Abstract
Introduction. The transcription factor NFE2-related factor 2 (Nrf2) is a central regulator of antioxidant and detoxification gene expression in response to electrophilic or oxidative stress. Nrf2 has recently been shown to cross-talk with metabolic pathways, and its gene deletion protected mice from high-fat-diet-(HFD-) induced obesity and insulin resistance. This study aimed to identify potential Nrf2-regulated genes of metabolic interest by comparing gene expression profiles of livers of wild-type (WT) versus Nrf2 knockout (Nrf2-KO) mice after a long-term HFD. Methods. WT and Nrf2-KO mice were fed an HFD for 180 days; total RNA was prepared from liver and used for microarray analysis and quantitative real-time RT-PCR (qRT-PCR). Results. The microarray analysis identified 601 genes that were differentially expressed between WT and Nrf2-KO mice after long-term HFD. Selected genes, including ones known to be involved in metabolic regulation, were prioritized for verification by qRT-PCR: Cyp7a1 and Fabp5 were significantly overexpressed in Nrf2-KO mice; in contrast, Car, Cyp2b10, Lipocalin 13, Aquaporin 8, Cbr3, Me1, and Nqo1 were significantly underexpressed in Nrf2-KO mice. Conclusion. Transcriptome profiling after HFD-induced obesity confirms that Nrf2 is implicated in liver metabolic gene networks. The specific genes identified here may provide insights into Nrf2-dependent mechanisms of metabolic regulation.
1. Introduction
Obesity, type 2 diabetes, and the metabolic syndrome are multifactorial diseases [1] that are considered an epidemic in westernized societies [2]; by increasing the risk of cardiovascular events, cancer, and other diseases, they have detrimental effects on life expectancy and quality [3, 4]. Although knowledge on the pathophysiology of obesity and diabetes is expanding, the identification of new molecular pathways involved in these disorders is necessary to better understand their pathogenesis and to identify potential drug targets. A transcription factor that has recently been implicated in obesity and metabolic dysregulation is Nrf2 (NFE2-related factor 2), encoded by NFE2L2 (nuclear erythroid factor 2-like 2) [5].
Nrf2 is a transcription factor of the “cap n'collar” family that has a central role in maintaining cellular homeostasis in response to oxidative and electrophilic stress [6–9]. Under basal conditions Nrf2 is localized mainly in the cytoplasm where it binds to the Kelch-like ECH-associating protein (Keap1) and is thereby targeted for ubiquitination and proteasomal degradation. Upon exposure to oxidative and electrophilic stress, Nrf2 escapes Keap1-mediated degradation and accumulates in the nucleus where it binds to cis elements in the regulatory domains (antioxidant response elements, AREs) of antioxidant and detoxification genes, inducing their expression [10].
Nrf2 has been described to have a protective function against a number of pathologies that are caused or aggravated by oxidative stress such as cancer, pulmonary disease, and neurodegenerative or inflammatory conditions [11, 12]. Recently, a role of Nrf2 in obesity has also been discovered. Using mainly the Nrf2 knockout (Nrf2-KO) mice as a model, it has been shown by our group and by others that deletion of Nfe2l2 protected mice from diet-induced obesity and insulin resistance [13–16]. In these studies, a variety of diet types has been used: high-fat diet with 60 kcal% fat [13, 16], high-fat diet with 41 kcal% fat [14], and high-fat western diet with 39.7 kcal% fat [15]. The exact mechanisms underlying the protective effect of Nrf2 deletion in high-fat diet-induced obesity remain to be elucidated. However, there is evidence that the cross-talk of Nrf2 with other metabolic factors such as peroxisome proliferator-activated receptor gamma (PPARγ) [14] or fibroblast growth factor 21 (FGF21) [13] may, at least partially, explain this phenotype. In a recent study, we described the phenotypic comparison of WT versus Nrf2-KO mice under high-fat diet (HFD, 60 kcal% fat) or a control diet (standard diet, St.D.) for 180 days [13]. Briefly, under St.D. no difference was observed in body weight gain, glucose tolerance, or insulin tolerance between the two genotypes. While under HFD, both genotypes initially gained weight at about the same rate, the Nrf2-KO mice reached a plateau earlier than WT, and after about 90 days on HFD weighed significantly lower than WT (about 15% lower). Already after 30 days on HFD, the Nrf2-KO mice were significantly more glucose tolerant than WT, and after 180 days they were also significantly more insulin sensitive (as evidenced by intraperitoneal (i.p.) glucose tolerance test and i.p. insulin tolerance test) [13].
Gene expression profiling studies in Nrf2-KO mice under metabolic stress have not yet been reported. The present study used microarray analysis to investigate hepatic genes and gene networks that are regulated directly or indirectly by Nrf2 in mice on a long-term (180 days) HFD regimen.
2. Materials and Methods
2.1. Mice
All animal procedures were approved by the institutional review board of the University of Patras Medical School and were in accordance with E.C. Directive 86/609/EEC. C57BL6J Nrf2+/− mice, originally developed by Professor M. Yamamoto, were obtained from RIKEN BRC (Tsukuba, Japan). Wild type (WT) and Nrf2-KO mice were generated by mating Nrf2+/− male and female mice; the offspring were genotyped as previously described [17]. Male WT and Nrf2-KO mice (9-10 weeks old) were fed ad libitum an St.D. (10 kcal% fat) or an HFD (60 kcal% fat, Research Diets, New Brunswick, NJ) for 180 days (n = 8 for each group). Mice were housed in the animal facility of the University of Patras Medical School in temperature-, light-, and humidity-controlled rooms with a 12 h light/dark cycle.
2.2. Liver RNA Isolation
Liver was excised from mice and was submerged immediately in RNA later solution (Ambion, Foster City, CA). Total RNA was isolated from liver samples from WT or Nrf2-KO mice using the TRIzol reagent (Life Technologies, Carlsbad, CA), following the manufacturer's instructions, and further purified using the RNeasy mini-kit (Qiagen, Hilden, Germany). RNA yield and quality were determined with a NanoDrop 1000 Spectrophotometer (NanoDrop Technologies, Montchanin, DE) and a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). For microarray analysis, pooled RNA from either WT or Nrf2-KO mice was used. For qRT-PCR purposes, RNA from individual liver samples was used to generate cDNA.
2.3. Microarray Experimentation and Analysis
The microarray experiments were performed using total pooled RNA from liver samples of 8 WT or 8 Nrf2-KO mice fed an HFD for 180 days. Four technical replicates were used for each genotype. Microarray experiments were carried out as one-color hybridizations on murine 4plex arrays from Agilent. The Agilent Whole Mouse Genome Microarray 4x44K slides were used (8 slides in total), each slide including 39430 probes. The labelling reaction of total RNA was performed using the Low Input Quick Amp Labelling Kit (Agilent) using 100 ng of total RNA as starting material, according to the manufacturer's instructions. cRNA synthesis was regarded successful provided that ≥1.65 μg of cRNA with a Cy3-incorporation rate ≥8.0 pmol/μg cRNA were synthesized. Fragmentation and hybridization of cRNA was performed as follows: 1.65 μg of Cy3-labelled cRNA were fragmented according to the manufacturer's instructions, and 1.425 μg of fragmented cRNA were hybridized. The hybridization was performed at 65°C for 17 h in an Agilent hybridization oven. Agilent arrays were then washed, scanned, and processed according to the supplier's protocol. After scanning at 5 μm resolution with a DNA microarray laser scanner (Agilent), features were extracted with image analysis tool version A.8.3.1 using default protocols and settings (Agilent). Primary data analysis was performed using Agilent's Feature Extraction Software (version 10.7.3.1). ATLAS Biolabs (Berlin, Germany) performed labelling and hybridization of samples as well as generation of the primary data.
The raw microarray data were initially background corrected, normalized using quantile normalization, and further log2 transformed. Significantly up- or downregulated genes were identified using Significance Analysis of Microarrays (SAM) in the software platform MeV 4.8 (TM4 Microarray Software Suite) [18, 19]. SAM assigns a score to each gene on the basis of a change in gene expression relative to the standard deviation of repeated measurements. For genes with scores greater than an adjustable threshold, SAM uses permutations of the repeated measurements to estimate the percentage of genes identified by chance (the false discovery rate, FDR). Analysis parameters (Delta) were set to result in FDR ≤ 1% (a stringent criterion).
2.4. GEO Accession Numbers
Microarray data discussed in this publication are MIAME compliant and have been deposited in NCBI gene expression omnibus with the following accession number: GSE33575 (GSM830131 through GSM830138).
2.5. Gene Ontology (GO) and Enrichment Analysis
Gene ontology (GO) analysis is helpful for the deduction of conclusions from microarray data. GO is a database with curated annotations for known genes, that is, gene biological processes, molecular functions, and cellular components. GO analysis was performed, using the Genesis 1.7.2 software and the WebGestalt toolkit (http://bioinfo.vanderbilt.edu/webgestalt/), as previously reported [20, 21]. The hypergeometric test with Bonferroni correction was used for enrichment evaluation analysis. The R function adjP was used in order to adjust the nominal P values of the large number of categories at the same time. The significance level for the adjusted P value was set at 0.01, and the minimum number of genes for a category was set at 2.
2.6. Ingenuity Pathway Analysis
Differentially expressed genes (DEGs) were investigated for network interrelation by ingenuity pathway analysis (IPA) software (Ingenuity Systems, Redwood City, CA). IPA scans the set of input genes to identify networks by using the ingenuity pathway knowledge base for interactions between identified “focus genes.” In this study, the liver DEGs between WT and Nrf2-KO mice and hypothetical interacting genes (stored in the knowledge base in IPA software) were used to generate a set of networks with a maximum network size of 35 genes/proteins. Networks are displayed graphically as genes/gene products (nodes) and the biological relationships between the nodes (edges). All edges are from canonical information stored in the ingenuity pathways knowledge base. In addition, IPA computes a score for each network according to the fit of the user's set of significant genes. The score indicates the likelihood that the focus genes in a network from ingenuity's knowledge base are found together due to random chance. A score of 3, as the cutoff for identifying gene networks, indicates that there is only a 1/1000 chance that the focus genes shown in a network are due to random chance; therefore, a score ≥3 indicates a 99.9% confidence level.
2.7. Quantitative Real-Time PCR
Total RNA from individual liver samples was used for cDNA synthesis after a DNAse digestion step (Turbo DNase, Life Technologies) so as to prevent genomic DNA contamination. cDNA was synthesized using the superscript first-strand synthesis system (Life Technologies), and quantitative real-time PCRs were performed in triplicate 20 μL reaction volumes on a StepOnePlus Instrument (Applied Biosystems, Foster City, CA) using Fast SYBR Green Master Mix (Applied Biosystems). Relative mRNA levels were calculated by the comparative threshold cycle method using TBP (TATA-binding protein) as the housekeeping gene. PCR efficiency was determined from a standard curve, and the Pfaffl method was used to calculate fold changes [22]. The correct size of the PCR products was confirmed by electrophoresis on a 2.5% agarose gel stained with ethidium bromide. Purity of the amplified products was assessed by melting curve analysis using the StepOne Software version 2.1 (Applied Biosystems). The primers used for Cyp7a1 (cytochrome P450, family 7, subfamily A, polypeptide 1), Fabp5 (fatty acid binding protein 5), Car (constitutive androstane receptor), Cyp2b10 (cytochrome P450, family 2, subfamily B, polypeptide 10), Lipocalin 13, Aquaporin 8, Cbr3 (carbonyl reductase 3), Me1 (malic enzyme 1), and Nqo1 (NADPH dehydrogenase quinone 1) were obtained from the PrimerBank (Center for Computational and Integrative Biology, Harvard Medical School, Massachusetts, USA) [23–25]. All primer sequences are shown in Table S1 (see Supplementary Material available online at http://dx.doi.org/10.1155/2013/340731).
2.8. Statistical Analysis
In microarrays, normality of the data distribution was checked by the Kolmogorov-Smirnov test. Differences in gene expression levels between WT and Nrf2-KO mice in liver were explored using SAM and the t-test. Numerical values were expressed as the mean ± standard deviation (SD). Statistical significance was set at the 95% confidence level (P < 0.05), and the fold change cutoff was set at 2. For statistical analysis of qRT-PCR data, one-way ANOVA followed by Tukey's test was used; qRT-PCR data were expressed as the mean ± SD. The number of biological or technical replicates used is described in the corresponding results. Statistical significance was set at the 95% confidence level (P < 0.05). The statistical package GraphPad Prism was used for calculations (GraphPad Software, La Jolla, CA).
3. Results
3.1. Differentially Expressed Liver Genes between Nrf2-KO and WT Mice Fed an HFD for 180 Days
SAM analysis, based on strict statistical criteria (fold change >2; median FDR < 0.01; 90th percentile FDR = 0.32), identified 601 liver differentially expressed genes (DEGs) between Nrf2-KO and WT mice after 180 days on HFD. Of these genes, 428 were significantly overexpressed (Table S2) and 173 were significantly underexpressed (Table S3) in Nrf2-KO versus WT mice. The 601 DEGs were clustered using a two-dimensional hierarchical clustering with Euclidean distance. Figure S1 depicts the heatmap of the genes that were over- (Figure S1A) or underexpressed (Figure S1B) in Nrf2-KO versus WT mice.
3.2. Gene Ontology (GO) Analysis of Differentially Expressed Genes
To obtain insights into the functions of the 601 DEGs, gene ontology (GO) analysis was performed. The main processes that these genes are involved in are categorized as follows: (1) immune response; (2) inflammatory response; (3) carbohydrate and pattern binding; (4) G protein and chemokine receptor binding; (5) glutathione transferase activity; (6) peptidase inhibitor activity; (7) cell surface, plasma membrane, and extracellular region genes; and (8) ion homeostasis. Table 1 lists the differentially expressed genes implicated in each of the aforementioned functions.
Table 1.
Differentially expressed genes implicated in the enriched gene ontology processes.
| Biological process—defense response—GO:0006952 | |
| adj P = 0.0001 | |
| Nupr1 | Nuclear protein 1 |
| Aoah | Acyloxyacyl hydrolase |
| Cd28 | CD28 antigen |
| C8b | Complement component 8, beta polypeptide |
| Lat | Linker for activation of T cells |
| Calca | Calcitonin/calcitonin-related polypeptide, alpha |
| Prg2 | Proteoglycan 2, bone marrow |
| Orm3 | Orosomucoid 3 |
| Clec2h | C-type lectin domain family 2, member h |
| Camp | Cathelicidin antimicrobial peptide |
| Wfdc12 | WAP four-disulfide core domain 12 |
| Chi3l3 | Chitinase 3-like 3 |
| Ccl5 | Chemokine (C-C motif) ligand 5 |
| Cd24a | CD24a antigen |
| Cfd | Complement factor D (adipsin) |
| Ccl12 | Chemokine (C-C motif) ligand 12 |
| Wfdc15b | WAP four-disulfide core domain 15B |
| Penk | Preproenkephalin |
| Sh2d1a | SH2 domain protein 1A |
| Ccl4 | Chemokine (C-C motif) ligand 4 |
| Mpo | Myeloperoxidase |
| Ccl8 | Chemokine (C-C motif) ligand 8 |
| Hamp | Hepcidin antimicrobial peptide |
| Hamp2 | Hepcidin antimicrobial peptide 2 |
| Cxcl9 | Chemokine (C-X-C motif) ligand 9 |
| Adora1 | Adenosine A1 receptor |
|
| |
| Biological process—immune response—GO:0006955 | |
| adj P = 0.0001 | |
| Cd8b1 | CD8 antigen, beta chain 1 |
| Thy1 | Thymus cell antigen 1, theta |
| Ltb | Lymphotoxin B |
| Eomes | Eomesodermin homolog (Xenopus laevis) |
| Cd28 | CD28 antigen |
| Igj | Immunoglobulin joining chain |
| H2-Aa | Histocompatibility 2, class II antigen A, alpha |
| Themis | Thymocyte selection associated |
| C8b | Complement component 8, beta polypeptide |
| Lat | Linker for activation of T cells |
| Cd8a | CD8 antigen, alpha chain |
| Prg2 | Proteoglycan 2, bone marrow |
| Fasl | Fas ligand (TNF superfamily, member 6) |
| Lck | Lymphocyte protein tyrosine kinase |
| Ccl5 | Chemokine (C-C motif) ligand 5 |
| Cd24a | CD24a antigen |
| Btla | B and T lymphocyte associated |
| Cfd | Complement factor D (adipsin) |
| Ccl12 | Chemokine (C-C motif) ligand 12 |
| Bcl2a1d | B-cell leukemia/lymphoma 2 related protein A1d |
| Sh2d1a | SH2 domain protein 1A |
| Ccl4 | chemokine (C-C motif) ligand 4 |
| Mpo | Myeloperoxidase |
| Ccl8 | Chemokine (C-C motif) ligand 8 |
| Cxcl9 | Chemokine (C-X-C motif) ligand 9 |
| Serpina3g | Serine (or cysteine) peptidase inhibitor, clade A, member 3G |
|
| |
| Biological process—immune system process—GO:0002376 | |
| adj P = 0.0005 | |
| Cd8b1 | CD8 antigen, beta chain 1 |
| Egr1 | Early growth response 1 |
| S100a9 | S100 calcium binding protein A9 (calgranulin B) |
| Thy1 | Thymus cell antigen 1, theta |
| Ltb | Lymphotoxin B |
| Cd28 | CD28 antigen |
| Cd8a | CD8 antigen, alpha chain |
| Prg2 | Proteoglycan 2, bone marrow |
| Fasl | Fas ligand (TNF superfamily, member 6) |
| Orm3 | Orosomucoid 3 |
| Lck | Lymphocyte protein tyrosine kinase |
| Ccl5 | Chemokine (C-C motif) ligand 5 |
| Btla | B and T lymphocyte associated |
| Cd24a | CD24a antigen |
| Ccl12 | Chemokine (C-C motif) ligand 12 |
| Ccl4 | Chemokine (C-C motif) ligand 4 |
| Bcl11b | B-cell leukemia/lymphoma 11B |
| Cd34 | CD34 antigen |
| Cxcl9 | Chemokine (C-X-C motif) ligand 9 |
| Adora1 | Adenosine A1 receptor |
| Eomes | Eomesodermin homolog (Xenopus laevis) |
| H2-Aa | Histocompatibility 2, class II antigen A, alpha |
| Igj | Immunoglobulin joining chain |
| Themis | Thymocyte selection associated |
| C8b | Complement component 8, beta polypeptide |
| Lat | Linker for activation of T cells |
| Cd5 | CD5 antigen |
| Cfd | Complement factor D (adipsin) |
| Bcl2a1d | B-cell leukemia/lymphoma 2 related protein A1d |
| Sh2d1a | SH2 domain protein 1A |
| Mpo | Myeloperoxidase |
| Ccl8 | Chemokine (C-C motif) ligand 8 |
| Serpina3g | Serine (or cysteine) peptidase inhibitor, clade A, member 3G |
| Cd3d | CD3 antigen, delta polypeptide |
|
| |
| Biological process—inflammatory response—GO:0006954 | |
| adj P = 0.0010 | |
| Nupr1 | Nuclear protein 1 |
| Aoah | Acyloxyacyl hydrolase |
| Cd28 | CD28 antigen |
| C8b | Complement component 8, beta polypeptide |
| Lat | Linker for activation of T cells |
| Calca | Calcitonin/calcitonin-related polypeptide, alpha |
| Orm3 | Orosomucoid 3 |
| Chi3l3 | Chitinase 3-like 3 |
| Ccl5 | Chemokine (C-C motif) ligand 5 |
| Cd24a | CD24a antigen |
| Cfd | Complement factor D (adipsin) |
| Ccl12 | Chemokine (C-C motif) ligand 12 |
| Ccl4 | Chemokine (C-C motif) ligand 4 |
| Ccl8 | Chemokine (C-C motif) ligand 8 |
| Cxcl9 | Chemokine (C-X-C motif) ligand 9 |
| Adora1 | Adenosine A1 receptor |
|
| |
| Biological process—cellular di-, tri-valent inorganic cation homeostasis—GO:0030005 | |
| adj P = 0.0035 | |
| Lck | Lymphocyte protein tyrosine kinase |
| Cd24a | CD24a antigen |
| Ltf | Lactotransferrin |
| Gpr12 | G-protein coupled receptor 12 |
| Mt2 | Metallothionein 2 |
| Hamp | Hepcidin antimicrobial peptide |
| Hamp2 | Hepcidin antimicrobial peptide 2 |
| Mfi2 | Antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5 |
| Calca | Calcitonin/calcitonin-related polypeptide, alpha |
| Slc39a5 | Solute carrier family 39 (metal ion transporter), member 5 |
|
| |
| Biological process—cellular cation homeostasis—GO:0030003 | |
| adj P = 0.0054 | |
| Lck | Lymphocyte protein tyrosine kinase |
| Cd24a | CD24a antigen |
| Ltf | Lactotransferrin |
| Gpr12 | G-protein coupled receptor 12 |
| Mt2 | Metallothionein 2 |
| Hamp | Hepcidin antimicrobial peptide |
| Hamp2 | Hepcidin antimicrobial peptide 2 |
| Mfi2 | Antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5 |
| Calca | Calcitonin/calcitonin-related polypeptide, alpha |
| Slc39a5 | Solute carrier family 39 (metal ion transporter), member 5 |
|
| |
| Biological process—response to wounding—GO:0009611 | |
| adj P = 0.0054 | |
| Slc1a3 | Solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| Nupr1 | Nuclear protein 1 |
| Aoah | Acyloxyacyl hydrolase |
| Cd28 | CD28 antigen |
| C8b | Complement component 8, beta polypeptide |
| Lat | Linker for activation of T cells |
| Calca | Calcitonin/calcitonin-related polypeptide, alpha |
| Tff1 | Trefoil factor 1 |
| Orm3 | Orosomucoid 3 |
| Chi3l3 | Chitinase 3-like 3 |
| Ccl5 | Chemokine (C-C motif) ligand 5 |
| Cd24a | CD24a antigen |
| Cfd | Complement factor D (adipsin) |
| Ccl12 | Chemokine (C-C motif) ligand 12 |
| Ccl4 | Chemokine (C-C motif) ligand 4 |
| Ccl8 | Chemokine (C-C motif) ligand 8 |
| Cxcl9 | Chemokine (C-X-C motif) ligand 9 |
| Adora1 | Adenosine A1 receptor |
|
| |
| Biological process—di-, tri-valent inorganic cation homeostasis—GO:0055066 | |
| adj P = 0.0054 | |
| Lck | Lymphocyte protein tyrosine kinase |
| Cd24a | CD24a antigen |
| Ltf | Lactotransferrin |
| Gpr12 | G-protein coupled receptor 12 |
| Mt2 | Metallothionein 2 |
| Hamp | Hepcidin antimicrobial peptide |
| Hamp2 | Hepcidin antimicrobial peptide 2 |
| Mfi2 | Antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5 |
| Calca | Calcitonin/calcitonin-related polypeptide, alpha |
| Slc39a5 | Solute carrier family 39 (metal ion transporter), member 5 |
|
| |
| Biological process—T cell activation—GO:0042110 | |
| adj P = 0.0067 | |
| Lck | Lymphocyte protein tyrosine kinase |
| Egr1 | Early growth response 1 |
| Cd5 | CD5 antigen |
| Btla | B and T lymphocyte associated |
| Cd24a | CD24a antigen |
| Bcl2a1d | B-cell leukemia/lymphoma 2 related protein A1d |
| Cd28 | CD28 antigen |
| H2-Aa | Histocompatibility 2, class II antigen A, alpha |
| Themis | Thymocyte selection associated |
| Bcl11b | B-cell leukemia/lymphoma 11B |
| Cd3d | CD3 antigen, delta polypeptide |
| Cd8a | CD8 antigen, alpha chain |
|
| |
| Biological process—defense response to fungus—GO:0050832 | |
| adj P = 0.0067 | |
| Hamp | Hepcidin antimicrobial peptide |
| Hamp2 | Hepcidin antimicrobial peptide 2 |
| Mpo | Myeloperoxidase |
|
| |
| Biological process—T cell differentiation—GO:0030217 | |
| adj P = 0.0067 | |
| Lck | Lymphocyte protein tyrosine kinase |
| Egr1 | Early growth response 1 |
| Bcl2a1d | B-cell leukemia/lymphoma 2 related protein A1d |
| Cd28 | CD28 antigen |
| H2-Aa | Histocompatibility 2, class II antigen A, alpha |
| Themis | Thymocyte selection associated |
| Bcl11b | B-cell leukemia/lymphoma 11B |
| Cd3d | CD3 antigen, delta polypeptide |
| Cd8a | CD8 antigen, alpha chain |
|
| |
| Molecular function—carbohydrate binding—GO:0030246 | |
| adj P = 0.0029 | |
| Vcan | Versican |
| Klrg1 | Killer cell lectin-like receptor subfamily G, member 1 |
| Ltf | Lactotransferrin |
| Gpnmb | Glycoprotein (transmembrane) nmb |
| Reg1 | Regenerating islet-derived 1 |
| Lgals2 | Lectin, galactose-binding, soluble 2 |
| Sftpd | Surfactant associated protein D |
| Prg2 | Proteoglycan 2, bone marrow |
| Clec2h | C-type lectin domain family 2, member h |
| Chi3l3 | Chitinase 3-like 3 |
| Cd24a | CD24a antigen |
| Ncam1 | Neural cell adhesion molecule 1 |
| Abp1 | Amiloride binding protein 1 (amine oxidase, Copper-containing) |
| Mpo | Myeloperoxidase |
| Klrd1 | Killer cell lectin-like receptor, subfamily D, member 1 |
| Crispld2 | Cysteine-rich secretory protein LCCL domain containing 2 |
| Ccl8 | Chemokine (C-C motif) ligand 8 |
|
| |
| Molecular function—peptidase inhibitor activity—GO:0030414 | |
| adj P = 0.0029 | |
| Wfdc12 | WAP four-disulfide core domain 12 |
| Serpina9 | Serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 9 |
| Serpinb1a | Serine (or cysteine) peptidase inhibitor, clade B, member 1a |
| Bcl2a1d | B-cell leukemia/lymphoma 2 related protein A1d |
| Wfdc2 | WAP four-disulfide core domain 2 |
| Wfdc15b | WAP four-disulfide core domain 15B |
| Cst7 | Cystatin F (leukocystatin) |
| Spink4 | Serine peptidase inhibitor, Kazal type 4 |
| Serpina3g | Serine (or cysteine) peptidase inhibitor, clade A, member 3G |
| Spink3 | Serine peptidase inhibitor, Kazal type 3 |
| Serpina12 | Serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 |
|
| |
| Molecular function—glutathione transferase activity—GO:0004364 | |
| adj P = 0.0032 | |
| Gstt3 | Glutathione S-transferase, theta 3 |
| Gstm1 | Glutathione S-transferase, mu 1 |
| Gstm4 | Glutathione S-transferase, mu 4 |
| Gsta2 | Glutathione S-transferase, alpha 2 (Yc2) |
| Gstm3 | Glutathione S-transferase, mu 3 |
|
| |
| Molecular function—endopeptidase inhibitor activity—GO:0004866 | |
| adj P = 0.0032 | |
| Wfdc12 | WAP four-disulfide core domain 12 |
| Serpina9 | Serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 9 |
| Serpinb1a | Serine (or cysteine) peptidase inhibitor, clade B, member 1a |
| Bcl2a1d | B-cell leukemia/lymphoma 2 related protein A1d |
| Wfdc2 | WAP four-disulfide core domain 2 |
| Cst7 | Cystatin F (leukocystatin) |
| Spink4 | Serine peptidase inhibitor, Kazal type 4 |
| Serpina3g | Serine (or cysteine) peptidase inhibitor, clade A, member 3G |
| Spink3 | Serine peptidase inhibitor, Kazal type 3 |
| Serpina12 | Serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 |
|
| |
| Molecular function—G-protein-coupled receptor binding—GO:0001664 | |
| adj P = 0.0033 | |
| Ccl8 | Chemokine (C-C motif) ligand 8 |
| Ccl5 | Chemokine (C-C motif) ligand 5 |
| Cxcl9 | Chemokine (C-X-C motif) ligand 9 |
| Adrb3 | Adrenergic receptor, beta 3 |
| Ccl12 | Chemokine (C-C motif) ligand 12 |
| Calca | Calcitonin/calcitonin-related polypeptide, alpha |
| Ccl4 | Chemokine (C-C motif) ligand 4 |
|
| |
| Molecular function—serine-type endopeptidase inhibitor activity—GO:0004867 | |
| adj P = 0.0065 | |
| Wfdc12 | WAP four-disulfide core domain 12 |
| Serpina9 | Serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 9 |
| Serpinb1a | Serine (or cysteine) peptidase inhibitor, clade B, member 1a |
| Serpina3g | Serine (or cysteine) peptidase inhibitor, clade A, member 3G |
| Spink4 | Serine peptidase inhibitor, Kazal type 4 |
| Spink3 | Serine peptidase inhibitor, Kazal type 3 |
| Serpina12 | Serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 |
| Wfdc2 | WAP four-disulfide core domain 2 |
|
| |
| Molecular function—chemokine activity—GO:0008009 | |
| adj P = 0.0097 | |
| Ccl8 | Chemokine (C-C motif) ligand 8 |
| Ccl5 | Chemokine (C-C motif) ligand 5 |
| Cxcl9 | Chemokine (C-X-C motif) ligand 9 |
| Ccl12 | Chemokine (C-C motif) ligand 12 |
| Ccl4 | Chemokine (C-C motif) ligand 4 |
|
| |
| Molecular function—chemokine receptor binding—GO:0042379 | |
| adj P = 0.0097 | |
| Ccl8 | Chemokine (C-C motif) ligand 8 |
| Ccl5 | Chemokine (C-C motif) ligand 5 |
| Cxcl9 | Chemokine (C-X-C motif) ligand 9 |
| Ccl12 | Chemokine (C-C motif) ligand 12 |
| Ccl4 | Chemokine (C-C motif) ligand 4 |
|
| |
| Molecular function—pattern binding—GO:0001871 | |
| adj P = 0.0097 | |
| Vcan | Versican |
| Chi3l3 | Chitinase 3-like 3 |
| Ncam1 | Neural cell adhesion molecule 1 |
| Ltf | Lactotransferrin |
| Gpnmb | Glycoprotein (transmembrane) nmb |
| Abp1 | Amiloride binding protein 1 (amine oxidase, copper-containing) |
| Crispld2 | Cysteine-rich secretory protein LCCL domain containing 2 |
| Mpo | Myeloperoxidase |
| Ccl8 | Chemokine (C-C motif) ligand 8 |
|
| |
| Molecular function—polysaccharide binding—GO:0030247 | |
| adj P = 0.0097 | |
| Vcan | Versican |
| Chi3l3 | Chitinase 3-like 3 |
| Ncam1 | Neural cell adhesion molecule 1 |
| Ltf | Lactotransferrin |
| Gpnmb | Glycoprotein (transmembrane) nmb |
| Abp1 | Amiloride binding protein 1 (amine oxidase, copper-containing) |
| Crispld2 | Cysteine-rich secretory protein LCCL domain containing 2 |
| Mpo | Myeloperoxidase |
| Ccl8 | Chemokine (C-C motif) ligand 8 |
|
| |
| Cellular component—extracellular region—GO:0005576 | |
| adj P = 5.75e − 09 | |
| Col5a2 | Collagen, type V, alpha 2 |
| Vcan | Versican |
| Slc1a3 | Solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| Dmbt1 | Deleted in malignant brain tumors 1 |
| Ltb | Lymphotoxin B |
| Itgbl1 | Integrin, beta-like 1 |
| Mgp | Matrix Gla protein |
| Cst7 | Cystatin F (leukocystatin) |
| Fgl2 | Fibrinogen-like protein 2 |
| Gzma | Granzyme A |
| Prom1 | Prominin 1 |
| Ltbp2 | Latent transforming growth factor beta binding protein 2 |
| Afp | Alpha fetoprotein |
| Calca | Calcitonin/calcitonin-related polypeptide, alpha |
| Tff1 | Trefoil factor 1 |
| Fasl | Fas ligand (TNF superfamily, member 6) |
| Camp | Cathelicidin antimicrobial peptide |
| Orm3 | Orosomucoid 3 |
| Ccl5 | Chemokine (C-C motif) ligand 5 |
| Ccl12 | Chemokine (C-C motif) ligand 12 |
| Abp1 | Amiloride binding protein 1 (amine oxidase, copper-containing) |
| Penk | Preproenkephalin |
| Wfdc15b | WAP four-disulfide core domain 15B |
| Ccl4 | Chemokine (C-C motif) ligand 4 |
| Adipoq | Adiponectin, C1Q and collagen domain containing |
| Hamp | Hepcidin antimicrobial peptide |
| Cxcl9 | Chemokine (C-X-C motif) ligand 9 |
| Il17rb | Interleukin 17 receptor B |
| Mmp8 | Matrix metallopeptidase 8 |
| Svep1 | Sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| Chi3l1 | Chitinase 3-like 1 |
| Gcg | Glucagon |
| Serpina9 | Serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 9 |
| Oosp1 | Oocyte secreted protein 1 |
| Ltf | Lactotransferrin |
| Sema3g | Sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G |
| Wfdc2 | WAP four-disulfide core domain 2 |
| C8b | Complement component 8, beta polypeptide |
| Lcn13 | Lipocalin 13 |
| Esm1 | Endothelial cell-specific molecule 1 |
| Dkk4 | Dickkopf homolog 4 (Xenopus laevis) |
| Gm128 | Predicted gene 128 |
| Sftpd | Surfactant associated protein D |
| Muc4 | Mucin 4 |
| Chi3l3 | Chitinase 3-like 3 |
| Wfdc12 | WAP four-disulfide core domain 12 |
| Tff2 | Trefoil factor 2 (spasmolytic protein 1) |
| Tff3 | Trefoil factor 3, intestinal |
| Cpxm2 | Carboxypeptidase X 2 (M14 family) |
| Cfd | Complement factor D (adipsin) |
| Nts | Neurotensin |
| Crispld2 | Cysteine-rich secretory protein LCCL domain containing 2 |
| Cd2 | CD2 antigen |
| Mmp7 | Matrix metallopeptidase 7 |
| Ccl8 | Chemokine (C-C motif) ligand 8 |
| Cgref1 | Cell growth regulator with EF hand domain 1 |
| Hamp2 | Hepcidin antimicrobial peptide 2 |
| Gpc1 | Glypican 1 |
| Dpt | Dermatopontin |
| Mfi2 | Antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5 |
| Spink4 | Serine peptidase inhibitor, Kazal type 4 |
| Spink3 | Serine peptidase inhibitor, Kazal type 3 |
| Serpina12 | Serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 12 |
| Fam3b | Family with sequence similarity 3, member B |
|
| |
| Cellular component—plasma membrane—GO:0005886 | |
| adj P = 1.09e − 07 | |
| Cd8b1 | CD8 antigen, beta chain 1 |
| Thy1 | Thymus cell antigen 1, theta |
| Adrb3 | Adrenergic receptor, beta 3 |
| Sirpb1 | Signal-regulatory protein beta 1 |
| Cd28 | CD28 antigen |
| Treh | Trehalase (brush-border membrane glycoprotein) |
| Cd8a | CD8 antigen, alpha chain |
| Slc39a5 | Solute carrier family 39 (metal ion transporter), member 5 |
| Klrc1 | Killer cell lectin-like receptor subfamily C, member 1 |
| Clec2h | C-type lectin domain family 2, member h |
| Gpr68 | G protein-coupled receptor 68 |
| Gpr171 | G protein-coupled receptor 171 |
| Btla | B and T lymphocyte associated |
| Cd24a | CD24a antigen |
| Krt19 | Keratin 19 |
| Lair1 | Leukocyte-associated Ig-like receptor 1 |
| Gldn | Gliomedin |
| Gpr110 | G protein-coupled receptor 110 |
| Plxdc2 | Plexin domain containing 2 |
| Adora1 | Adenosine A1 receptor |
| Muc3 | Mucin 3, intestinal |
| Dsg1c | Desmoglein 1 gamma |
| Il17rb | Interleukin 17 receptor B |
| Mas1 | MAS1 oncogene |
| Abcc3 | ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| Lor | Loricrin |
| Gbp5 | Guanylate binding protein 5 |
| H2-Aa | Histocompatibility 2, class II antigen A, alpha |
| Espn | Espin |
| Glycam1 | Glycosylation dependent cell adhesion molecule 1 |
| Cd3g | CD3 antigen, gamma polypeptide |
| Lat | Linker for activation of T cells |
| Ildr1 | Immunoglobulin-like domain containing receptor 1 |
| Cxcr6 | Chemokine (C-X-C motif) receptor 6 |
| Plek2 | Pleckstrin 2 |
| Gpr65 | G-protein coupled receptor 65 |
| Aqp8 | Aquaporin 8 |
| Kcnn4 | Potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| Slco1a1 | Solute carrier organic anion transporter family, member 1a1 |
| Pdcd1 | Programmed cell death 1 |
| Mapt | Microtubule-associated protein tau |
| Cd5 | CD5 antigen |
| Clstn3 | Calsyntenin 3 |
| Gpr12 | G-protein coupled receptor 12 |
| Cd2 | CD2 antigen |
| Gbp1 | Guanylate binding protein 1 |
| Vsig2 | V-set and immunoglobulin domain containing 2 |
| Gpc1 | Glypican 1 |
| Gna14 | Guanine nucleotide binding protein, alpha 14 |
| Slc1a3 | Solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| Itgbl1 | Integrin, beta-like 1 |
| Ltb | Lymphotoxin B |
| Arhgap10 | Rho GTPase activating protein 10 |
| Gbp2 | Guanylate binding protein 2 |
| Prom1 | Prominin 1 |
| Car9 | Carbonic anhydrase 9 |
| Pip5k1a | Phosphatidylinositol-4-phosphate 5-kinase, type 1 alpha |
| Rab25 | RAB25, member RAS oncogene family |
| Fasl | Fas ligand (TNF superfamily, member 6) |
| Tjp3 | Tight junction protein 3 |
| Lck | Lymphocyte protein tyrosine kinase |
| Gpr174 | G protein-coupled receptor 174 |
| Slc16a13 | Solute carrier family 16 (monocarboxylic acid transporters), member 13 |
| Gnat2 | Guanine nucleotide binding protein, alpha transducing 2 |
| Ncam1 | Neural cell adhesion molecule 1 |
| Igsf9 | Immunoglobulin superfamily, member 9 |
| Klrd1 | Killer cell lectin-like receptor, subfamily D, member 1 |
| Art2b | ADP-ribosyltransferase 2b |
| Dsg1b | Desmoglein 1 beta |
| Cd34 | CD34 antigen |
| Oscp1 | Organic solute carrier partner 1 |
| Itk | IL2-inducible T-cell kinase |
| Gpr128 | G protein-coupled receptor 128 |
| Rasgrp2 | RAS, guanyl releasing protein 2 |
| Slc26a3 | Solute carrier family 26, member 3 |
| Cdh1 | Cadherin 1 |
| Il2rb | Interleukin 2 receptor, beta chain |
| Gpnmb | Glycoprotein (transmembrane) nmb |
| Ms4a4b | Membrane-spanning 4-domains, subfamily A, member 4B |
| C8b | Complement component 8, beta polypeptide |
| Grin3b | Glutamate receptor, ionotropic, NMDA3B |
| Clic6 | Chloride intracellular channel 6 |
| Grm8 | Glutamate receptor, metabotropic 8 |
| Mfi2 | Antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5 |
| Cd3d | CD3 antigen, delta polypeptide |
| Slc30a10 | Solute carrier family 30, member 10 |
| Pcdh20 | Protocadherin 20 |
|
| |
| Cellular component—cell surface—GO:0009986 | |
| adj P = 4.72e − 07 | |
| Cd8b1 | CD8 antigen, beta chain 1 |
| Slc1a3 | Solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| Thy1 | Thymus cell antigen 1, theta |
| Il2rb | Interleukin 2 receptor, beta chain |
| Cd28 | CD28 antigen |
| H2-Aa | Histocompatibility 2, class II antigen A, alpha |
| Prom1 | Prominin 1 |
| H2-M2 | Histocompatibility 2, M region locus 2 |
| Cd8a | CD8 antigen, alpha chain |
| Fasl | Fas ligand (TNF superfamily, member 6) |
| Pdcd1 | Programmed cell death 1 |
| Klrc1 | Killer cell lectin-like receptor subfamily C, member 1 |
| Cd24a | CD24a antigen |
| Btla | B and T lymphocyte associated |
| Cd5 | CD5 antigen |
| Ncam1 | Neural cell adhesion molecule 1 |
| Klrd1 | Killer cell lectin-like receptor, subfamily D, member 1 |
| Cd2 | CD2 antigen |
| Cd34 | CD34 antigen |
| Il17rb | Interleukin 17 receptor B |
|
| |
| Cellular component—external side of plasma membrane—GO:0009897 | |
| adj P = 1.43e − 06 | |
| Cd8b1 | CD8 antigen, beta chain 1 |
| Cd5 | CD5 antigen |
| Btla | B and T lymphocyte associated |
| Cd24a | CD24a antigen |
| Thy1 | Thymus cell antigen 1, theta |
| Ncam1 | Neural cell adhesion molecule 1 |
| Il2rb | Interleukin 2 receptor, beta chain |
| Cd28 | CD28 antigen |
| H2-Aa | Histocompatibility 2, class II antigen A, alpha |
| Cd2 | CD2 antigen |
| Klrd1 | Killer cell lectin-like receptor, subfamily D, member 1 |
| Cd34 | CD34 antigen |
| Cd8a | CD8 antigen, alpha chain |
| Fasl | Fas ligand (TNF superfamily, member 6) |
| Pdcd1 | Programmed cell death 1 |
| Klrc1 | Killer cell lectin-like receptor subfamily C, member 1 |
|
| |
| Cellular component—plasma membrane part—GO:0044459 | |
| adj P = 1.08e − 05 | |
| Cd8b1 | CD8 antigen, beta chain 1 |
| Thy1 | Thymus cell antigen 1, theta |
| Itgbl1 | Integrin, beta-like 1 |
| Cd28 | CD28 antigen |
| Prom1 | Prominin 1 |
| Cd8a | CD8 antigen, alpha chain |
| Fasl | Fas ligand (TNF superfamily, member 6) |
| Slc39a5 | Solute carrier family 39 (metal ion transporter), member 5 |
| Tjp3 | Tight junction protein 3 |
| Klrc1 | Killer cell lectin-like receptor subfamily C, member 1 |
| Clec2h | C-type lectin domain family 2, member h |
| Gnat2 | Guanine nucleotide binding protein, alpha transducing 2 |
| Cd24a | CD24a antigen |
| Btla | B and T lymphocyte associated |
| Ncam1 | Neural cell adhesion molecule 1 |
| Igsf9 | Immunoglobulin superfamily, member 9 |
| Art2b | ADP-ribosyltransferase 2b |
| Klrd1 | Killer cell lectin-like receptor, subfamily D, member 1 |
| Dsg1b | Desmoglein 1 beta |
| Cd34 | CD34 antigen |
| Muc3 | Mucin 3, intestinal |
| Dsg1c | Desmoglein 1 gamma |
| Il17rb | Interleukin 17 receptor B |
| Rasgrp2 | RAS, guanyl releasing protein 2 |
| Slc26a3 | Solute carrier family 26, member 3 |
| Abcc3 | ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| Lor | Loricrin |
| Cdh1 | Cadherin 1 |
| Il2rb | Interleukin 2 receptor, beta chain |
| Gpnmb | Glycoprotein (transmembrane) nmb |
| H2-Aa | Histocompatibility 2, class II antigen A, alpha |
| Espn | Espin |
| Cd3g | CD3 antigen, gamma polypeptide |
| Ms4a4b | Membrane-spanning 4-domains, subfamily A, member 4B |
| C8b | Complement component 8, beta polypeptide |
| Lat | Linker for activation of T cells |
| Grin3b | Glutamate receptor, ionotropic, NMDA3B |
| Aqp8 | Aquaporin 8 |
| Kcnn4 | Potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| Slco1a1 | Solute carrier organic anion transporter family, member 1a1 |
| Pdcd1 | Programmed cell death 1 |
| Cd5 | CD5 antigen |
| Grm8 | Glutamate receptor, metabotropic 8 |
| Cd2 | CD2 antigen |
| Vsig2 | V-set and immunoglobulin domain containing 2 |
| Cd3d | CD3 antigen, delta polypeptide |
| Gna14 | Guanine nucleotide binding protein, alpha 14 |
|
| |
| Cellular component—extracellular region part—GO:0044421 | |
| adj P = 0.0043 | |
| Col5a2 | Collagen, type V, alpha 2 |
| Vcan | Versican |
| Slc1a3 | Solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| Dmbt1 | Deleted in malignant brain tumors 1 |
| Ltb | Lymphotoxin B |
| Prom1 | Prominin 1 |
| Afp | Alpha fetoprotein |
| Calca | Calcitonin/calcitonin-related polypeptide, alpha |
| Sftpd | Surfactant associated protein D |
| Fasl | Fas ligand (TNF superfamily, member 6) |
| Orm3 | Orosomucoid 3 |
| Ccl5 | Chemokine (C-C motif) ligand 5 |
| Tff3 | Trefoil factor 3, intestinal |
| Cpxm2 | Carboxypeptidase X 2 (M14 family) |
| Cfd | Complement factor D (adipsin) |
| Ccl12 | Chemokine (C-C motif) ligand 12 |
| Ccl4 | Chemokine (C-C motif) ligand 4 |
| Crispld2 | Cysteine-rich secretory protein LCCL domain containing 2 |
| Adipoq | Adiponectin, C1Q and collagen domain containing |
| Mmp7 | Matrix metallopeptidase 7 |
| Ccl8 | Chemokine (C-C motif) ligand 8 |
| Gpc1 | Glypican 1 |
| Dpt | Dermatopontin |
| Cxcl9 | Chemokine (C-X-C motif) ligand 9 |
| Mmp8 | Matrix metallopeptidase 8 |
| Fam3b | Family with sequence similarity 3, member B |
|
| |
| Cellular component—intrinsic to plasma membrane—GO:0031226 | |
| adj P = 0.0093 | |
| Slc26a3 | Solute carrier family 26, member 3 |
| Thy1 | Thymus cell antigen 1, theta |
| Itgbl1 | Integrin, beta-like 1 |
| Abcc3 | ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| Gpnmb | Glycoprotein (transmembrane) nmb |
| Ms4a4b | Membrane-spanning 4-domains, subfamily A, member 4B |
| Prom1 | Prominin 1 |
| C8b | Complement component 8, beta polypeptide |
| Lat | Linker for activation of T cells |
| Grin3b | Glutamate receptor, ionotropic, NMDA3B |
| Aqp8 | Aquaporin 8 |
| Kcnn4 | Potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| Slco1a1 | Solute carrier organic anion transporter family, member 1a1 |
| Clec2h | C-type lectin domain family 2, member h |
| Cd24a | CD24a antigen |
| Btla | B and T lymphocyte associated |
| Grm8 | Glutamate receptor, metabotropic 8 |
| Cd2 | CD2 antigen |
| Vsig2 | V-set and immunoglobulin domain containing 2 |
| Il17rb | Interleukin 17 receptor B |
3.3. qRT-PCR Verification of Microarray Results
For validation of the microarray data, a group of genes known to be directly or indirectly implicated in lipid or carbohydrate metabolism were selected for quantification with quantitative real-time RT-PCR (qRT-PCR). These genes were Cyp7a1 (cytochrome P450, family 7, subfamily A, polypeptide 1), which is the rate limiting enzyme in bile acid synthesis from cholesterol; Fabp5 (fatty acid binding protein 5); Car (Nr1I3) (constitutive androstane receptor); Cyp2b10 (cytochrome P450,family 2, subfamily B, polypeptide 10), which is a Car target; Lipocalin 13; Aquaporin 8; Cbr3 (carbonyl reductase 3); and Me1 (malic enzyme 1). Nqo1 (NAD(P)H dehydrogenase, quinone 1) was selected as a prototypical Nrf2 target gene. These genes were quantified not only in the liver of WT and Nrf2-KO mice under HFD for 180 days, but also in the liver of WT and Nrf2-KO mice under standard diet (St.D.) for the same time period. The relative gene expression levels are depicted in Figure 1. Cyp7a1 and Fabp5 were found to be over-expressed in the livers of Nrf2-KO mice after HFD feeding compared to WT mice, while the rest of the genes were under-expressed. There was excellent agreement between the microarray and the qRT-PCR data (Pearson correlation coefficient = 0.919; P value <0.001) (Figure 2).
Figure 1.
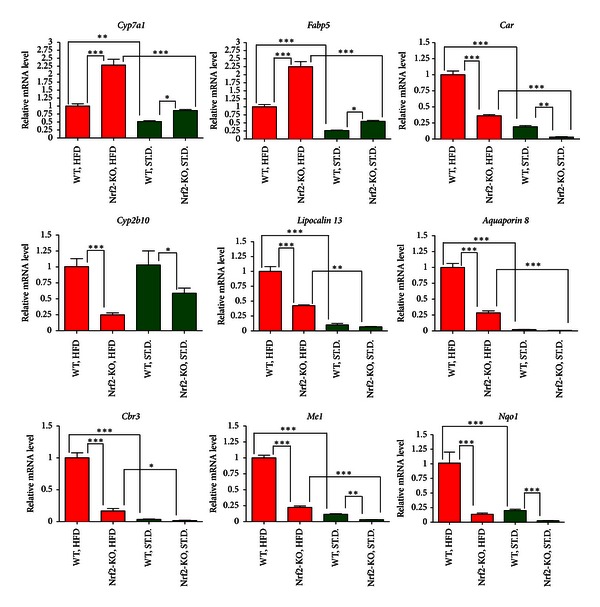
Relative mRNA levels based on qRT-PCR analysis. Cyp7a1, Fabp5, Car, Cyp2b10, Lipocalin 13, Aquaporin8, Cbr3, Me1, and Nqo1 were selected for quantification with qRT-PCR in WT and Nrf2-KO mice under standard diet (St.D.) or high-fat diet (HFD) for 180 days (n = 8 for each genotype in either diet type). The qRT-PCR was performed in triplicate wells for each sample. Bars show means ± SD. *P < 0.01, **P < 0.001, ***P < 0.0001.
Figure 2.
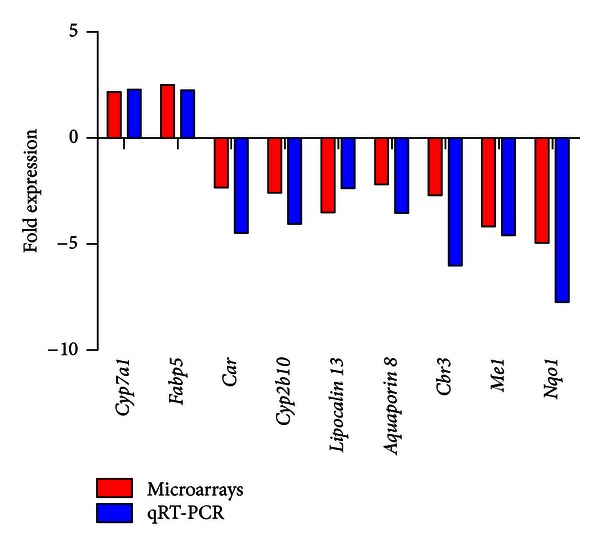
Correlation between microarray and qRT-PCR data. Nine genes identified by microarray analysis differentially expressed in the liver between Nrf2-KO and WT mice after 180 days on HFD were selected for qRT-PCR quantification. The red bars show the fold difference in the mRNA expression of a gene as calculated by microarray analysis, and the blue bars show the relevant fold difference as calculated by qRT-PCR analysis. Very good agreement was observed between the microarrays and qRT-PCR (Pearson correlation coefficient = 0.919; P = 0.0004).
3.4. Canonical Pathways and Networks Impacted by Nrf2 under HFD Feeding
Ingenuity pathway analysis (IPA) was used to rank gene networks by order of consistency of the microarray results with relationships confirmed by previously published results. Figure S2 presents a network which comprises Nrf2 and was generated by the microarray data. Nrf2 and genes that are under-expressed in the Nrf2-KO mice are shown in green; genes that are over-expressed in the Nrf2-KO mice are shown in red. It is obvious that all of the under-expressed genes in this network have been described to be directly regulated by Nrf2: carboxylesterase 1 g (Ces1g) [26]; glutathione S-transferase mu 5 (Gstm5)[27]; glutathione S-transferase alpha 5 (Gsta5) [28]; and NAD(P)H dehydrogenase quinone 1 (Nqo1) [29]. In contrast, none of the genes that are over-expressed in the Nrf2-KO mice is known to be directly regulated by Nrf2. These genes are calcitonin-related polypeptide beta (Calcb); collagen type V alpha 2 (Col5a2); cytochrome P450 family 2 subfamily C polypeptide 8 (Cyp2c8); growth factor independent 1 (Gfi1); H2-M2 histocompatibility 2, M region, locus 2 (H2-m2); interferon gamma (Ifng); solute carrier family 14 (urea transporter) member 1 (Slc14a1); solute carrier family 26 member 3 (Slc26a3); solute carrier family 9 (sodium hydrogen exchanger) member 3 (Slc9a3); serine peptidase inhibitor, Kazal type 4 (Spink4); sulfotransferase family 1E, estrogen preferring, member 1 (Su1lte1); and trefoil factor 1 (Tff1).
IPA analysis identified the statistically significant canonical pathways in the gene list. A corrected Fischer's exact test P value <0.05 was used as the threshold of significance (Figure 3). The number of genes (n) that were differentially expressed in each canonical pathway is shown below along with the P value and the ratio. In the “xenobiotic metabolism signaling” pathway (n = 7; P value = 3.95E − 06; ratio = 0.027), Cyp2c8 and Sult1e1 were over-expressed, whereas Gstm5, Ces1g, Gsta5, Nqo1, Nfe2l2 (as expected), and sulfotransferase family cytosolic 2A dehydroepiandrosterone-preferring member 1 (Sult2a1) were under-expressed. In the “aryl hydrocarbon receptor signaling” pathway (n = 6; P value = 5.88E − 06; ratio = 0.038), Tff1 and Fas ligand (Faslg) were over-expressed, whereas Gstm5, Gsta5, Nqo1, and Nfe2l2 were underexpressed. In the “LPS/IL-1 mediated inhibition of RXR function” pathway (n = 5; P value = 5.49E − 04; ratio = 0.022), Cyp2c8 and Sult1e1 were over-expressed, whereas Gstm5, Gsta5, and Sult2a1 were underexpressed. In the “metabolism of xenobiotics by cytochrome P450” pathway (n = 4; P value = 5.65E − 04; ratio = 0.02), Cyp2c8 was over-expressed and Cyp2b13/Cyp2b9, Gstm5, and Gsta5 were underexpressed. In the “sulfur metabolism” pathway (n = 2; P value = 1.96E − 03; ratio = 0.034), Sult1e1 was overexpressed, whereas Sult2a1 was underexpressed.
Figure 3.
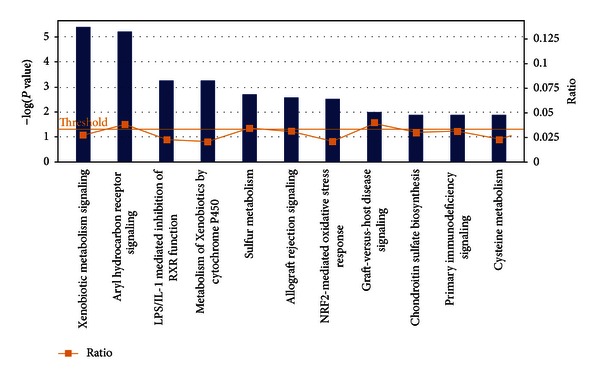
Significantly enriched canonical pathways identified by IPA. The diagram shows significantly overrepresented canonical pathways. A multiple testing corrected P value was calculated using the Benjamini-Hochberg method to control the rate of false discoveries in statistical hypothesis testing. The ratio value represents the number of molecules in a given pathway that meet cutoff criteria, divided by the total number of molecules associated with the respective biological function.
4. Discussion
The findings of our previous study that male Nrf2-KO mice were at least partially protected from HFD-induced (60 kcal% fat) obesity and were more insulin sensitive and more glucose tolerant compared to their WT counterparts [13] are consistent with previous reports using comparable but different treatment parameters (40 kcal% fat diet or modified high-fat-diets) [14–16]. The purpose of this study was to identify hepatic genes differentially expressed between WT and Nrf2-KO mice after long-term (180 days) high-fat-diet-(HFD-) induced obesity. Such information could provide insights into the recently appreciated implication of Nrf2 in the development of obesity and metabolic syndrome. To this end, microarray-based transcriptome analysis was performed, employing strict statistical criteria.
The microarray-based gene expression analysis in these mice generated a total of 601 genes that were differentially expressed between the two genotypes: 478 genes were over-expressed in the Nrf2-KO mice, and 173 were underexpressed. These genes are not only implicated in functions that are typical of Nrf2 target genes, such as immune response [30], inflammation [31–33], and glutathione transferase activity [34, 35], but some gene groups were also found to be associated with carbohydrate and pattern binding (interacting selectively and noncovalently with a repeating or polymeric structure, such as a polysaccharide or peptidoglycan); G protein and chemokine receptor binding; peptidase inhibitor activity; cell surface, plasma membrane, and extracellular region genes; and ion homeostasis. These results further reinforce the notion that Nrf2 should not be regarded solely as a central antioxidant transcription factor, but it may be also implicated directly or indirectly in various tissue-specific homeostatic and/or physiological processes [11].
Among a total 601 differentially expressed genes (DEGs), a subset was selected for validation by qRT-PCR quantification based on their relevance to metabolic pathways. The metabolic pathways and the respective representative genes chosen were bile acid synthesis from cholesterol (Cyp7a1), free fatty acid binding and transport (Fabp5), glucose metabolism (Lipocalin 13), glycerol transport (Aquaporin 8), fatty acid biosynthesis (Me1), and energy homeostasis (Car and its target gene Cyp2b10). Nqo1 mRNA levels were quantified as Nqo1 is considered a prototypical Nrf2 target gene.
The qRT-PCR-based mRNA quantification of specific genes of metabolic interest that were either over-expressed (Cyp7a1 and Fabp5) or under-expressed (Car, Cyp2b10, Lipocalin 13, Aquaporin 8, Cbr3, and Me1) in Nrf2-KO mice compared to WT under HFD revealed potential candidate genes that may be implicated in the development of the different metabolic phenotype of Nrf2-KO mice. As shown in Figure 1, the differential expression of some of these genes was also evident under the St.D. regimen (with the exception of Aquaporin 8, Lipocalin 13, and Cbr3), indicating that these genes may be regulated by Nrf2 under basal conditions as well. HFD for 180 days increased the expression of these genes (with the exception of Cyp2b10 in both genotypes and of Nqo1 in the Nrf2-KO mice), and the fold difference between the two genotypes was generally accentuated. This observation may indicate that the possible regulation (direct or indirect) of these genes by Nrf2 becomes more prominent under stress conditions (HFD-induced obesity). Given that Nqo1 is a prototypical Nrf2 target, its over-expression after HFD in WT mice suggests an increase in the transcriptional activity of Nrf2 by HFD, as supported by the fact that Nqo1 induction was not observed in the Nrf2-KO mice under HFD. The possible functional importance of each of the other validated DEGs is discussed below.
Cyp7a1 is the rate-limiting enzyme in bile acid synthesis from cholesterol. In agreement with previous studies [36], we show that Cyp7a1 mRNA levels increased significantly in both genotypes after HFD feeding (Figure 1). The Cyp7a1 mRNA levels also differed between the two genotypes, with the Nrf2-KO mice showing higher levels under St.D. (about 60% higher) and much higher levels under HFD (about 120% higher) compared to WT. In a previous study, a short-term (30 days) HFD did not accentuate the basal difference between the two genotypes [36], probably because an HFD feeding for a shorter period exposed the animals to lower metabolic and oxidative stress, such that the differences caused by Nrf2 deletion were not as pronounced. Moreover, given that small heterodimer partner (Shp) represses Cyp7a1 expression [37], and Nrf2 induces Shp gene expression [38], a reasonable hypothesis could be that Nrf2 represses Cyp7a1 expression through Shp. This repression of Cyp7a1 expression is abrogated by Nrf2 deletion, leading to increased levels of Cyp7a1 in Nrf2-KO mice compared to WT, which may partially protect them from obesity [39]. To clarify these molecular mechanisms, future experiments should involve Shp and Nrf2 single and double KO mice.
Fabp5 is a member of the fatty acid-binding proteins which binds free fatty acids and regulates lipid metabolism and transport; it was first identified as being upregulated in psoriasis tissue [40]. In this study, Fabp5 was increased after HFD feeding in both genotypes (Figure 1), which is in agreement with previous studies that have shown strong up-regulation of Fabp5 by western-type diet or HFD [41, 42]. Fabp5 also exhibited higher mRNA levels in Nrf2-KO mice under either St.D. or HFD; a study using proteomic analysis showed similar results [43]. The specific physiological significance of Fabp5 elevation in Nrf2-KO mice remains to be elucidated. Further experiments with Nrf2 over-expression or silencing and with concurrent measurement of Nrf2 levels in hepatocytes are warranted to elucidate the possible regulation of Fabp5 by Nrf2.
Car (Nr1I3), initially characterized as a sensor of xenobiotics that regulates responses to toxicants [44], has recently been implicated in the control of energy and metabolism [45]. Car has been ascribed a function as an antiobesity receptor, because treatment of mice with a Car agonist partially prevented HFD-induced obesity in mice, and partially reversed obesity in mice that were already obese [46, 47]. In the present study, mRNA levels of Car, along with those of its primary target gene, Cyp2b10 [48], were lower in Nrf2-KO mice compared to WT under either St.D. or HFD (Figure 1). In this case, the lower expression of Car cannot justify the ameliorated metabolic phenotype of Nrf2-KO compared to WT after long-term HFD. Nevertheless, the observation that Car and Cyp2b10 mRNA levels are lower in Nrf2-KO mice than WT is in accordance with previous studies [49, 50]. Car mRNA was found to be increased in both genotypes after the HFD regimen. But Cyp2b10 that is considered a Car target gene does not follow the same trend. This may indicate a difference in the mRNA turnover of Cyp2b10 that may or may not be reflected in the protein levels. Further investigations, such as cell culture studies with manipulation of Nrf2 levels/activity and measurement of Car levels/activity, are necessary to clarify the mechanisms that underlie the possible regulation of Car by Nrf2.
Lipocalin 13 is a lipocalin family member involved in glucose metabolism, and its deficiency is associated with obesity [51]. Herein, lipocalin 13 liver mRNA levels were found to be lower in Nrf2-KO mice than in WT after long-term HFD; no difference was observed between the two genotypes under St.D. (Figure 1). As the existing data on the role of lipocalin 13 in obesity are scarce, this differential lipocalin 13 mRNA expression between the two genotypes after the HFD feeding for 180 days warrants further elucidation.
Aquaporin family members are mainly water channels, but some of them have also been found to transport glycerol and to be involved in the development of obesity [52]. Aquaporin 8 is expressed in liver [53], and in the present experimental model it exhibited lower mRNA expression in the liver of Nrf2-KO mice compared to WT after HFD. No difference was found between the two genotypes under St.D., but aquaporin 8 was markedly induced in both genotypes after HFD with its levels being lower in the Nrf2-KO mice (Figure 1). As aquaporins may be implicated in the transport of glycerol (a product of the catabolism of triacylglycerols), a possible indirect regulation of aquaporin 8 by Nrf2 may be of metabolic interest.
Cbr3 catalyzes the reduction of carbonyl compounds (highly reactive lipid aldehydes) to the corresponding alcohols (inactive compounds) [54]. A recent clinical study [55] showed that a genetic variation in Cbr3 gene in humans correlates with type 2 diabetes and this effect can potentially be attributed to the catalysis of the conversion of prostaglandin E2 to prostaglandin F2α. In the present study, Cbr3 liver mRNA levels were about 5 times lower in Nrf2-KO mice after HFD compared to WT. Although WT mice tended to have greater Cbr3 mRNA levels under standard diet, this difference was not statistically significant. Given that recent studies have shown that the Cbr3 promoter comprises antioxidant response element (ARE) sequences that are recognized by Nrf2 to induce Cbr3 expression [50, 56, 57], it would be interesting to test whether the Nrf2-regulated Cbr3 expression can contribute to the observed phenotype in the Nrf2-KO mice after HFD feeding.
Me1 is an enzyme that generates NADPH for fatty acid biosynthesis. In this study, Me1 showed decreased mRNA levels in Nrf2-KO mice under St.D. or HFD; this is consistent with previous gene expression profiling studies that describe Me1 as a Nrf2-dependent gene [58–60]. It has been previously shown that Nrf2 can redirect glucose and glutamine to anabolic pathways in cancer cells, and Me-1 is implicated in these pathways [60]; however, these results in cancer cells cannot necessarily be safely extrapolated to nontransformed hepatocytes [61].
A limitation of this study is that microarray analysis was performed only in mice under HFD and not also on standard diet (St.D.). Thus, it is not possible to delineate among the 601 DEGs those genes that are differentially expressed irrespective of the treatment versus those that demonstrate diet-induced differential expression, except for the subset of genes that were validated by qRT-PCR, which analyzed gene expression in mice under both St.D. and HFD. Another limitation of this study is that the hepatic gene expression in this whole body knock-out model may be affected indirectly by endocrine factors that are secreted from other tissues (e.g., adipose tissue, muscle) that are also deficient in Nrf2. Therefore, some of the differentially expressed genes we detect in the liver may be affected by extrahepatic factors. The use of a liver-specific knock-out model could resolve this issue.
5. Conclusions
In conclusion, the current study showed that Nrf2 deletion significantly altered the hepatic gene expression profile after long-term HFD, yielding a set of 601 DEGs that can be the focus of further studies on the role of Nrf2 in obesity. The majority of these DEGs are involved in pathways relevant to the defense against oxidative and electrophilic stress, already known to be regulated by Nrf2. However, certain genes such as Cyp7a1, Fabp5, Car, Cbr3, and Me1 can have specific metabolic effects and appear to be directly or indirectly regulated by Nrf2; these genes may be implicated in the less obese and more insulin sensitive metabolic phenotype of the Nrf2-KO mice. Novel mechanistic understanding and therapeutic interventions for obesity/metabolic syndrome may arise from the elucidation of the cross-talk of Nrf2 with metabolic pathways regulated by these genes.
Supplementary Material
The supplementary material comprises of two figures and three tables. A heatmap of the differentially expressed genes between WT and Nrf2-KO mice after HFD feeding is depicted in figure S1. Figure S2 shows a gene network of some of the differentially expressed genes that also comprises Nrf2. The primer sequences used in this study are provided in Table S1. The 428 genes that were over-expressed as well the 173 genes that were under-expressed in the liver of the Nrf2-KO mice compared to their WT counterparts after 180 days on HFD, are presented in Tables S2 and S3 correspondingly.
Conflict of Interests
All the authors declare that they have no conflict of interests.
Authors' Contribution
Dionysios V. Chartoumpekis and Panos G. Ziros contributed equally to this work. Dionysios V. Chartoumpekis, Panos G. Ziros, Gerasimos P. Sykiotis, and Ioannis G. Habeos conceived and designed the experiments. Dionysios V. Chartoumpekis, Panos G. Ziros, Ralista P. Iskrenova, and Ioannis G. Habeos performed the experiments. Apostolos Zaravinos, Dionysios V. Chartoumpekis, and Ioannis G. Habeos analyzed data. Apostolos Zaravinos, Agathoklis I. Psyrogiannis, and Venetsana E. Kyriazopoulou contributed to these experiments with the reagents, materials, and analysis tools. Dionysios V. Chartoumpekis, Apostolos Zaravinos, Agathoklis I. Psyrogiannis, Venetsana E. Kyriazopoulou, Gerasimos P. Sykiotis, and Ioannis G. Habeos wrote the paper.
Acknowledgments
This work was partially supported by a grant from the Hellenic Endocrine Society (I. G. Habeos) and by the University of Patras internal funding programme “Constantin Carathéodory” (I. G. Habeos). The addresses 2 and 4 are the present addresses of Dionysios V. Chartoumpekis and Apostolos Zaravinos.
References
- 1.Haffner S, Taegtmeyer H. Epidemic obesity and the metabolic syndrome. Circulation. 2003;108(13):1541–1545. doi: 10.1161/01.CIR.0000088845.17586.EC. [DOI] [PubMed] [Google Scholar]
- 2.James PT, Leach R, Kalamara E, Shayeghi M. The worldwide obesity epidemic. Obesity Research. 2001;9(supplement 4):228S–233S. doi: 10.1038/oby.2001.123. [DOI] [PubMed] [Google Scholar]
- 3.Calle EE, Kaaks R. Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nature Reviews Cancer. 2004;4(8):579–591. doi: 10.1038/nrc1408. [DOI] [PubMed] [Google Scholar]
- 4.Bergman RN, Kim SP, Hsu IR, et al. Abdominal obesity: role in the pathophysiology of metabolic disease and cardiovascular risk. American Journal of Medicine. 2007;120(2, supplement 1):3–8. doi: 10.1016/j.amjmed.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 5.Sykiotis GP, Habeos IG, Samuelson AV, Bohmann D. The role of the antioxidant and longevity-promoting Nrf2 pathway in metabolic regulation. Current Opinion in Clinical Nutrition and Metabolic Care. 2011;14(1):41–48. doi: 10.1097/MCO.0b013e32834136f2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calkins MJ, Johnson DA, Townsend JA, et al. The Nrf2/ARE pathway as a potential therapeutic target in neurodegenerative disease. Antioxidants and Redox Signaling. 2009;11(3):497–508. doi: 10.1089/ars.2008.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi M, Yamamoto M. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Advances in Enzyme Regulation. 2006;46(1):113–140. doi: 10.1016/j.advenzreg.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Sykiotis GP, Bohmann D. Stress-activated cap’n’collar transcription factors in aging and human disease. Science Signaling. 2010;3(112):p. re3. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annual Review of Pharmacology and Toxicology. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 10.Osburn WO, Kensler TW. Nrf2 signaling: an adaptive response pathway for protection against environmental toxic insults. Mutation Research. 2008;659(1-2):31–39. doi: 10.1016/j.mrrev.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee JM, Li J, Johnson DA, et al. Nrf2, a multi-organ protector? FASEB Journal. 2005;19(9):1061–1066. doi: 10.1096/fj.04-2591hyp. [DOI] [PubMed] [Google Scholar]
- 12.Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends in Molecular Medicine. 2004;10(11):549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 13.Chartoumpekis DV, Ziros PG, Psyrogiannis AI, et al. Nrf2 represses FGF21 during long-term high-fat diet-induced obesity in mice. Diabetes. 2011;60(10):2465–2473. doi: 10.2337/db11-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pi J, Leung L, Xue P, et al. Deficiency in the nuclear factor E2-related factor-2 transcription factor results in impaired adipogenesis and protects against diet-induced obesity. Journal of Biological Chemistry. 2010;285(12):9292–9300. doi: 10.1074/jbc.M109.093955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang YK, Wu KC, Liu J, Klaassen CD. Nrf2 deficiency improves glucose tolerance in mice fed a high-fat diet. Toxicology and Applied Pharmacology. 2012;264(3):305–314. doi: 10.1016/j.taap.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meher AK, Sharma PR, Lira VA, et al. Nrf2 deficiency in myeloid cells is not sufficient to protect mice from high-fat diet-induced adipose tissue inflammation and insulin resistance. Free Radical Biology & Medicine. 2012;52(9):1708–1715. doi: 10.1016/j.freeradbiomed.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itoh K, Chiba T, Takahashi S, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochemical and Biophysical Research Communications. 1997;236(2):313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 18.Saeed AI, Bhagabati NK, Braisted JC, et al. TM4 microarray software suite. Methods in Enzymology. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 19.Saeed AI, Sharov V, White J, et al. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 2003;34(2):374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 20.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Research. 2005;33(2):W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaravinos A, Lambrou GI, Boulalas I, Delakas D, Spandidos DA. Identification of common differentially expressed genes in urinary bladder cancer. PLoS ONE. 2011;6(4) doi: 10.1371/journal.pone.0018135.e18135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29(9, article e45) doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spandidos A, Wang X, Wang H, Dragnev S, Thurber T, Seed B. A comprehensive collection of experimentally validated primers for Polymerase Chain Reaction quantitation of murine transcript abundance. BMC Genomics. 2008;9(article 633) doi: 10.1186/1471-2164-9-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spandidos A, Wang X, Wang H, Seed B. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Research. 2010;38(1):D792–D799. doi: 10.1093/nar/gkp1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Research. 2003;31(24, article e154) doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Cheng X, Aleksunes L, Klaassen CD. Transcription factor-mediated regulation of carboxylesterase enzymes in livers of mice. Drug Metabolism and Disposition. 2012;40(6):1191–1197. doi: 10.1124/dmd.111.043877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayes JD, Chanas SA, Henderson CJ, et al. The Nrf2 transcription factor contributes both to the basal expression of glutathione S-transferases in mouse liver and to their induction by the chemopreventive synthetic antioxidants, butylated hydroxyanisole and ethoxyquin. Biochemical Society Transactions. 2000;28(2):33–41. doi: 10.1042/bst0280033. [DOI] [PubMed] [Google Scholar]
- 28.Yates MS, Kwak MK, Egner PA, et al. Potent protection against aflatoxin-induced tumorigenesis through induction of Nrf2-regulated pathways by the triterpenoid 1-[2-cyano-3-,12-dioxooleana-1, 9(11)-dien-28-oyl]imidazole. Cancer Research. 2006;66(4):2488–2494. doi: 10.1158/0008-5472.CAN-05-3823. [DOI] [PubMed] [Google Scholar]
- 29.Nioi P, McMahon M, Itoh K, Yamamoto M, Hayes JD. Identification of a novel NRF2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochemical Journal. 2003;374(2):337–348. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thimmulappa RK, Lee H, Rangasamy T, et al. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. Journal of Clinical Investigation. 2006;116(4):984–995. doi: 10.1172/JCI25790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Itoh K, Mochizuki M, Ishii Y, et al. Transcription factor Nrf2 regulates inflammation by mediating the effect of 15-deoxy-Δ12,14-prostaglandin j(2) Molecular and Cellular Biology. 2004;24(1):36–45. doi: 10.1128/MCB.24.1.36-45.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Innamorato NG, Rojo AI, García-Yagüe ÁJ, Yamamoto M, De Ceballos ML, Cuadrado A. The transcription factor nrf2 is a therapeutic target against brain inflammation. Journal of Immunology. 2008;181(1):680–689. doi: 10.4049/jimmunol.181.1.680. [DOI] [PubMed] [Google Scholar]
- 33.Li N, Nel AE. Role of the Nrf2-mediated signaling pathway as a negative regulator of inflammation: implications for the impact of particulate pollutants on asthma. Antioxidants and Redox Signaling. 2006;8(1-2):88–98. doi: 10.1089/ars.2006.8.88. [DOI] [PubMed] [Google Scholar]
- 34.Chanas SA, Jiang Q, McMahon M, et al. Loss of the Nrf2 transcription factor causes a marked reduction in constitutive and inducible expression of the glutathione S-transferase Gsta1, Gsta2, Gstm1, Gstm2, Gstm3 and Gstm4 genes in the livers of male and female mice. Biochemical Journal. 2002;365(2):405–416. doi: 10.1042/BJ20020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Research. 2002;62(18):5196–5203. [PubMed] [Google Scholar]
- 36.Tanaka Y, Aleksunes LM, Yeager RL, et al. NF-E2-related factor 2 inhibits lipid accumulation and oxidative stress in mice fed a high-fat diet. Journal of Pharmacology and Experimental Therapeutics. 2008;325(2):655–664. doi: 10.1124/jpet.107.135822. [DOI] [PubMed] [Google Scholar]
- 37.Zhang YKJ, Guo GL, Klaassen CD. Diurnal variations of mouse plasma and hepatic bile acid concentrations as well as expression of biosynthetic enzymes and transporters. PLoS ONE. 2011;6(2) doi: 10.1371/journal.pone.0016683.e16683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang J, Tabbi-Anneni I, Gunda V, Wang L. Transcription factor Nrf2 regulates SHP and lipogenic gene expression in hepatic lipid metabolism. American Journal of Physiology. 2010;299(6):G1211–G1221. doi: 10.1152/ajpgi.00322.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yiu WF, Kwan PL, Wong CY, et al. Attenuation of fatty liver and prevention of hypercholesterolemia by extract of Curcuma longa through regulating the expression of CYP7A1, LDL-receptor, HO-1, and HMG-CoA reductase. Journal of Food Science. 2011;76(3):H80–H89. doi: 10.1111/j.1750-3841.2011.02042.x. [DOI] [PubMed] [Google Scholar]
- 40.Slegenthaler G, Hotz R, Chatellard-Gruaz D, Jaconi S, Saurat JH. Characterization and expression of a novel human fatty acid-binding protein: the epidermal type (E-FABP) Biochemical and Biophysical Research Communications. 1993;190(2):482–487. doi: 10.1006/bbrc.1993.1073. [DOI] [PubMed] [Google Scholar]
- 41.Hoekstra M, Stitzinger M, Van Wanrooij EJA, et al. Microarray analysis indicates an important role for FABP5 and putative novel FABPs on a Western-type diet. Journal of Lipid Research. 2006;47(10):2198–2207. doi: 10.1194/jlr.M600095-JLR200. [DOI] [PubMed] [Google Scholar]
- 42.Toye AA, Dumas ME, Blancher C, et al. Subtle metabolic and liver gene transcriptional changes underlie diet-induced fatty liver susceptibility in insulin-resistant mice. Diabetologia. 2007;50(9):1867–1879. doi: 10.1007/s00125-007-0738-5. [DOI] [PubMed] [Google Scholar]
- 43.Kitteringham NR, Abdullah A, Walsh J, et al. Proteomic analysis of Nrf2 deficient transgenic mice reveals cellular defence and lipid metabolism as primary Nrf2-dependent pathways in the liver. Journal of Proteomics. 2010;73(8):1612–1631. doi: 10.1016/j.jprot.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei P, Zhang J, Egan-Haftey M, Liang S, Moore DD. The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature. 2000;407(6806):920–923. doi: 10.1038/35038112. [DOI] [PubMed] [Google Scholar]
- 45.Wada T, Gao J, Xie W. PXR and CAR in energy metabolism. Trends in Endocrinology and Metabolism. 2009;20(6):273–279. doi: 10.1016/j.tem.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 46.Gao J, He J, Zhai Y, Wada T, Xie W. The constitutive androstane receptor is an anti-obesity nuclear receptor that improves in sulin sensitivity. Journal of Biological Chemistry. 2009;284(38):25984–25992. doi: 10.1074/jbc.M109.016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dong B, Saha PK, Huang W, et al. Activation of nuclear receptor CAR ameliorates diabetes and fatty liver disease. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(44):18831–18836. doi: 10.1073/pnas.0909731106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Honkakoski P, Zelko I, Sueyoshi T, Negishi M. The nuclear orphan receptor CAR-retinoid X receptor heterodimer activates the phenobarbital-responsive enhancer module of the CYP2B gene. Molecular and Cellular Biology. 1998;18(10):5652–5658. doi: 10.1128/mcb.18.10.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anwar-Mohamed A, Degenhardt OS, El Gendy MAM, Seubert JM, Kleeberger SR, El-Kadi AOS. The effect of Nrf2 knockout on the constitutive expression of drug metabolizing enzymes and transporters in C57Bl/6 mice livers. Toxicology in Vitro. 2011;25(4):785–795. doi: 10.1016/j.tiv.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 50.Wu KC, Cui JY, Klaassen CD. Effect of graded Nrf2 activation on phase-I and -II drug metabolizing enzymes and transporters in mouse liver. PLoS One. 2012;7(7) doi: 10.1371/journal.pone.0039006.e39006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho KW, Zhou Y, Sheng L, Rui L. Lipocalin-13 regulates glucose metabolism by both insulin-dependent and insulin-independent mechanisms. Molecular and Cellular Biology. 2011;31(3):450–457. doi: 10.1128/MCB.00459-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wintour EM, Henry BA. Glycerol transport: an additional target for obesity therapy? Trends in Endocrinology and Metabolism. 2006;17(3):77–78. doi: 10.1016/j.tem.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 53.García F, Kierbel A, Larocca MC, et al. The water channel aquaporin-8 is mainly intracellular in rat hepatocytes, and its plasma membrane insertion is stimulated by cyclic AMP. Journal of Biological Chemistry. 2001;276(15):12147–12152. doi: 10.1074/jbc.M009403200. [DOI] [PubMed] [Google Scholar]
- 54.Pilka ES, Niesen FH, Lee WH, et al. Structural basis for substrate specificity in human monomeric carbonyl reductases. PLoS ONE. 2009;4(10, article e7113) doi: 10.1371/journal.pone.0007113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang YC, Liu PH, Tsai YC, et al. Genetic variation in the carbonyl reductase 3 gene confers risk of type 2 diabetes and insulin resistance: a potential regulator of adipogenesis. Journal of Molecular Medicine. 2012;90(7):847–858. doi: 10.1007/s00109-012-0898-8. [DOI] [PubMed] [Google Scholar]
- 56.Cheng Q, Kalabus JL, Zhang J, Blanco JG. A conserved antioxidant response element (ARE) in the promoter of human carbonyl reductase 3 (CBR3) mediates induction by the master redox switch Nrf2. Biochemical Pharmacology. 2012;83(1):139–148. doi: 10.1016/j.bcp.2011.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ebert B, Kisiela M, Malátková P, El-Hawari Y, Maser E. Regulation of human carbonyl reductase 3 (CBR3; SDR21C2) expression by Nrf2 in cultured cancer cells. Biochemistry. 2010;49(39):8499–8511. doi: 10.1021/bi100814d. [DOI] [PubMed] [Google Scholar]
- 58.Hu R, Xu C, Shen G, et al. Gene expression profiles induced by cancer chemopreventive isothiocyanate sulforaphane in the liver of C57BL/6J mice and C57BL/6J/Nrf2 (-/-) mice. Cancer Letters. 2006;243(2):170–192. doi: 10.1016/j.canlet.2005.11.050. [DOI] [PubMed] [Google Scholar]
- 59.Hu R, Xu C, Shen G, et al. Identification of Nrf2-regulated genes induced by chemopreventive isothiocyanate PEITC by oligonucleotide microarray. Life Sciences. 2006;79(20):1944–1955. doi: 10.1016/j.lfs.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 60.Mitsuishi Y, Taguchi K, Kawatani Y, et al. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22(1):66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 61.Lazar MA, Birnbaum MJ. Physiology. De-meaning of metabolism. Science. 2012;336(6089):1651–1652. doi: 10.1126/science.1221834. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The supplementary material comprises of two figures and three tables. A heatmap of the differentially expressed genes between WT and Nrf2-KO mice after HFD feeding is depicted in figure S1. Figure S2 shows a gene network of some of the differentially expressed genes that also comprises Nrf2. The primer sequences used in this study are provided in Table S1. The 428 genes that were over-expressed as well the 173 genes that were under-expressed in the liver of the Nrf2-KO mice compared to their WT counterparts after 180 days on HFD, are presented in Tables S2 and S3 correspondingly.


