Abstract
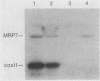
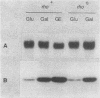
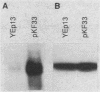
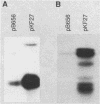
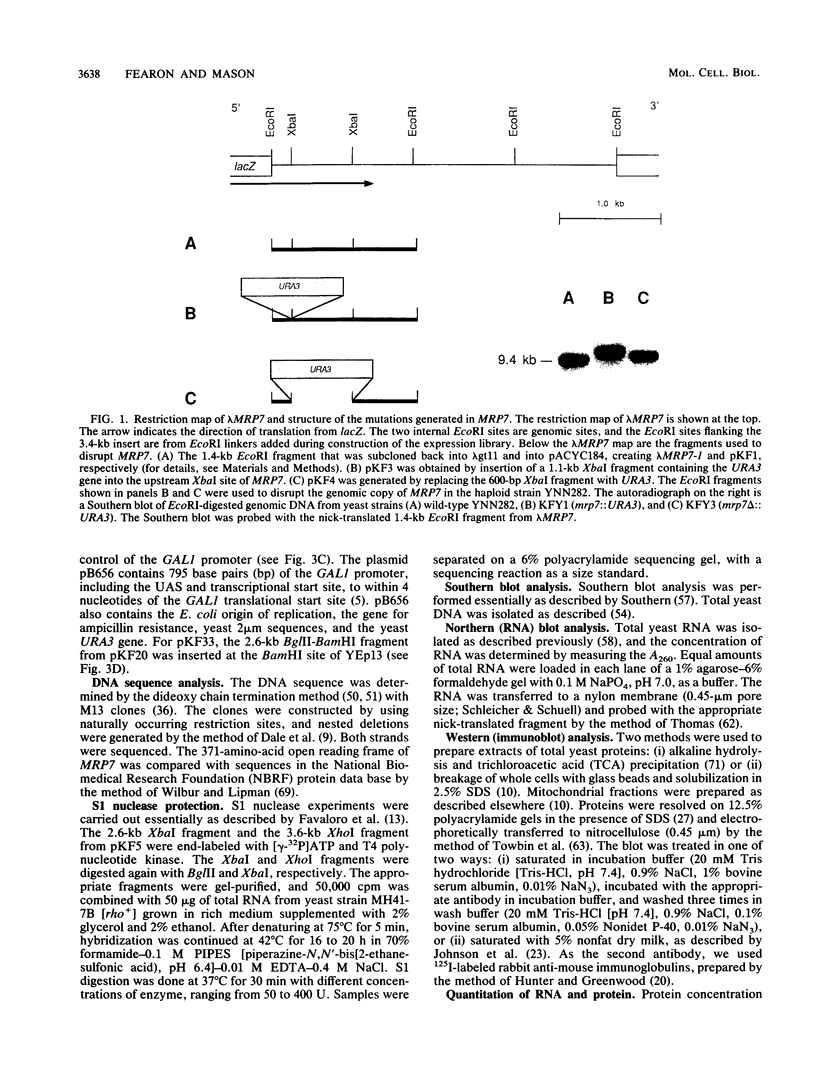
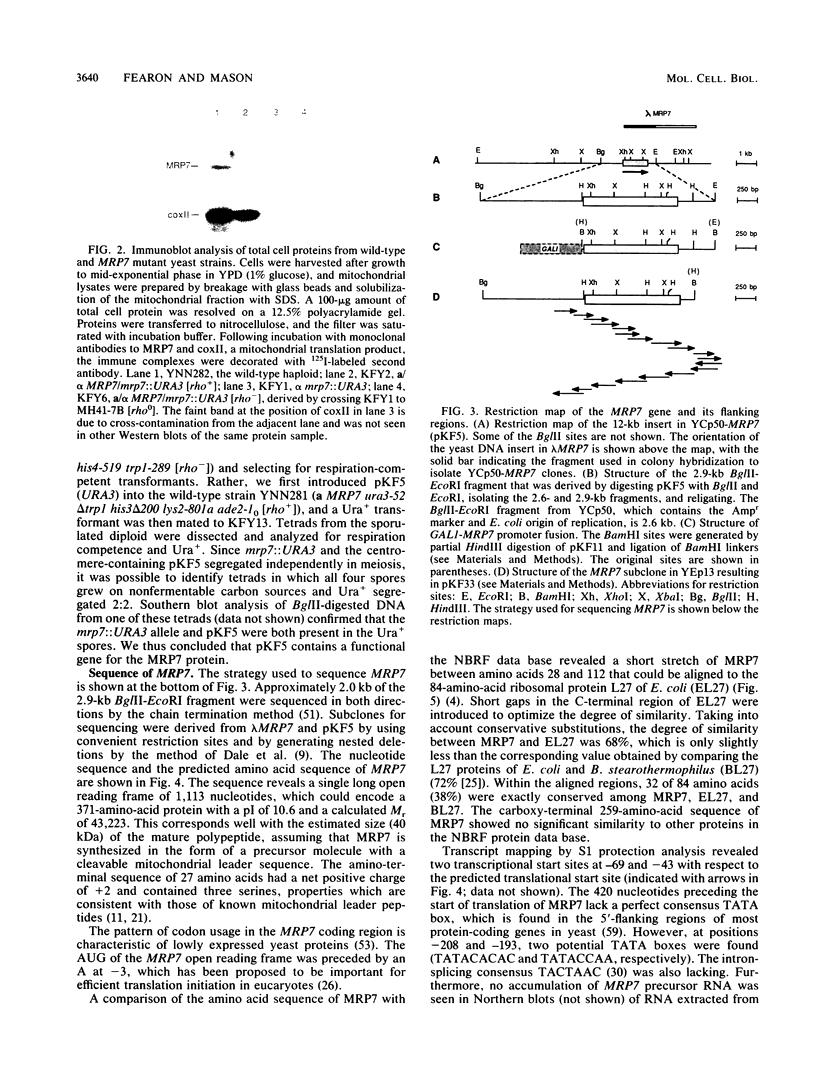
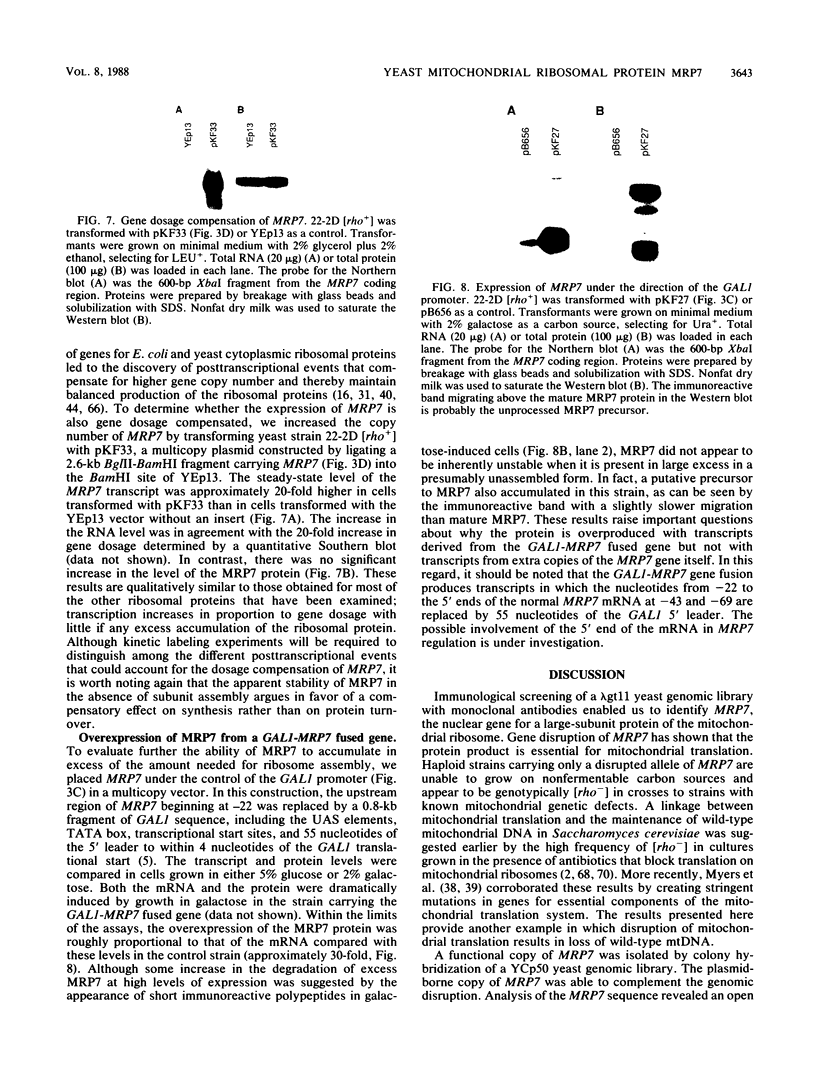
The gene for MRP7, a 40-kilodalton protein of the large subunit of the yeast mitochondrial ribosome, was identified in a lambda gt11 expression library by immunological screening with a monoclonal antibody to MRP7. An intact copy of MRP7 was then isolated from a yeast genomic library by colony hybridization. Gene disruption showed that MRP7 protein was essential for ribosomal function. Sequencing of MRP7 revealed a coding region for a basic (pI 10.6), 43.2-kilodalton protein containing 371 amino acid residues. Amino acid residues 28 to 112 of the deduced MRP7 sequence aligned with the 84 residues of the Escherichia coli ribosomal protein L27, but no significant similarity was detected between the carboxy-terminal 259 amino acids of MRP7 and other protein sequences in existing computer data bases. Within the aligned region, there was 49% amino acid identity between MRP7 and L27, compared with the 57% identity observed between L27 and its homolog in Bacillus stearothermophilus. The steady-state levels of the MRP7 protein and its mRNA were monitored in response to catabolite repression and to increased dosage of the MRP7 gene. The response to catabolite repression was characterized by a ninefold change in the level of the protein and little, if any, change in the level of the mRNA. In cells carrying the MRP7 gene on a high-copy-number plasmid, the mRNA was increased 20-fold, but there was no significant increase in MRP7 protein. Furthermore, MRP7 mRNA and protein accumulated at normal levels in [rho0] cells, which are devoid of 21S rRNA, indicating that the protein is relatively stable in the absence of ribosome assembly. Together, these results suggest that MRP7 is regulated posttranscriptionally, probably at the level of protein synthesis rather than protein turnover.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abovich N., Gritz L., Tung L., Rosbash M. Effect of RP51 gene dosage alterations on ribosome synthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Dec;5(12):3429–3435. doi: 10.1128/mcb.5.12.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Mende L., Arfsten U. The primary structure of protein L27 from the peptidyl-tRNA binding site of Escherichia coli ribosomes. FEBS Lett. 1975 Nov 1;59(1):96–99. doi: 10.1016/0014-5793(75)80349-8. [DOI] [PubMed] [Google Scholar]
- Citron B. A., Donelson J. E. Sequence of the Saccharomyces GAL region and its transcription in vivo. J Bacteriol. 1984 Apr;158(1):269–278. doi: 10.1128/jb.158.1.269-278.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curgy J. J. The mitoribosomes. Biol Cell. 1985;54(1):1–38. doi: 10.1111/j.1768-322x.1985.tb00377.x. [DOI] [PubMed] [Google Scholar]
- Dabbs E. R., Hasenbank R., Kastner B., Rak K. H., Wartusch B., Stöffler G. Immunological studies of Escherichia coli mutants lacking one or two ribosomal proteins. Mol Gen Genet. 1983;192(3):301–308. doi: 10.1007/BF00392166. [DOI] [PubMed] [Google Scholar]
- Dabeva M. D., Post-Beittenmiller M. A., Warner J. R. Autogenous regulation of splicing of the transcript of a yeast ribosomal protein gene. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5854–5857. doi: 10.1073/pnas.83.16.5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale R. M., McClure B. A., Houchins J. P. A rapid single-stranded cloning strategy for producing a sequential series of overlapping clones for use in DNA sequencing: application to sequencing the corn mitochondrial 18 S rDNA. Plasmid. 1985 Jan;13(1):31–40. doi: 10.1016/0147-619x(85)90053-8. [DOI] [PubMed] [Google Scholar]
- Daum G., Böhni P. C., Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Biol Chem. 1982 Nov 10;257(21):13028–13033. [PubMed] [Google Scholar]
- Douglas M. G., McCammon M. T., Vassarotti A. Targeting proteins into mitochondria. Microbiol Rev. 1986 Jun;50(2):166–178. doi: 10.1128/mr.50.2.166-178.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Faye G., Sor F. Analysis of mitochondrial ribosomal proteins of Saccharomyces cerevisiae by two dimensional polyacrylamide gel electrophoresis. Mol Gen Genet. 1977 Sep 21;155(1):27–34. doi: 10.1007/BF00268557. [DOI] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- HUNTER W. M., GREENWOOD F. C. Preparation of iodine-131 labelled human growth hormone of high specific activity. Nature. 1962 May 5;194:495–496. doi: 10.1038/194495a0. [DOI] [PubMed] [Google Scholar]
- Hanahan D., Meselson M. Plasmid screening at high colony density. Gene. 1980 Jun;10(1):63–67. doi: 10.1016/0378-1119(80)90144-4. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Cohen E. H. Sequences responsible for transcription termination on a gene segment in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Aug;4(8):1515–1520. doi: 10.1128/mcb.4.8.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Kelly J. D., Cohen E. H. Transcription terminates in yeast distal to a control sequence. Cell. 1983 Jun;33(2):607–614. doi: 10.1016/0092-8674(83)90441-5. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Chow C. K. The complete amino acid sequences of ribosomal proteins L17, L27, and S9 from Bacillus stearothermophilus. Eur J Biochem. 1984 Mar 1;139(2):225–234. doi: 10.1111/j.1432-1033.1984.tb07998.x. [DOI] [PubMed] [Google Scholar]
- Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983 Mar;47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lake J. A. Evolving ribosome structure: domains in archaebacteria, eubacteria, eocytes and eukaryotes. Annu Rev Biochem. 1985;54:507–530. doi: 10.1146/annurev.bi.54.070185.002451. [DOI] [PubMed] [Google Scholar]
- Lake J. A., Strycharz W. A. Ribosomal proteins L1, L17 and L27 from Escherichia coli localized at single sites on the large subunit by immune electron microscopy. J Mol Biol. 1981 Dec 25;153(4):979–992. doi: 10.1016/0022-2836(81)90462-9. [DOI] [PubMed] [Google Scholar]
- Langford C. J., Gallwitz D. Evidence for an intron-contained sequence required for the splicing of yeast RNA polymerase II transcripts. Cell. 1983 Jun;33(2):519–527. doi: 10.1016/0092-8674(83)90433-6. [DOI] [PubMed] [Google Scholar]
- Lindahl L., Zengel J. M. Ribosomal genes in Escherichia coli. Annu Rev Genet. 1986;20:297–326. doi: 10.1146/annurev.ge.20.120186.001501. [DOI] [PubMed] [Google Scholar]
- Maicas E., Pluthero F. G., Friesen J. D. The accumulation of three yeast ribosomal proteins under conditions of excess mRNA is determined primarily by fast protein decay. Mol Cell Biol. 1988 Jan;8(1):169–175. doi: 10.1128/mcb.8.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters B. S., Stohl L. L., Clayton D. A. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell. 1987 Oct 9;51(1):89–99. doi: 10.1016/0092-8674(87)90013-4. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Morrison C. A., Garrett R. A., Zeichhardt H., Stöffler G. Proteins occurring at, or near, the subunit interface of E. coli ribosomes. Mol Gen Genet. 1973 Dec 31;127(4):359–368. doi: 10.1007/BF00267106. [DOI] [PubMed] [Google Scholar]
- Myers A. M., Crivellone M. D., Tzagoloff A. Assembly of the mitochondrial membrane system. MRP1 and MRP2, two yeast nuclear genes coding for mitochondrial ribosomal proteins. J Biol Chem. 1987 Mar 5;262(7):3388–3397. [PubMed] [Google Scholar]
- Myers A. M., Pape L. K., Tzagoloff A. Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 1985 Aug;4(8):2087–2092. doi: 10.1002/j.1460-2075.1985.tb03896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M., Gourse R., Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- Partaledis J. A., Mason T. L. Structure and regulation of a nuclear gene in Saccharomyces cerevisiae that specifies MRP13, a protein of the small subunit of the mitochondrial ribosome. Mol Cell Biol. 1988 Sep;8(9):3647–3660. doi: 10.1128/mcb.8.9.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson N. J., Fried H. M., Warner J. R. Yeast use translational control to compensate for extra copies of a ribosomal protein gene. Cell. 1982 Jun;29(2):347–355. doi: 10.1016/0092-8674(82)90151-9. [DOI] [PubMed] [Google Scholar]
- Rosbash M., Harris P. K., Woolford J. L., Jr, Teem J. L. The effect of temperature-sensitive RNA mutants on the transcription products from cloned ribosomal protein genes of yeast. Cell. 1981 Jun;24(3):679–686. doi: 10.1016/0092-8674(81)90094-5. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Fink G. R. KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell. 1987 Mar 27;48(6):1047–1060. doi: 10.1016/0092-8674(87)90712-4. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Royden C. S., Pirrotta V., Jan L. Y. The tko locus, site of a behavioral mutation in D. melanogaster, codes for a protein homologous to prokaryotic ribosomal protein S12. Cell. 1987 Oct 23;51(2):165–173. doi: 10.1016/0092-8674(87)90144-9. [DOI] [PubMed] [Google Scholar]
- Röhl R., Nierhaus K. H. Assembly map of the large subunit (50S) of Escherichia coli ribosomes. Proc Natl Acad Sci U S A. 1982 Feb;79(3):729–733. doi: 10.1073/pnas.79.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Tuohy T. M., Mosurski K. R. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986 Jul 11;14(13):5125–5143. doi: 10.1093/nar/14.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M., Elledge S., Davis R. W. Rapid mapping of antigenic coding regions and constructing insertion mutations in yeast genes by mini-Tn10 "transplason" mutagenesis. Proc Natl Acad Sci U S A. 1986 Feb;83(3):730–734. doi: 10.1073/pnas.83.3.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sprague G. F., Jr, Jensen R., Herskowitz I. Control of yeast cell type by the mating type locus: positive regulation of the alpha-specific STE3 gene by the MAT alpha 1 product. Cell. 1983 Feb;32(2):409–415. doi: 10.1016/0092-8674(83)90460-9. [DOI] [PubMed] [Google Scholar]
- Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987 May 8;49(3):295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- Takata R. Genetic studies of the ribosomal proteins in Escherichia coli. XI. Mapping of the genes for L21, L27, S15 and S21 by using hybrid bacteria and over-production of these proteins in the merodiploid strains. Mol Gen Genet. 1978 Apr 6;160(2):151–155. doi: 10.1007/BF00267476. [DOI] [PubMed] [Google Scholar]
- Tatchell K., Chaleff D. T., DeFeo-Jones D., Scolnick E. M. Requirement of either of a pair of ras-related genes of Saccharomyces cerevisiae for spore viability. Nature. 1984 Jun 7;309(5968):523–527. doi: 10.1038/309523a0. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A., Myers A. M. Genetics of mitochondrial biogenesis. Annu Rev Biochem. 1986;55:249–285. doi: 10.1146/annurev.bi.55.070186.001341. [DOI] [PubMed] [Google Scholar]
- Warner J. R., Mitra G., Schwindinger W. F., Studeny M., Fried H. M. Saccharomyces cerevisiae coordinates accumulation of yeast ribosomal proteins by modulating mRNA splicing, translational initiation, and protein turnover. Mol Cell Biol. 1985 Jun;5(6):1512–1521. doi: 10.1128/mcb.5.6.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weislogel P. O., Butow R. A. Low temperature and chloramphenicol induction of respiratory deficiency in a cold-sensitive mutant of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1970 Sep;67(1):52–58. doi: 10.1073/pnas.67.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson D. H., Maroudas N. G., Wilkie D. Induction of the cytoplasmic petite mutation in Saccharomyces cerevisiae by the antibacterial antibiotics erythromycin and chloramphenicol. Mol Gen Genet. 1971;111(3):209–223. doi: 10.1007/BF00433106. [DOI] [PubMed] [Google Scholar]
- Yaffe M. P., Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Efficient isolation of genes by using antibody probes. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1194–1198. doi: 10.1073/pnas.80.5.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Yeast RNA polymerase II genes: isolation with antibody probes. Science. 1983 Nov 18;222(4625):778–782. doi: 10.1126/science.6356359. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]
- elBaradi T. T., van der Sande C. A., Mager W. H., Raué H. A., Planta R. J. The cellular level of yeast ribosomal protein L25 is controlled principally by rapid degradation of excess protein. Curr Genet. 1986;10(10):733–739. doi: 10.1007/BF00405095. [DOI] [PubMed] [Google Scholar]