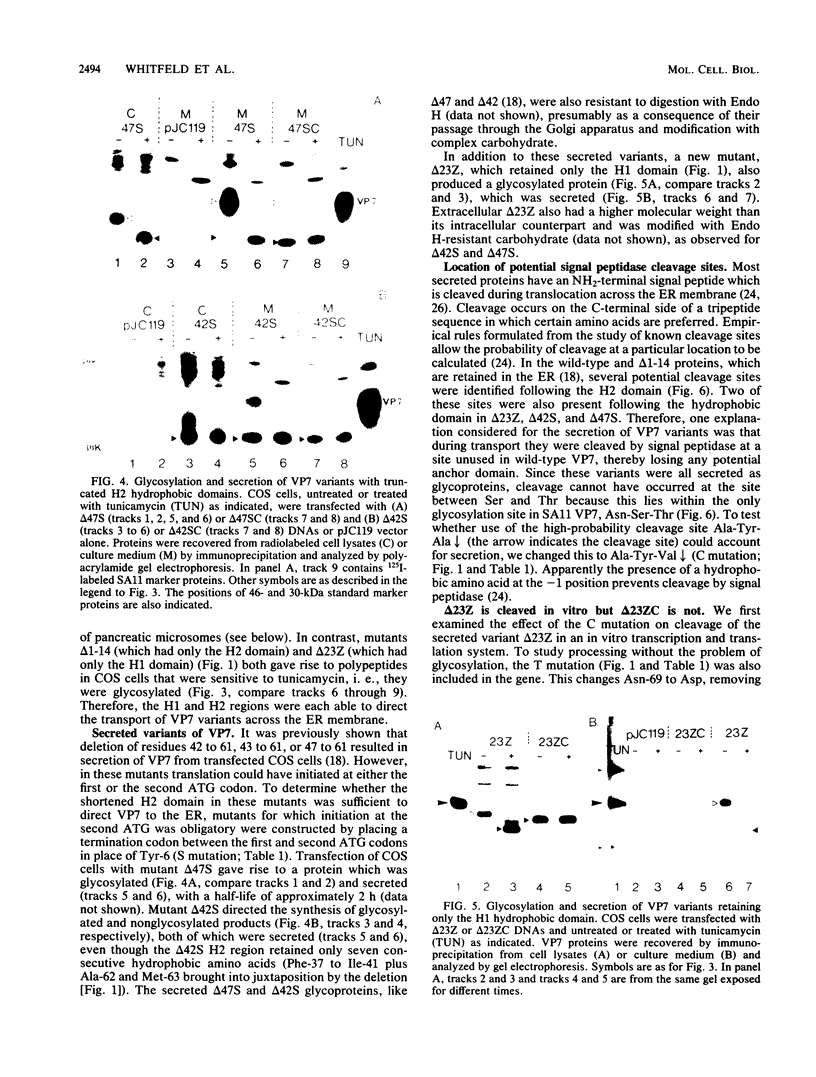
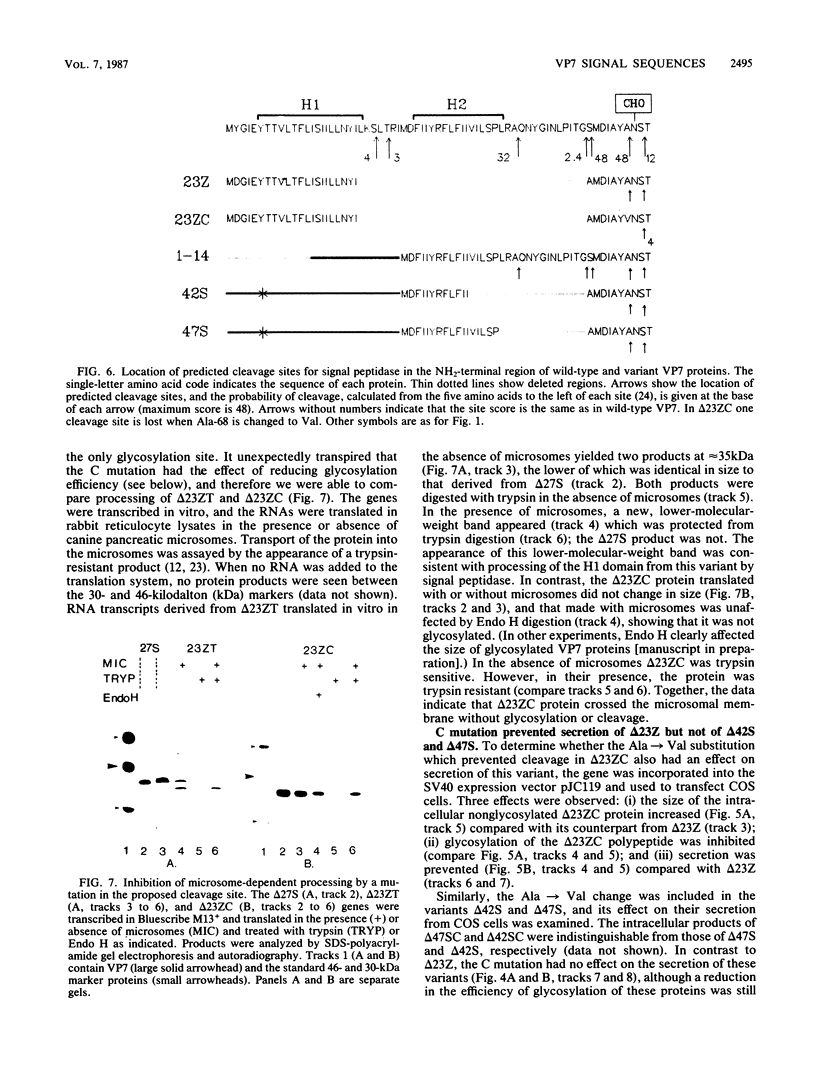
The Simian 11 rotavirus glycoprotein VP7 is directed to the endoplasmic reticulum (ER) of the cell and retained as an integral membrane protein. The gene coding for VP7 predicts two potential initiation codons, each of which precedes a hydrophobic region of amino acids (H1 and H2) with the characteristics of a signal peptide. Using the techniques of gene mutagenesis and expression, we have determined that either hydrophobic domain alone can direct VP7 to the ER. A protein lacking both hydrophobic regions was not transported to the ER. Some polypeptides were directed across the ER membrane and then into the secretory pathway of the cell. For a variant retaining only the H1 domain, secretion was cleavage dependent, since an amino acid change which prevented cleavage also stopped secretion. However, secretion of two other deletion mutants lacking H1 and expressing truncated H2 domains was unaffected by this mutation, suggesting that these proteins were secreted without cleavage of their NH2-terminal hydrophobic regions or secreted after cleavage at a site(s) not predicted by current knowledge.
Full text
PDF

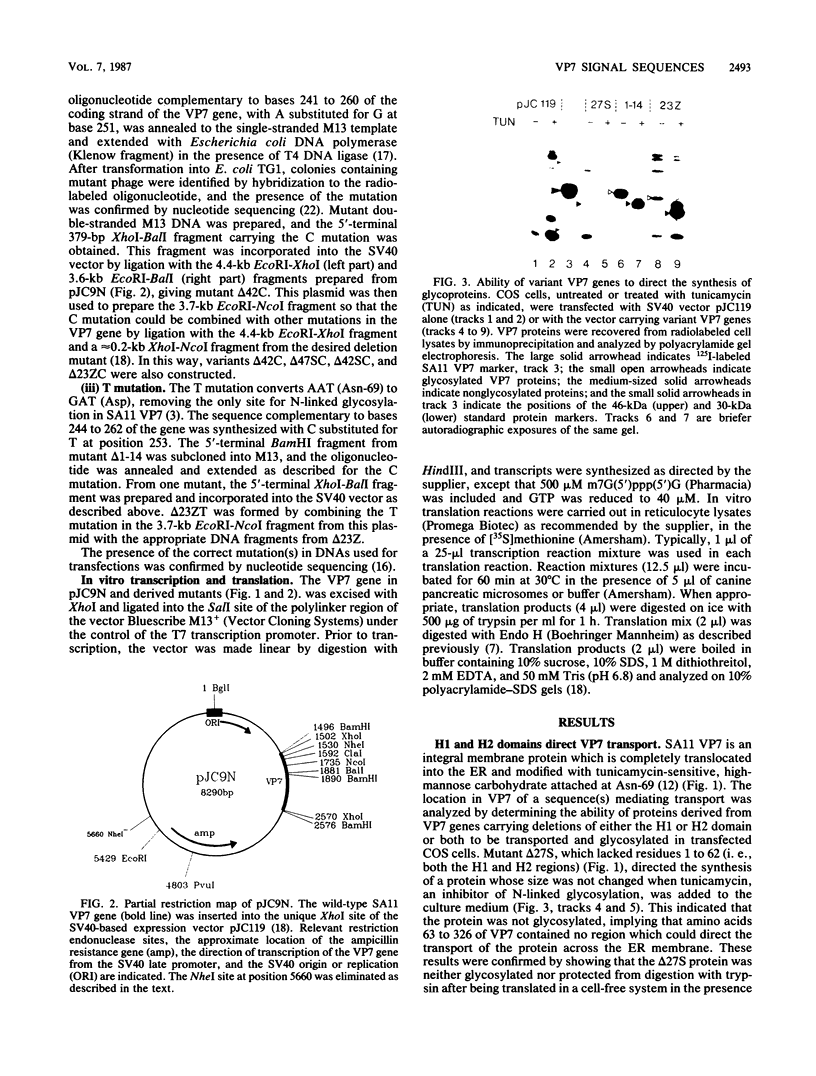


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams G. A., Rose J. K. Structural requirements of a membrane-spanning domain for protein anchoring and cell surface transport. Cell. 1985 Jul;41(3):1007–1015. doi: 10.1016/s0092-8674(85)80081-7. [DOI] [PubMed] [Google Scholar]
- Andrew M. E., Boyle D. B., Coupar B. E., Whitfeld P. L., Both G. W., Bellamy A. R. Vaccinia virus recombinants expressing the SA11 rotavirus VP7 glycoprotein gene induce serotype-specific neutralizing antibodies. J Virol. 1987 Apr;61(4):1054–1060. doi: 10.1128/jvi.61.4.1054-1060.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos T. J., Davis A. R., Nayak D. P. NH2-terminal hydrophobic region of influenza virus neuraminidase provides the signal function in translocation. Proc Natl Acad Sci U S A. 1984 Apr;81(8):2327–2331. doi: 10.1073/pnas.81.8.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Both G. W., Mattick J. S., Bellamy A. R. Serotype-specific glycoprotein of simian 11 rotavirus: coding assignment and gene sequence. Proc Natl Acad Sci U S A. 1983 May;80(10):3091–3095. doi: 10.1073/pnas.80.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W. K., Penaranda M. E., Crawford S. E., Estes M. K. Two glycoproteins are produced from the rotavirus neutralization gene. Virology. 1986 Jun;151(2):243–252. doi: 10.1016/0042-6822(86)90046-2. [DOI] [PubMed] [Google Scholar]
- Engelman D. M., Steitz T. A. The spontaneous insertion of proteins into and across membranes: the helical hairpin hypothesis. Cell. 1981 Feb;23(2):411–422. doi: 10.1016/0092-8674(81)90136-7. [DOI] [PubMed] [Google Scholar]
- Ericson B. L., Graham D. Y., Mason B. B., Hanssen H. H., Estes M. K. Two types of glycoprotein precursors are produced by the simian rotavirus SA11. Virology. 1983 Jun;127(2):320–332. doi: 10.1016/0042-6822(83)90147-2. [DOI] [PubMed] [Google Scholar]
- Estes M. K., Palmer E. L., Obijeski J. F. Rotaviruses: a review. Curr Top Microbiol Immunol. 1983;105:123–184. doi: 10.1007/978-3-642-69159-1_3. [DOI] [PubMed] [Google Scholar]
- Glass R. I., Keith J., Nakagomi O., Nakagomi T., Askaa J., Kapikian A. Z., Chanock R. M., Flores J. Nucleotide sequence of the structural glycoprotein VP7 gene of Nebraska calf diarrhea virus rotavirus: comparison with homologous genes from four strains of human and animal rotaviruses. Virology. 1985 Mar;141(2):292–298. doi: 10.1016/0042-6822(85)90260-0. [DOI] [PubMed] [Google Scholar]
- Gunn P. R., Sato F., Powell K. F., Bellamy A. R., Napier J. R., Harding D. R., Hancock W. S., Siegman L. J., Both G. W. Rotavirus neutralizing protein VP7: antigenic determinants investigated by sequence analysis and peptide synthesis. J Virol. 1985 Jun;54(3):791–797. doi: 10.1128/jvi.54.3.791-797.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L. V., Compans R. W., Davis A. R., Bos T. J., Nayak D. P. Surface expression of influenza virus neuraminidase, an amino-terminally anchored viral membrane glycoprotein, in polarized epithelial cells. Mol Cell Biol. 1985 Sep;5(9):2181–2189. doi: 10.1128/mcb.5.9.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabcenell A. K., Atkinson P. H. Processing of the rough endoplasmic reticulum membrane glycoproteins of rotavirus SA11. J Cell Biol. 1985 Oct;101(4):1270–1280. doi: 10.1083/jcb.101.4.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986 Jan 31;44(2):283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poruchynsky M. S., Tyndall C., Both G. W., Sato F., Bellamy A. R., Atkinson P. H. Deletions into an NH2-terminal hydrophobic domain result in secretion of rotavirus VP7, a resident endoplasmic reticulum membrane glycoprotein. J Cell Biol. 1985 Dec;101(6):2199–2209. doi: 10.1083/jcb.101.6.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose J. K., Bergmann J. E. Altered cytoplasmic domains affect intracellular transport of the vesicular stomatitis virus glycoprotein. Cell. 1983 Sep;34(2):513–524. doi: 10.1016/0092-8674(83)90384-7. [DOI] [PubMed] [Google Scholar]
- Roth M. G., Compans R. W., Giusti L., Davis A. R., Nayak D. P., Gething M. J., Sambrook J. Influenza virus hemagglutinin expression is polarized in cells infected with recombinant SV40 viruses carrying cloned hemagglutinin DNA. Cell. 1983 Jun;33(2):435–443. doi: 10.1016/0092-8674(83)90425-7. [DOI] [PubMed] [Google Scholar]
- Sabatini D. D., Kreibich G., Morimoto T., Adesnik M. Mechanisms for the incorporation of proteins in membranes and organelles. J Cell Biol. 1982 Jan;92(1):1–22. doi: 10.1083/jcb.92.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess M., Lodish H. F. An internal signal sequence: the asialoglycoprotein receptor membrane anchor. Cell. 1986 Jan 17;44(1):177–185. doi: 10.1016/0092-8674(86)90496-4. [DOI] [PubMed] [Google Scholar]
- Wickner W. T., Lodish H. F. Multiple mechanisms of protein insertion into and across membranes. Science. 1985 Oct 25;230(4724):400–407. doi: 10.1126/science.4048938. [DOI] [PubMed] [Google Scholar]
- Zerial M., Melancon P., Schneider C., Garoff H. The transmembrane segment of the human transferrin receptor functions as a signal peptide. EMBO J. 1986 Jul;5(7):1543–1550. doi: 10.1002/j.1460-2075.1986.tb04395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983 Jun 1;133(1):17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985 Jul 5;184(1):99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]