Abstract
Axillary bud outgrowth determines shoot architecture and is under the control of endogenous hormones and a fine-tuned gene-expression network, which probably includes small RNAs (sRNAs). Although it is well known that sRNAs act broadly in plant development, our understanding about their roles in vegetative bud outgrowth remains limited. Moreover, the expression profiles of microRNAs (miRNAs) and their targets within axillary buds are largely unknown. Here, we employed sRNA next-generation sequencing as well as computational and gene-expression analysis to identify and quantify sRNAs and their targets in vegetative axillary buds of the biofuel crop sugarcane (Saccharum spp.). Computational analysis allowed the identification of 26 conserved miRNA families and two putative novel miRNAs, as well as a number of trans-acting small interfering RNAs. sRNAs associated with transposable elements and protein-encoding genes were similarly represented in both inactive and developing bud libraries. Conversely, sequencing and quantitative reverse transcription-PCR results revealed that specific miRNAs were differentially expressed in developing buds, and some correlated negatively with the expression of their targets at specific stages of axillary bud development. For instance, the expression patterns of miR159 and its target GAMYB suggested that they may play roles in regulating abscisic acid-signalling pathways during sugarcane bud outgrowth. Our work reveals, for the first time, differences in the composition and expression profiles of diverse sRNAs and targets between inactive and developing vegetative buds that, together with the endogenous balance of specific hormones, may be important in regulating axillary bud outgrowth.
Key words: microRNAs, phytormones, small RNAs, sugarcane, vegetative axillary buds
Introduction
Plant architecture is defined as a three-dimensional organization of the plant body including shoot development pattern, shape, and position of leaves and flower organs (Reinhardt and Kuhlemeier, 2002). The aerial plant architecture is basically determined by shoot branching and tillering, which, in turn, are coordinated by the formation of the shoot apical meristem during the embryonic phase and subsequently by the formation of axillary meristems (Schmitz and Theres, 2005; Leyser, 2009). Axillary meristems eventually develop into axillary buds that can arrest their growth and become dormant/inactive in several plant species (Ongaro et al., 2008). After the release of shoot apical meristem dominance, dormant/inactive axillary buds may resume their growth and produce entire new axes (Shimizu-Sato and Mori, 2001; Leyser, 2009). Thus, axillary bud outgrowth is crucial to control shoot architecture and biomass production. High biomass production is a desirable characteristic for biofuel crops such as sugarcane.
Sugarcane (Saccharum spp.) is characterized by a highly complex polyploid genome with publicly available expressed sequence tag (EST) and Genomic Survey Sequences Databases that assist genetic studies on this tropical crop. Commercial cultivars are usually propagated using stem cuttings that contain three to four axillary buds. After planting, buds grow out to form primary shoots and develop roots to support the newly developing plant. Therefore, efficient bud outgrowth is also important in establishing and propagating new sugarcane plantlets in the field. Notable progress has been done to elucidate the molecular and physiological bases of vegetative bud outgrowth in model plants such as Arabidopsis, rice, and pea (Leyser, 2009; Doust, 2007), but in sugarcane this developmental process is poorly characterized.
Bud outgrowth is a complex phenomenon that relies on modifications in cell growth and proliferation, accompanied by drastic changes in the production and perception of hormones, including auxins, cytokinins, gibberellins, abscisic acid (ABA), and strigolactones (Leyser, 2009; Wang and Li, 2008). Reinitiation of bud growth is determined mainly by the plant’s genetic programme, which probably includes small RNAs (sRNAs). Interestingly, several members of gene families associated with shoot development are targets for regulation by sRNAs (Chuck et al., 2007; Koyama et al., 2007; Schwarz et al., 2008; Jiao et al., 2010; Wang et al., 2010).
sRNAs (19–25 nt) are classified into two major categories: small interfering RNAs (siRNAs) and microRNAs (miRNAs). Both are recognized as important regulators of gene expression and epigenetic regulation in diverse plant species (Voinnet, 2009; Khraiwesh et al., 2010). miRNA loci are transcribed by RNA polymerase II into primary transcripts, which are processed by nuclear RNase III-like enzyme Dicer-like (DCL) proteins, such as DCL1. Mature miRNAs are incorporated into Argonaute complexes that target degradation or translational repression of mRNAs. As a result, miRNAs play important roles in plant development, including cell patterning and organ development, as well as hormone signalling (Voinnet, 2009).
siRNAs are a complex pool of sRNAs in plants, which includes trans-acting siRNAs (tasiRNAs) and repeat-associated siRNAs (rasiRNAs) (Vaucheret, 2006; Vazquez, 2006). Most siRNAs target the same locus from which they were derived, except for phased 21 nt tasiRNAs, which are produced by TAS loci and target mRNAs from different loci, similar to miRNAs (Allen et al., 2005). Phased trans-acting 21 nt RNAs can also be generated by other loci in plant genomes, probably triggered by miRNA-directed cleavage of the transcripts (Zhai et al., 2011). rasiRNAs are generated from and target repetitive sequences including transposable elements and satellite and microsatellite repeats, as well as centromere heterochromatic repeats (Chen et al., 2006; Domingues et al., 2012). Recently, it has been shown that a particular class of transposons, miniature inverted-repeat transposable elements (MITEs), can generate miRNA-like rasiRNAs that have roles during plant stress responses and hormone signalling (Yan et al., 2011).
sRNAs are often identified by computational analysis and/or experimental approaches such as cloning and sequencing (Wang et al., 2009; Wei et al., 2009; Calvino et al., 2011). However, sRNA data are scarce for sugarcane, partly because its genome sequence is presently unavailable. As a result, few miRNAs have been identified in this important biofuel crop thus far. Recently, our research group computationally identified 19 distinct miRNA precursors (Zanca et al., 2010), whilst other groups have cloned and sequenced a number of small RNAs associated with the drought response in sugarcane (Ferreira et al., 2012; Thiebaut et al., 2012). Additionally, at present, it is unknown whether important regulators like miRNAs play roles during vegetative axillary bud outgrowth. To enrich our knowledge of sRNAs associated with axillary bud development, we generated sRNA libraries from sugarcane dormant/inactive and outgrown buds. We found a select group of sRNAs expressed in both inactive and developing buds, including a number of rasiRNAs and 21 nt tasiRNAs. We identified numerous known microRNAs, as well as two novel miRNA candidates. Several miRNA targets were computationally predicted, and the expression of five was monitored during bud outgrowth by quantitative reverse transcription-PCR (qRT-PCR). As with their mRNA targets, we examined the expression profiles of selected microRNAs using stem–loop qRT-PCR. Further in situ hybridization analysis on selected miRNAs revealed distinct spatial localization patterns within the axillary buds. Additionally, we quantified ABA and catabolite levels in sugarcane inactive and developing buds. Our work supplies novel insights into the dynamic developmental processes underlying vegetative axillary bud development, which are orchestrated at least in part by sRNA-mediated gene regulation.
Materials and methods
Plant material
Six-month-old sugarcane plants of the hybrid SP80-3280 were used to obtain vegetative axillary buds. For all experiments, we excised axillary buds from the fourth to the eighth node of sugarcane culms, which showed similar outgrowth rates (data not shown). We also collected axillary bud tissues from mature plants of Saccharum officinarum (accession Muntok, Java) and Saccharum spontaneum (accession SES205A) grown in the field. Developing axillary buds at 2 and 5 days after planting (DAP) were obtained from stem cuttings planted into an appropriate substratum and grown under greenhouse conditions. Vegetative apexes were collected from 3-week-old in vitro plants of the hybrid SP80-3280. Vegetative tissues of Sorghum bicolor (BTx623), 1-month-old plantlets of Oryza sativa (ssp. japonica cv. Nipponbare) and Brachypodium disthacyon (Bd21), 2-week-old Solanum lycopersicum (cv. Micro-Tom) seedlings, and 2-week-old seedlings of Arabidopsis thaliana (Columbia) were also harvested. Arabidopsis seedlings were grown in a growth chamber with a 16h/8h light/dark photoperiod at 200 μmol m–2 s–1.
Public sequence data sets
Sugarcane EST sequences were retrieved from the Sugarcane Gene Index (release 3.0, July 2011), and sugarcane bacterial artificial chromosome (BAC) sequences were downloaded from GenBank. All recorded miRNAs from plants were obtained from miRBase (version 17; http://www.mirbase.org/). We also employed BLAST searches to identify sRNAs sequences similar to tRNAs [GtRNAdb: http://gtrnadb.ucsc.edu/ (Chan and Lowe, 2009) and rRNAs (Rfam database, http://rfam.sanger.ac.uk/ Gardner et al., 2011)]. Complete genomic sequences of sorghum, rice, and Arabidopsis were downloaded from Phytozome (http://www.phytozome.net/), GenBank, and TAIR (http://www.arabidopsis.org/), respectively. Their transcriptomes were obtained from The Gene Index Project (http://compbio.dfci.harvard.edu/tgi/).
sRNA library construction and bioinformatic analysis
Total RNA from 0 DAP inactive and 2 DAP developing axillary buds was resolved on a 15% TBE/urea polyacrylamide gel to obtain the sRNA fraction of 16–30 nt. Briefly, 5’-adenylated single-stranded adapter was first ligated to the 3’ end of the RNA using T4 RNA ligase without ATP, followed by a second single-stranded adapter ligation at the 5’ end of the RNA using T4 RNA ligase in the presence of ATP. The resulting products were fractioned on a 10% TBE/urea polyacrylamide gel and used for cDNA synthesis and PCR amplification. Illumina sequencing was performed as described previously (Blevins et al., 2011). Raw sequences were retrieved in a FASTQ formatted file and the adapter sequences were removed using Perl Scripts. After trimming of adapter sequences, the inserts were sorted into separate files according to their lengths. Subsequently, we grouped all similar sequences and counted the read contributions from each library for each non-redundant sRNA sequence. We used the program MAQ (http://maq.sourceforge.net/) to map 19–25 nt RNA reads against several databases utilized in this study as described above. MAQ is a p rogram that rapidly aligns short reads to reference sequences. The sRNA library data from axillary buds were deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE38894.
To identify EST sequences similar to MITEs, we firstly blasted the Sugarcane Gene Index sequences against the Gramineae Repeat Database (http://plantrepeats.plantbiology.msu.edu/index.html) using the following parameters: 80% of EST coverage and Expect (E) value <e–5. The recovering sequences were blasted against each other and those showing ≥90% nucleotide identity were regarded as one MITE-containing transcript. sRNAs from both libraries were blasted against the MITE-like EST sequences using the following parameters: no mismatches allowed, word size of seven, and E value of 1000.
tasiRNAs derived from sugarcane transcripts were identified using the computational approach described by Chen et al. (2007). This approach is non-biased and applicable to predict transcripts yielding phased sRNAs, especially when analysing data from organisms with incomplete genomic information, such as sugarcane. Sugarcane 21 nt sRNAs and EST sequences were retrieved from the respective databases as described above. The coordinates of the sRNAs were then analysed using the TAS prediction algorithm based on Perl. ESTs with P <0.001 were considered as potential tasiRNA-generating transcripts.
Identification of novel and known miRNAs expressed in sugarcane axillary buds
The sRNAs sequences were mapped using MAQ against miRbase version 17 and currently available sugarcane miRNAs sequences. To minimize the noise, we did not consider, for further analyses, low-abundance miRNAs (with a total number of reads <15). We also mapped the sRNAs against non-coding sequences of the Sugarcane Gene Index for identification of novel miRNAs in sugarcane. Precursor sequences of approximately 620 nt were extracted (300 nt upstream and 300 nt downstream from the BLAST hits) and used for hairpin structure predictions using the MFOLD3.2 algorithm. The number of structures, free energy, miRNA-like helicity, number of arms per structure, size of helices within arms, and size/symmetry of internal loops within arms were analysed by our in-house MIRcheck-based script (Zanca et al., 2010), followed by manual inspection. To distinguish new miRNAs from rasiRNAs, we blasted precursor and mature sequences of sRNAs against the Gramineae Repeat Database (Ouyang and Buell, 2004). Finally, to identify differentially expressed miRNAs between 0 and 2 DAP sRNA deep-sequencing data, we firstly normalized the data by the number of the total mapped reads (transcripts per million) and then applied statistical significance using Fisher’s exact test with Bonferroni correction and adjusted P values <0.01 as a criteria. For target prediction, we used approaches described previously (Zanca et al., 2010; Dai and Zhao, 2011).
RNA extraction and stem–loop pulsed RT-PCR
Total RNA was extracted using Trizol reagent (Invitrogen, USA) according to the manufacturer’s instructions and treated with DNAse I (Invitrogen) to remove any residue of genomic DNA. DNAse-treated RNA (1.5 μg) was reverse transcribed to generate the first-strand cDNA according to the method of Varkonyi-Gasic et al. (2007). Oligo(dT) primer was also added to the reaction for the detection of target mRNAs and internal controls. cDNA dilutions were used for PCRs containing 1.0 μl of cDNA, 1.5mM magnesium sulfate, 0.25mM of each dNTP, 10 pmol of each primer, and 1U of Taq DNA polymerase (Promega, USA). The reactions were done with the following cycling conditions: 94°C for 2min, and an appropriate number of cycles of 94 °C for 20 s, 60 °C for 30 s, and 72 °C for 45 s. All reactions were repeated twice with two biological samples. Primer sequences are described in Supplementary Table S1 at JXB online.
Stem–loop pulsed qRT-PCR
First-strand cDNA was transcribed as described above. A SYBR Green PCR was performed using a 7500/7500 fast Sequence Detection System (Applied Biosystems, USA). Briefly, 5 μl of 1:80 (v/v) cDNA dilutions was added to 12.5 μl of Platinum SYBR Green qPCR superMix-UDG (Invitrogen), 3 pmol of each primer and ddH2O to a final volume of 25 μl. The reactions were amplified for 2min at 50 °C and 2min at 95 ºC, followed by 40 cycles of 95 °C for 15 s and 60 °C for 30 s. The sugarcane 25S rRNA gene (TC148086) was used as an internal control (Iskandar et al., 2004). PCR products for each primer set were subjected to melt-curve analysis, confirming the presence of only one peak on thermal dissociation generated by the thermal-denaturing protocol. The PCR efficiency for each primer set was evaluated through standard curve constructed from the amplification values of serial dilutions of cDNA. Three replicates were analysed for each biological sample along with template-free reactions as negative controls. The threshold cycle (C T) was determined automatically by the instrument, and the fold change of each gene was calculated using the equation 2−ΔΔCT (Livak and Schmittgen, 2001). Primer sequences are described in Supplementary Table S1.
sRNA in situ hybridization
We used 0 and 5 DAP vegetative axillary buds harvested from 6-month-old sugarcane hybrid SP80-3280 plants for sRNA in situ hybridizations as described by Javelle and Timmermans (2012). Locked nucleic acid (LNA) probes with sequences complementary to selected miRNAs and negative controls (Scramble-miR) were synthesized by Exiqon (USA) and labelled with digoxigenin using a DIG Oligonucleotide 3’-end Labeling kit (Roche Applied Science, USA). Ten picomoles of each probe was used for each slide, and hybridization and washing steps were performed at 55 °C.
Comparative sequence analysis
Comparative analysis of possible orthologues of SsDCL1 (TC143136) was done by constructing a phylogenetic tree containing highly similar plant sequences. Phylogenetic relationships of aligned protein sequences were constructed using the neighbour-joining method in MEGA4 software (Tamura et al., 2007). The accession numbers of the Arabidopsis and rice DCL sequences have been published elsewhere (Nogueira et al., 2009).
ABA and catabolite quantification
Vegetative axillary buds of each developmental stage (0, 2, and 5 DAP) were harvested from 6-month-old sugarcane hybrid SP80-3280 plants. The procedures for tissue sample extraction, purification, and further quantification of ABA and catabolites were performed as described previously using an ultra-performance liquid chromatography tandem mass spectrometry method (Chiwocha et al., 2003).
Results
Overview of sRNAs expressed in vegetative axillary buds
Vegetative axillary bud outgrowth determines shoot architecture and is under the control of endogenous hormones and a fine-tuned gene-expression network. To unravel additional components of regulatory networks operating within axillary buds, we sought to identify sRNA populations associated with bud outgrowth. We generated two bar-coded sequencing libraries using total RNA extracted from sugarcane inactive buds (0 DAP) and from outgrown axillary buds (2 DAP; Fig. 1). After excluding poor-quality reads, those without inserts, and those with inserts smaller than 19 nt, we identified reads matching known cellular sRNAs of 19–25 nt of various classes (Supplementary Table S2 at JXB online).
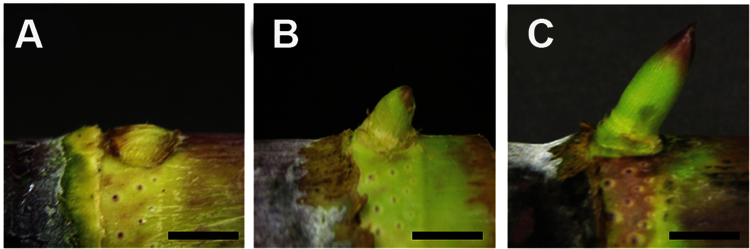
Fig. 1.
Sugarcane axillary bud outgrowth. Sugarcane hybrid SP80-3280 main stem containing inactive or 0 DAP buds (A), and stem cuttings containing developing buds at 2 DAP (B) and 5 DAP (C). Tissues from 0 and 2 DAP developing buds were harvested for constructing the sRNA libraries, gene-expression analyses, and hormone profiling. Tissues from 5 DAP developing buds were used for gene-expression analyses and hormone profiling. Bars, 1.0cm. (This figure is available in colour at JXB online.)
The distribution of sequences in different size classes of sRNAs was comparable in inactive and developing buds (Supplementary Fig. S1A at JXB online), which suggests that multiple sRNA pathways are similarly active in cells not undergoing substantial proliferation (in 0 DAP inactive buds) when compared with cells from 2 DAP outgrown buds. As generally described, the majority of the sRNA sequences in both libraries are 24 nt (4 918 279 reads), followed by 21 nt (2 108 665 reads) and 22 nt (1 478 716 reads) sequences. To understand better the possible roles of this complex pool of sRNAs, we initially evaluated the distribution of the two major classes of axillary bud-associated sRNAs (21 and 24 nt sRNAs) on publicly available sugarcane BACs that are fully annotated (Jannoo et al., 2007; Garsmeur et al., 2011). We observed a similar pattern of distribution of these two classes of sRNAs between homoeologous haplotypes from Saccharum officinarum and Saccharum spontaneum, the ancient polyploid progenitors of sugarcane modern commercial hybrids (Dillon et al., 2007). Several sequences from the 21 and 24 nt classes of sRNAs matched transposable elements (TEs) housed into intergenic regions or within protein-coding regions (Supplementary Fig. S1B). This indicated that most of these sRNAs are rasiRNAs and that these TEs might be transcriptionally active in ancient species as well as in modern hybrids.
Numerous 20–24 nt sRNAs from axillary buds match MITEs (Supplementary Fig. S1B), providing evidence for active sRNA biogenesis derived from these types of TE in the sugarcane genome. MITEs often exist in high copy number in genomes and some are transcribed and capable of generating sRNAs (Kuang et al., 2009). MITE-derived sRNAs may have important roles in sRNA-mediated gene regulation and the evolution of several species. Moreover, they might represent the evolutionary link between miRNA and siRNAs (Piriyapongsa and Jordan, 2008; Zanca et al., 2010; Li et al., 2011). To identify transcribed sugarcane MITEs that perfectly matched sRNAs from axillary buds, we first searched for MITEs in the EST database (see Materials and methods). We identified 23 ESTs similar to known MITEs (the majority belonging to the Tourist-like superfamily; data not shown), nine of which had perfect matches with axillary bud-expressed sRNAs. These MITEs were named as MiSc1–MiSc9 (Supplementary Table S3 at JXB online), similar to the nomenclature provided by Kuang et al. (2009).
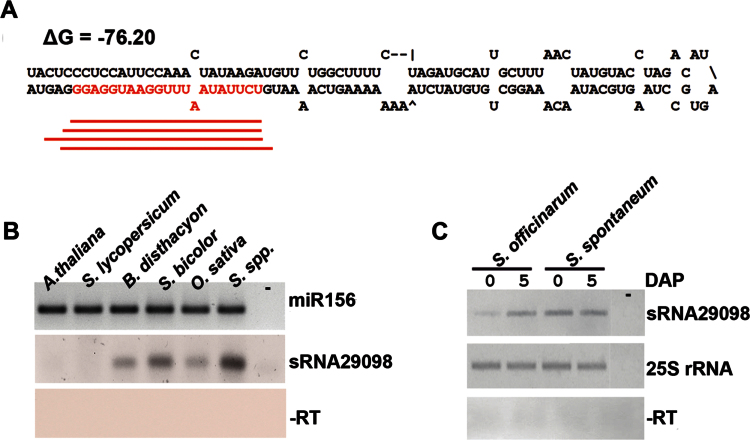
Whilst most transcribed sugarcane MITEs (Tourist-like superfamily) had hits with 22–24 nt sRNAs, MiSc8 and MiSc9 seem to generate mostly 20 and 21 nt sRNAs (Supplementary Table S3). MiSc9 displayed a weak similarity to Stowaway-like element and formed a harpin-like structure from which sRNAs from different sizes were apparently excised at specific locations like canonical miRNAs (Fig. 2A). We evaluated the accumulation pattern of the most represented 21 nt sRNA (namely sRNA2908; Supplementary Table S3) that initiated with a 5’-terminal U residue and perfectly matched MiSc9 stem–loop via stem–loop RT-PCR (Varkonyi-Gasic et al., 2007). sRNA29098 transcripts were readily detectable, indicating that it was an authentic sRNA. Moreover, it accumulated in all interrogated monocots but not in the eudicots Arabidopsis and tomato, indicating it is potentially generated only by the genome of monocots (Fig. 2B). As sRNA29098 perfectly matched Saccharum spontaneum BAC (position around 90 000bp; Supplementary Fig. S1B), we tested whether this sRNA accumulated in Saccharum officinarum and Saccharum spontaneum. sRNA29098 transcripts accumulated in axillary buds from ancient species and modern hybrids (Fig. 2B, C). Based upon the fact that sRNA29098 and its variants matched a specific location in the stem–loop (Fig. 2A), reminiscent of DCL1 cleavage products, we propose that they originated from MiSc9 via miRNA or miRNA-like biogenesis pathways.
Fig. 2.
MITE-derived sRNAs are expressed in inactive and developing vegetative buds. (A) Predicted stem–loop structure of MiSc9 shows the sRNA29098 sequence in red. Red lines represent the sRNA reads shown in Supplementary Table S3. (B) Stem–loop pulsed RT-PCR to detect sRNA29098 transcripts in tissues of sugarcane (Saccharum spp.), sorghum (Sorghum bicolor), rice (O. sativa), brachypodium (B. disthacyon), tomato (Solanum lycopersicum), and Arabidopsis (A. thaliana). The highly conserved miR156 was used as an internal control. (C) Stem–loop pulsed RT-PCR of sRNA29098 in inactive (0 DAP) and developing (5 DAP) axillary buds of Saccharum officinarum and Saccharum spontaneum. The sugarcane 25S rRNA gene (TC148086) was used as a loading control. Reactions without RT (–RT) and without cDNA (–) were used as negative controls.
Among the sugarcane ESTs with sRNA hits, it is possible that some represent transcripts capable of yielding 21 nt tasiRNAs (Allen et al., 2005). To investigate the presence of this type of siRNA in our axillary bud libraries, we used the computational approach described by Chen et al. (2007). We identified seven potential tasiRNA-generating ESTs (Table 1), two of which represent sugarcane TAS3 precursors (Shen et al., 2009). We detected a total of nine unique bud-expressed tasiRNAs from the two TAS3 precursors, one of which (5’-UCUUGACCUUGUAAGACCCAA-3’) is homologous to Arabidopsis tasiR-ARFs (Allen et al., 2005) and possibly targets sugarcane AUXIN RESPONSE FACTOR (ARF) genes (data not shown). These results suggested that the TAS3 tasiRNA pathway was operating in sugarcane axillary buds.
Table 1.
Putative tasiRNA-generating sugarcane ESTs.
| No. | EST | Start | Strand | Total no. siRNAsa | No. non-redundant tasiRNAsb | P value | Annotation |
|---|---|---|---|---|---|---|---|
| 1 | CA151010 | 22 | 1 | 27 | 5 | 2.67E–04 | Similar to sugarcane TAS3 precursor EU327139.1c |
| 2 | TC140837 | 449 | 1 | 21 | 5 | 3.77E–04 | Similar to sugarcane TAS3 precursor EU327139.1 |
| 3 | CA195273 | 305 | –1 | 8 | 6 | 1.97E–04 | Putative NBS-LRR disease resistance protein |
| 119 | 1 | 15 | 8 | 5.52E–05 | |||
| 4 | CA261958 | 407 | 1 | 80 | 11 | 1.81E–04 | Putative transposase protein |
| 5 | DV735856 | 216 | –1 | 37 | 11 | 7.75E–04 | Hypothetical protein |
| 93 | 1 | 36 | 11 | 9.45E–04 | |||
| 6 | DV548691 | 69 | 1 | 91 | 15 | 6.84E–04 | Saccharum hybrid cultivar R570 retrotransposon |
| 7 | TC119297 | 439 | 1 | 10 | 5 | 3.77E–04 | DNA-directed RNA polymerase subunit RPABC4-like |
a Total number of 21-nt tasiRNAs that perfectly match each sugarcane ESTs.
b Only ESTs perfectly matching at least five non-redundant tasiRNAs (P <0.001) were considered.
c EST identified by Shen et al. (2009).
Identification of novel and known miRNAs expressed in sugarcane vegetative axillary buds
Conserved families of miRNAs are found in many plant species and are known to have important functions in plant development (Voinnet, 2009). Conversely, the functions of most non-conserved and species-specific miRNAs remain to be determined. We identified two new 21 nt miRNA candidates (miRcand1 and miRcand2) that are generated by distinct precursors, both supported by singleton ESTs (Table 2 and Supplementary Table S4 at JXB online). Although miRNA* accumulation provides strong supporting evidence for the cleavage by DCLs during miRNA biogenesis (Voinnet, 2009), we could not detect any sequence in our axillary bud libraries for the novel miRNA candidates. This observation was not surprising as non-conserved miRNAs and their miRNA*s are generally expressed at low levels or in specific cell types or under specific growth conditions (Rajagopalan et al., 2006).
Table 2.
Known and novel sugarcane microRNA candidates.
| Number | miRNA familya | No. transcripts (TPM) from inactive bud (0 DAP)b | No. transcripts (TPM) from active bud (2 DAP)b | Annotationc |
|---|---|---|---|---|
| Known miRNAs | ||||
| 1 | miR156 | 2.5 | 5.8 | miRbase, EST |
| 2 | miR159* | 100244.6 | 46877.7 | miRbase, EST |
| 3 | miR160* | 31.5 | 227.8 | miRbase |
| 4 | miR162* | 88.5 | 117.7 | miRbase |
| 5 | miR164* | 6.1 | 34.6 | miRbase |
| 6 | miR166* | 912.6 | 2224.2 | miRbase, EST (CN607727)d |
| 7 | miR167* | 7.8 | 11.5 | miRbase, EST |
| 8 | miR168* | 1157.1 | 3937 | miRbase, EST |
| 9 | miR169* | 6.6 | 3.7 | miRbase, EST |
| 10 | miR171* | 22.4 | 450.8 | miRbase |
| 11 | miR172 | 1.2 | 3.3 | miRbase |
| 12 | miR319* | 2584.1 | 766.8 | miRbase, EST |
| 13 | miR393 | 7.8 | 4.7 | miRbase |
| 14 | miR394 | 8.7 | 14.0 | miRbase |
| 15 | miR395 | 3.0 | 7.5 | miRbase |
| 16 | miR396 | 91.0 | 74.9 | miRbase, EST |
| 17 | miR397* | 0.0 | 6.8 | miRbase |
| 18 | miR398* | 0.0 | 5.4 | miRbase |
| 19 | miR399 | 5.5 | 1.6 | miRbase |
| 20 | miR408* | 0.8 | 9.1 | miRbase, EST |
| 21 | miR444* | 0.5 | 6.8 | miRbase, EST |
| 22 | miR444b.2* | 0.9 | 3.5 | miRbase, EST |
| 23 | miR444d.3* | 2.3 | 6.8 | miRbase, EST |
| 24 | miR529 | 18.2 | 20.6 | miRbase |
| 25 | miR827* | 27.6 | 150.4 | miRbase, EST |
| 26 | miR1878* | 39.9 | 10.3 | miRbase |
| Novel miRNA candidates | ||||
| 1 | miRcand1* | 30.6 | 1.2 | EST (CA257509)d |
| 2 | miRcand2* | 29.4 | 530.6 | EST (CA222602)d |
a microRNAs with an asterisk showed significant differential expression (P <0.01) using the Bonferroni-corrected P value of Fisher’s exact test.
b The normalized expression of transcripts per million (TPM) was obtained by the following formula: (total read count of each miRNA/total count of high-quality small RNA reads)×1 000 000. The normalized values were rounded to the nearest tenth.
c Annotation of the reads based on miRbase version17 and/or matching with EST sequences.
d Identified in this work.
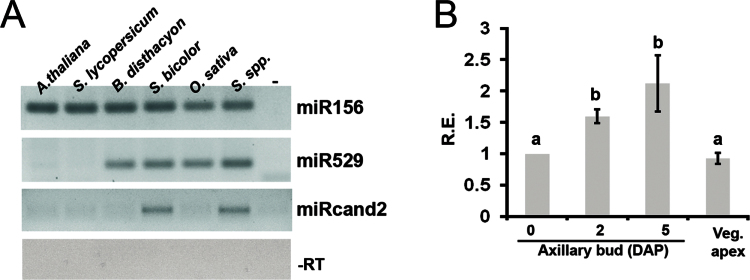
BLASTn analysis against all nucleotide sequences in the NCBI database revealed that no homologues for miRcand1 were found in other plant species, suggesting that this putative newly miRNA is sugarcane specific. In contrast, sugarcane miRcand2 seemed to have a homologue precursor in the sorghum genome. We identified a highly similar sorghum EST (CD227606; 85% nucleotide sequence identity) that was analysed further by our in-house MIRcheck-based script (Supplementary Fig. S2 at JXB online). The sorghum miRcand2 precursor was located at chromosome 5 (position 60199113–60199773). To verify experimentally whether miRcand2 is generated by other plant species, we monitored the accumulation of this 21 nt putative novel miRNA in some monocots and eudicots via stem–loop RT-PCR (Varkonyi-Gasic et al., 2007). miRcand2 was readily detectable by stem–loop RT-PCR, although only in sugarcane and sorghum tissues (Fig. 3A). The result corroborated the hypothesis that this newly microRNA candidate is restricted to the genomes of sugarcane and its closest relatives. Sugarcane miRcand2 was expressed in vegetative axillary buds as well as in the vegetative apex (apical meristem plus a few leaf primordia) of hybrid SP80-3280 at variable levels (Fig. 3B), indicating that it may have roles in regulating gene expression during sugarcane early developmental stages.
Fig. 3.
Axillary bud-expressed miRcand2 is restricted to the genome of sugarcane and its closest relative. (A) Stem–loop pulsed RT-PCR to detect miRcand2 transcripts in tissues of sugarcane (Saccharum spp.), sorghum (Sorghum bicolor), rice (O. sativa), brachypodium (B. disthacyon), tomato (Solanum lycopersicum), and Arabidopsis (A. thaliana). The highly conserved miR156 and the non-conserved miR529 were used as internal controls. Reactions without RT (–RT) and reaction without cDNA (–) were used as negative controls. (B) Detection of miRcand2 transcripts through stem–loop pulsed qRT-PCR in 0, 2, and 5 DAP developing axillary buds, as well as in the vegetative apex of sugarcane hybrid SP80-3280. All qRT-PCR experiments used inactive bud (0 DAP) as the reference sample (set to 1.0). Error bars indicate standard deviation of three biological replicates. Distinct letters indicate significant differences at P <0.05 (Student’s t-test). R.E., relative expression.
To gain more insights into the functions of these two novel miRNAs in sugarcane, we computationally predicted possible targets among the bulk of sugarcane EST sequences (see Materials and methods). As a result, predicted target genes were identified only for miRcand2, which were represented by ESTs encoding pentatricopeptide repeat (PPR)-like proteins (Supplementary Table S5 at JXB online). None the less, our 5’ rapid amplification of cDNA ends (5’RACE) experiments for these ESTs were inconclusive (data not shown). Therefore, at this point, miRcan2 cannot be confirmed as a functional novel miRNA targeting PPR genes in sugarcane. To identify known miRNAs in our bud libraries, we employed a homology-based approach using BLASTn comparison, miRBase (version 17.0) as a reference set, and sugarcane miRNA precursors (Zanca et al., 2010). We found several miRNA candidate sequences that exhibited perfect or near-perfect matches with at least 26 families of known miRNAs (Table 2 and Supplementary Table S4). Candidate miRNA reads were usually 21 nt and several could be mapped to sugarcane miRNA precursors (Supplementary Table S4). In addition to the sugarcane miRNA precursors already deposited in miRBase, we identified a novel miR166 precursor that was validated by our in-house MIRcheck-based script and matched several miR166 candidate reads (Supplementary Fig. S2 and Table S4). Moreover, 16 known miRNAs identified in the developing axillary buds (Table 2) were also expressed in leaf tissues of sugarcane plants exposed to drought stress (Ferreira et al., 2012).
Interestingly, we identified highly represented miRNAs in our libraries with variants in the 5’ or 3’ terminus, as well as internal mismatches. For instance, the miR159 family had 56 variants in both libraries (Supplementary Table S4). All polymorphic sequences corresponding to the mature miR159 were precisely excised at the same specific location of the sugarcane MIR159 precursors (Supplementary Fig. S3 at JXB online). Although we cannot rule out the presence of sequencing errors, the high number of polymorphic sequences is probably a reflection of the high level of SsMIR159 gene expression in axillary buds combined with the elevated number of alleles generally observed for most sugarcane loci (Cordeiro et al., 2000). Similar results have been observed for miRNAs from allotetraploid cottons (Pang et al., 2009). Whether or not these variants have any biological impact on target regulation in polyploid species remains to be seen.
Expression profiles of known miRNAs and targets during axillary bud outgrowth
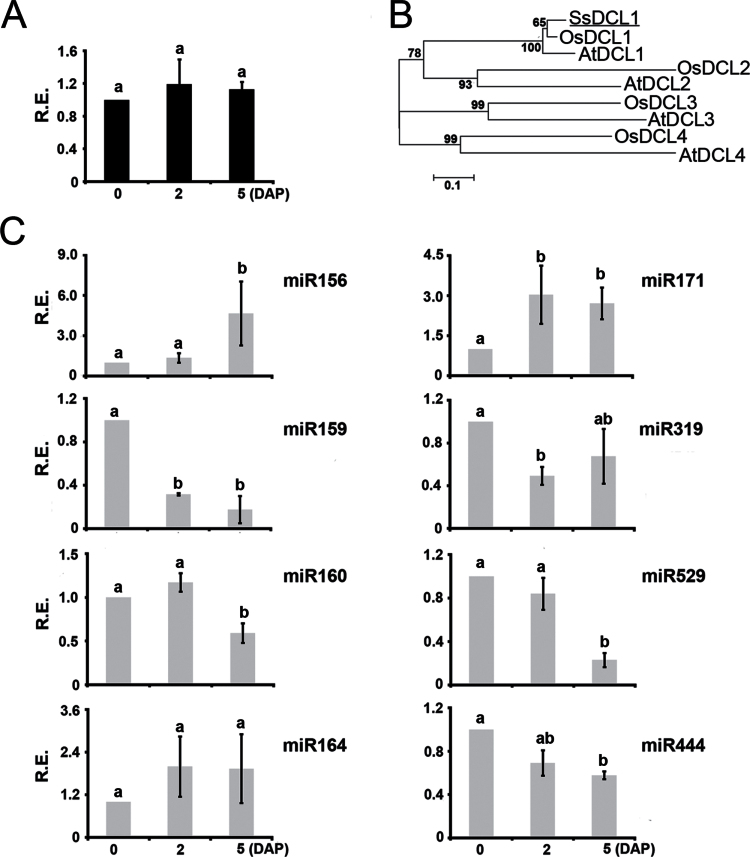
In general, sRNAs associated with TEs and protein-encoding genes were similarly represented in both inactive and developing bud libraries (data not shown). Conversely, most miRNAs (20/28) were differentially expressed (P <0.01) during bud outgrowth (Table 2). Post-transcriptional mechanisms that operate at the biogenesis level could modulate the spatiotemporal generation of miRNAs, perhaps through differential expression of the necessary biogenesis factors (Nogueira et al., 2009). This seems not to be the case for bud-associated miRNAs, as we detected similar levels of SsDCL1 (Saccharum spp. DCL1) transcripts in inactive and developing buds (Fig. 4A, B). Although we cannot exclude the possibility that other miRNA processing components are differentially expressed in developing buds, it is likely that the differential accumulation of miRNAs in axillary buds is mainly regulated at the transcriptional level.
Fig. 4.
DCL1 and known miRNAs are expressed during axillary bud outgrowth. (A) Detection of SsDCL1 transcripts through qRT-PCR in 0, 2, and 5 DAP developing axillary buds of sugarcane hybrid SP80-3280. (B) Inferred phylogenetic relationships among SsDCL1 orthologues in rice (Os) and Arabidopsis (At). The p-distances were calculated from amino acid alignments of conserved blocks, and tree topology was inferred with the neighbour-joining method. Only bootstrap values higher than 50% are shown for 1000 replicates. SsDCL1 is underlined. (C) Accumulation of selected mature miRNAs was quantified through stem–loop pulsed qRT-PCR in 0, 2, and 5 DAP axillary buds of sugarcane hybrid SP80-3280. All qRT-PCR experiments used inactive bud (0 DAP) as the reference sample (set to 1.0). Error bars indicate standard deviation of three biological replicates. Distinct letters indicate significant differences at P <0.05 (Student’s t-test). R.E., relative expression.
To complement the sRNA sequencing analysis and assemble a comprehensive repository of information on miRNAs associated with axillary bud outgrowth, we employed stem–loop real-time qRT-PCR using total RNA from new biological samples of sugarcane axillary buds. Stem–loop qRT-PCR is a standard technique to accurately quantify miRNAs (Varkonyi-Gasic et al., 2007). Initially, we compared the expression profiles of four sugarcane genes commonly used as reference genes in qRT-PCR experiments (Iskandar et al., 2004). Based on the qPCR results for three biological replicates, the 25S rRBA gene (TC148086) was the most stable gene (Supplementary Fig. S4). Therefore, it was selected as the reference gene for all qRT-PCR experiments in this study. To verify the expression profiles of miRNAs in inactive and developing buds (2 and 5 DAP buds), we selected six broadly conserved and two non-conserved miRNAs for further analysis. All were readily detected by stem–loop qRT-PCR (Fig. 4C), which indicated that these miRNAs are authentic sRNAs. Whilst miR159, miR319, and miR444 transcripts accumulated at higher levels in inactive buds, miR156 and miR171 were expressed at higher levels in outgrown buds. miR529 and miR160 transcripts accumulated at low levels specifically in 5 DAP developing buds (Fig. 4C). Some discrepancies between the frequency of miRNAs represented in the libraries and the stem–loop qRT-PCR data (Table 2 and Fig. 4C) might have resulted from insufficient depth of sequencing coverage or cloning bias.
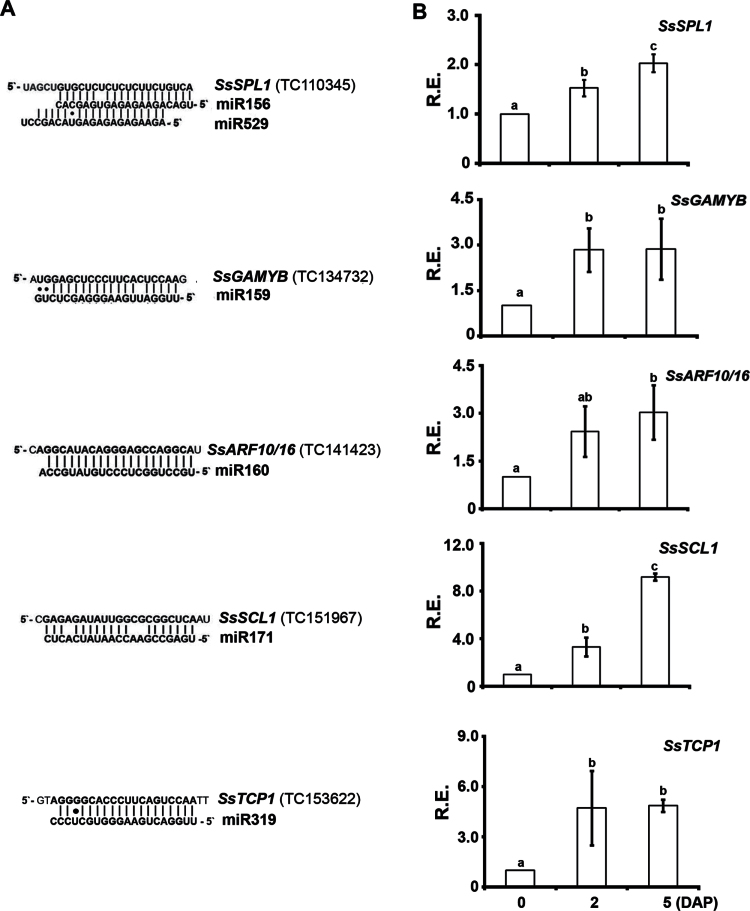
In this study, we computationally predicted more than 30 targets for sugarcane miRNAs. All sugarcane cDNAs predicted to be targets of known miRNAs were homologues of miRNA target genes in Arabidopsis and other plant species and may be associated with various developmental processes (Supplementary Table S5). We monitored the expression of five homologous miRNA targets by qRT-PCR: Saccharum spp. SQUAMOSA PROMOTER BINDING-LIKE PROTEIN1 (SsSPL1), SsGAMYB, SsTCP1, AUXIN RESPONSE FACTOR10/16 (SsARF10/16), and SCARECROW-LIKE1 (SsSCL1) (Fig. 5A). Interestingly, all the genes were upregulated at variable levels in developing buds when compared with inactive buds (Fig. 5B). Among them, SsGAMYB displayed an inverse relationship with miR159 expression at all developmental stages (Fig. 4C and Fig. 5B). In Arabidopsis, the levels of miR159 and GAMYB transcripts are oppositely regulated by the phytormone ABA (Reyes and Chua, 2007). To examine whether there was a correlation between ABA levels and miR159/SsGAMYB expression profiles, we quantified ABA and catabolites in sugarcane axillary buds. ABA levels were reduced from the inactive buds to 2 and 5 DAP buds (Supplementary Fig. S5). The sharp increase in the levels of dihydrophaseic acid suggested that bioactive ABA was initially biosynthesized in inactive buds and further catabolized in developing buds mainly through the 8’-hydroxylation pathway (which results in phaseic acid that is further reduced to dihydrophaseic acid). These data suggested that ABA content within axillary buds correlates positively with miR159 accumulation, whilst the opposite was observed for SsGAMYB transcript levels.
Fig. 5.
Expression profiles of selected miRNA targets during axillary bud outgrowth. (A) miRNA-complementary sites in the target mRNA and the miRNA. Watson–Crick pairing (vertical dashes) is indicated. (B) Detection of SsSPL1, SsGAMYB, SsTCP1, SsARF10/16, and SsSCL1 transcripts was performed by qRT-PCR in 0, 2, and 5 DAP axillary buds of sugarcane hybrid SP80-3280. All qRT-PCR experiments used inactive bud (0 DAP) as the reference sample (set to 1.0). Error bars indicate standard deviation of three biological replicates. Distinct letters indicate significant differences at P <0.05 (Student’s t-test). R.E., relative expression.
Expression patterns of miRNAs in sugarcane axillary buds
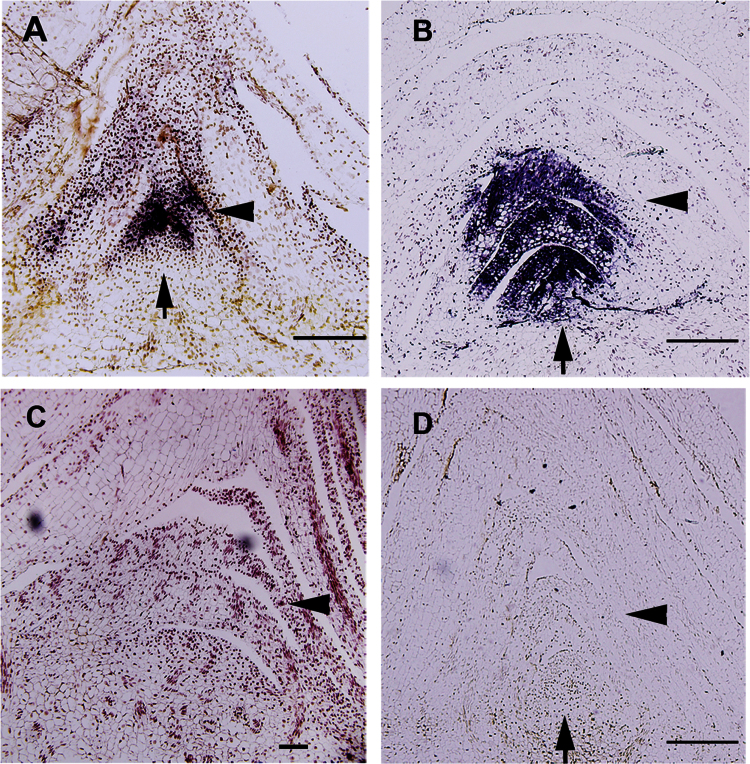
To gain further insights into the possible roles of sRNAs during sugarcane axillary bud development, we investigated the spatial and temporal patterns of miRNA accumulation by in situ hybridization. We selected two microRNAs for our analysis: a highly represented miRNA (miR159) and a low-level detected microRNA (miR156) based on our axillary bud sRNA libraries (Table 2). Consistently with its abundance in the sRNA libraries, miR159 display high expression levels in axillary buds (Fig. 6A, B). miR156 was broadly expressed in axillary meristems and leaf primordia at low levels (Fig. 6C), whilst miR159 showed high expression levels in vascular bundles of young leaf primordia (Fig. 6B). The expression pattern of miR159 was similar in inactive buds (Fig. 6A) and 5 DAP developing buds (Fig. 6B), suggesting that its transcript levels rather than spatiotemporal patterning were developmentally regulated during axillary bud outgrowth.
Fig. 6.
Spatiotemporal expression patterns of miRNAs in axillary buds of sugarcane. (A, B) A probe of a 3’-labelled LNA-modified oligonucleotide detecting miR159 was hybridized with longitudinal sections of 0 DAP (A) and 5 DAP (B) vegetative buds. (C) A similar probe detecting miR156 was hybridized with longitudinal sections of 0 DAP buds. (D) A scramble-miRNA 3’-labelled LNA probe was used as a negative control. Purple and brown staining show probe localization. Arrows represents meristem positions and arrowheads indicates young leaf primordia. Bars, 10 μm.
Discussion
Axillary bud outgrowth is crucial to control plant shoot architecture, which is important for biomass production of biofuel crops such as sugarcane. Although it is well known that bud outgrowth is controlled mainly by conserved networks that comprise a complex set of transcription factors (Leyser, 2009), the presence of additional layers of gene regulation embedded in these networks remains poorly understood. Therefore, the identification of sRNAs and elucidation of their functions in vegetative bud development will help us to understand better the genetic networks underlying plant shoot architecture.
To identify novel genetic factors that potentially play roles in vegetative bud outgrowth, we employed a deep-sequencing approach coupled with conventional RNA detection methods to profile sRNAs in inactive and developing axillary buds of the commercial sugarcane hybrid SP80-3280. We have provided the first draft of the sRNA transcriptome in sugarcane vegetative axillary buds, which reinforces the idea that the sRNA component is complex and diverse within the plant kingdom. Although high-throughput sRNA profiling is generally straightforward for model plants (Zhu et al., 2008), it is rather challenging for plant species such as the polyploid sugarcane for which the genome is highly complex and largely unknown. However, even using limited publicly available genomic and EST sources, we were able to identify known and novel miRNAs and show that several siRNAs expressed in buds match distinct TEs and that some are phased tasiRNAs originating from specific transcripts.
Diverse populations of sRNAs are expressed in inactive and developing axillary buds
In general, we found similar sRNA abundance when comparing inactive and growing axillary buds, suggesting that inactive buds are also actively generating sRNAs to control gene expression. This data is consistent with the idea that quiescent cells in inactive/dormant buds are metabolically and molecularly active (Stafstrom et al., 1998). For instance, several bud-expressed rasiRNAs matched intergenic and intron-embedded TEs, indicating that these TEs are either active or under RNA-dependent epigenetic regulation. Such epigenetic regulation may be crucial for the developmental programmes associated with outgrowth of vegetative axillary buds, similarly to chromatin modifications that take place during bud dormancy release in some woody perennial plants (Leida et al., 2012).
MITEs, a type of non-autonomous TE, may assert gene regulation via sRNA pathways (Kuang et al., 2009). Our identification of MiSc sRNAs indicated that MITEs could serve as both origins of sRNAs and targets for sRNA-mediated gene silencing during axillary bud outgrowth of modern sugarcane hybrids and ancient progenitor species. For instance, sugarcane genes bearing MITE insertions may be co-opted for using sRNA-based regulation, similar to regulation imposed by SINE-like direct repeats in the FWA promoter (Chan et al., 2004). Additionally, some MiSc rasiRNAs expressed in axillary buds are probably generated and may act similarly to miRNAs (Fig. 2), as has been shown recently for two rice MITE-derived sRNAs that positively regulate ABA signalling by post-transcriptionally regulating MAIF1 expression (Yan et al., 2011). All these mechanisms are expected to contribute to the complexity of epigenetic regulation during axillary bud outgrowth.
Four families comprising eight tasiRNA loci have been described in Arabidopsis, whereas hundreds of loci of unknown function generate phased sRNAs in grasses (Johnson et al., 2009). In sugarcane axillary buds, we identified tasiRNAs that seemed to be generated by a small number of transcripts (Table 1), including TAS3 and nucleotide-binding site–leucine-rich repeat (NBS-LRR) disease resistance protein-coding transcripts. Secondary tasiRNAs generated from these types of transcript are known to regulate gene expression in trans in several plant species (Allen et al., 2005; Zhai et al., 2011). Interestingly, we detected sequences similar to 21 nt miR390 (sRNA that triggers tasi-ARF production; Allen et al., 2005) and 22 nt miR2118 (sRNA that triggers the production of NBS-LRR tasiRNAs; Zhai et al., 2011). However, as there were fewer than 15 reads representing these miRNAs in our sRNA libraries, they were not included in further analyses (see Materials and methods). The tasi-ARF pathway has been shown to play a role in leaf development (Nogueira et al., 2007), whilst NBS-LRR tasiRNAs are involved in pathogen defence pathways in leguminous plants (Zhai et al., 2011). Our data suggest that sugarcane TAS3 and NBS-LRR gene-derived tasiRNAs are also part of the intricate regulatory networks operating during vegetative bud development. It will be interesting to test whether siRNAs from the additional transcripts (Table 1) act as trans-acting or cis-acting siRNAs in sugarcane.
Dynamic expression of miRNAs and targets during axillary vegetative bud outgrowth
Comparison of expression profiles between miRNAs and the corresponding target genes can reveal positive or negative correlations, which expand our understanding of how miRNAs are involved in sugarcane vegetative bud development. Specific microRNAs displayed dynamic expression profiles in inactive and developing vegetative buds (Fig. 4C), suggesting that they cooperate with their targets to achieve global gene-expression networks that function within axillary buds. Interestingly, the expression profiles of some miRNAs were positively correlated with their targets. For instance, levels of transcripts of miR171 and one of its potential targets in sugarcane, SsSCL1, increased as the bud grew out (Figs 4C and 5B). This may due to the consequence of feedback regulation (Baulcombe, 2004) or to the fact that this miRNA may function by translational repression (Lanet et al., 2009).
SsSPL1 is a member of the SQUAMOSA PROMOTER BINDING-LIKE PROTEIN (SPL) family of transcription factors, of which some members are associated with shoot development and plant architecture (Schwarz et al., 2008; Jiao et al., 2010). In monocots, some SPL mRNAs are targeted by both miR156 and miR529 (Fig. 5A; Chuck et al., 2010). Based on our qRT-PCR data, levels of miR529 and SsSPL1 transcripts were inversely correlated specifically in 5 DAP developing buds, consistent with SsSPL1 being negatively regulated by this miRNA. Interestingly, the expression profile of miR156 was positively correlated with that of SsSPL1 at all developmental stages (Figs 4C and 5B). Moreover, miR156 was less represented in both libraries when compared with miR529 (Table 2). It is possible that SsSPL1 expression in axillary buds is independent of both miRNAs in the early stages of development. However, at the late stages of bud outgrowth (5 DAP), SsSPL1 transcript levels may be regulated predominantly by miR529.
miRNA159 was the most enriched miRNA in our axillary bud libraries, followed by miR319 (Table 2). The specificity of these closely related miRNAs is achieved in part by their distinct expression, with miR319 expressed at much lower levels than miR159 in Arabidopsis tissues (Palatnik et al., 2007). Consistently, miR319 was underrepresented in our sRNAs libraries when compared with miR159 (Table 2). Interestingly, miRNA159 and miR319 accumulated at high levels in inactive buds (Table 2), whilst their targets (SsGAMYB and SsTCP1, respectively) were upregulated in developing buds (Figs 4C and 5).
In situ localization of miR159 in axillary buds revealed that it accumulated mainly in young leaf primordia (Fig. 6A, B) and could potentially regulate its targets in these tissues. miR159-regulated GAMYB-like genes encode R2R3 MYB domain transcription factors that are implicated in ABA signalling in germinating seeds (Reyes and Chua, 2007). Interestingly, the ABA content of vegetative axillary buds is closely correlated with bud dormancy/inactivity (Shimizu-Sato and Mori, 2001). Although speculative, we propose a scenario in which miR159 levels in sugarcane axillary buds are positively regulated by ABA, as has been shown in germinating seeds of Arabidopsis (Reyes and Chua, 2007). The differential accumulation of ABA in inactive and outgrown buds (Supplementary Fig. S5) may converge into a fine-tuned regulation of miR159 expression, which, in turn, would act as a molecular switch during the inactive stage of bud development, permitting higher expression of SsGAMYB only in growing axillary buds. Further functional studies are needed to clarify the roles of miR159-regulated pathways associated with axillary bud development.
In summary, we have presented a comprehensive survey of sRNAs in vegetative axillary buds of the tropical biofuel crop sugarcane. Collectively, the dynamic changes in miRNA accumulation and their targets probably promote signalling and metabolic pathways that are essential for bud outgrowth. Future research on the miR159 family and target regulation may increase our understanding of the biological role of this miRNA and its contribution to the early emergence of vegetative buds. Our work has opened a new avenue for functional studies on sRNA-mediated gene regulation in vegetative axillary bud outgrowth and consequently plant vegetative architecture.
Supplementary data
Supplementary material is available at JXB online.
Supplementary Fig. S1. sRNA distribution between axillary bud libraries and sRNA counts over annotated sugarcane genomic regions.
Supplementary Fig. S2. Predicted miRNA harpins identified in this study using EST sequences and the mFOLD program.
Supplementary Fig. S3. Multiple sequence alignments of sugarcane miR159 mature sequences.
Supplementary Fig. S4. Expression profiles of sugarcane reference genes evaluated through qRT-PCR in 0, 2, and 5 DAP developing axillary buds of sugarcane hybrid SP80-3280.
Supplementary Fig. S5. Concentrations of ABA and its catabolite dihydrophaseic acid (DPA) were monitored in 0, 2, and 5 DAP axillary buds and expressed as ng g–1 dry weight (ng/g DW).
Supplementary Table S1. Primers used in this study.
Supplementary Table S2. Statistics of redundant sRNA sequences from the individual libraries.
Supplementary Table S3. Sugarcane EST sequences similar to MITEs and their associated sRNA sequences.
Supplementary Table S4. Total reads of known and novel sugarcane miRNA candidates identified in the axillary bud libraries.
Supplementary Table S5. Predicted target sequences of known and novel sugarcane miRNAs identified in the axillary bud libraries.
Acknowledgements
We thank Dr Enio Oliveira (ESALQ/USP) for providing sugarcane plant material and Ana Bovolato for technical assistance. This work was supported by the State of Sao Paulo Research Foundation, FAPESP (grant no. 07/58289-5) and partially by the National Council for Scientific and Technological Development, CNPq (grant no. 474635/2008-2).
Glossary
Abbreviations:
- ABA
abscisic acid
- BAC
bacterial artificial chromosome
- DAP
days after planting
- EST
expressed sequence tag
- LNA
locked nucleic acid
- miRNA
microRNA
- MITE
miniature inverted-repeat transposable element
- NBS-LRR
nucleotide-binding site–leucine-rich repeat
- PPR
pentatricopeptide repeat
- qRT-PCR
quantitative reverse transcription-PCR
- sRNA
small RNA
- siRNA
small interfering RNA
- rasiRNA
repeat-associated siRNA
- tasiRNA
trans-acting siRNA
- TE
transposable element.
References
- Allen E, Xie ZX, Gustafson AM, Carrington JC. 2005. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221. [DOI] [PubMed] [Google Scholar]
- Baulcombe D. 2004. RNA silencing in plants. Nature 431, 356–363. [DOI] [PubMed] [Google Scholar]
- Blevins T, Rajeswaran R, Aregger M, Borah BK, Schepetilnikov M, Baerlocher L, Farinelli L, Meins F, Hohn T, Pooggin MM. 2011. Massive production of small RNAs from a non-coding region of Cauliflower mosaic virus in plant defense and viral counter-defense. Nucleic Acids Research 39, 5003–5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvino M, Bruggmann R, Messing J. 2011. Characterization of the small RNA component of the transcriptome from grain and sweet sorghum stems. BMC Genomics 12, 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, Lowe TM. 2009. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Research 37, D93–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SWL, Zilberman D, Xie ZX, Johansen LK, Carrington JC, Jacobsen SE. 2004. RNA silencing genes control de novo DNA methylation. Science 303, 1336–1336. [DOI] [PubMed] [Google Scholar]
- Chen HM, Li YH, Wu SH. 2007. Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis . Proceedings of the National Academy of Sciences, USA 104, 3318–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZY, Zhang JJ, Kong J, Li SQ, Fu Y, Li SB, Zhang H, Li YS, Zhu YG. 2006. Diversity of endogenous small non-coding RNAs in Oryza sativa . Genetica 128, 21–31. [DOI] [PubMed] [Google Scholar]
- Chiwocha SDS, Abrams SR, Ambrose SJ, Cutler AJ, Loewen M, Ross ARS, Kermode AR. 2003. A method for profiling classes of plant hormones and their metabolites using liquid chromatography-electrospray ionization tandem mass spectrometry: an analysis of hormone regulation of thermodormancy of lettuce (Lactuca sativa L.) seeds. The Plant Journal 35, 405–417. [DOI] [PubMed] [Google Scholar]
- Chuck G, Cigan A, Saeteurn K, Hake S. 2007. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nature Genetics 544–549. [DOI] [PubMed] [Google Scholar]
- Chuck G, Whipple C, Jackson D, Hake S. 2010. The maize SBP-box transcription factor encoded by tasselsheath4 regulates bract development and the establishment of meristem boundaries. Development 137, 1243. [DOI] [PubMed] [Google Scholar]
- Cordeiro GM, Taylor GO, Henry RJ. 2000. Characterisation of microsatellite markers from sugarcane (Saccharum sp.), a highly polyploid species. Plant Science 155, 161–168. [DOI] [PubMed] [Google Scholar]
- Dai XB, Zhao PX. 2011. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Research 39, W155–W159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon SL, Shapter FM, Henry RJ, Cordeiro G, Izquierdo L, Lee LS. 2007. Domestication to crop improvement: genetic resources for Sorghum and Saccharum (Andropogoneae). Annals of Botany 100, 975–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingues DS, Cruz GMQ, Metcalfe CJ, Nogueira FTS, Vicentini R, Alves CD, Van Sluys MA. 2012. Analysis of plant LTR-retrotransposons at the fine-scale family level reveals individual molecular patterns. BMC Genomics 13, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doust A. 2007. Grass architecture: genetic and environmental control of branching. Current Opinion in Plant Biology 21–25. [DOI] [PubMed] [Google Scholar]
- Ferreira TH, Gentile A, Vilela RD, Costa GGL, Dias LI, Endres L, Menossi M. 2012. MicroRNAs associated with drought response in the bioenergy crop sugarcane (Saccharum spp.). PloS One 7, e46703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner PP, Daub J, Tate J, et al. 2011. Rfam: Wikipedia, clans and the “decimal” release. Nucleic Acids Research 39, D141––D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsmeur O, Charron C, Bocs S, et al. 2011. High homologous gene conservation despite extreme autopolyploid redundancy in sugarcane. New Phytologist 189, 629–642. [DOI] [PubMed] [Google Scholar]
- Iskandar HM, Simpson RS, Casu RE, Bonnett GD, Maclean DJ, Manners JM. 2004. Comparison of reference genes for quantitative real-time polymerase chain reaction analysis of gene expression. Plant Molecular Biology Reporter 22, 325–337. [Google Scholar]
- Jannoo N, Grivet L, Chantret N, Garsmeur O, Glaszmann JC, Arruda P, D’Hont A. 2007. Orthologous comparison in a gene-rich region among grasses reveals stability in the sugarcane polyploid genome. The Plant Journal 50, 574–585. [DOI] [PubMed] [Google Scholar]
- Javelle M, Timmermans MCP. 2012. In situ localization of small RNAs in plants by using LNA probes. Nature Protocols 7, 533–541. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X. 2010. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nature Genetics. [DOI] [PubMed] [Google Scholar]
- Johnson C, Kasprzewska A, Tennessen K, Fernandes J, Nan GL, Walbot V, Sundaresan V, Vance V, Bowman LH. 2009. Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Research 22, 592–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khraiwesh B, Arif MA, Seumel GI, Ossowski S, Weigel D, Reski R, Frank W. 2010. Transcriptional control of gene expression by microRNAs. Cell 140, 111–122. [DOI] [PubMed] [Google Scholar]
- Koyama T, Furutani M, Tasaka M, Ohme-Takagi M. 2007. TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis . Plant Cell 473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang HH, Padmanabhan C, Li F, Kamei A, Bhaskar PB, Shu OY, Jiang JM, Buell CR, Baker B. 2009. Identification of miniature inverted-repeat transposable elements (MITEs) and biogenesis of their siRNAs in the Solanaceae: new functional implications for MITEs. Genome Research 19, 42–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanet E, Delannoy E, Sormani R, Floris M, Brodersen P, Crete P, Voinnet O, Robaglia C. 2009. Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell 21, 1762–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leida C, Conesa A, Llacer G, Badenes ML, Rios G. 2012. Histone modifications and expression of DAM6 gene in peach are modulated during bud dormancy release in a cultivar-dependent manner. New Phytologist 193, 67–80. [DOI] [PubMed] [Google Scholar]
- Leyser O. 2009. The control of shoot branching: an example of plant information processing. Plant Cell and Environment 694–703. [DOI] [PubMed] [Google Scholar]
- Li Y, Li CQ, Xia J, Jin YX. 2011. Domestication of transposable elements into microRNA genes in plants. Plos One 6, e19212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K, Schmittgen T. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Nogueira FTS, Chitwood DH, Madi S, Ohtsu K, Schnable PS, Scanlon MJ, Timmermans MCP. 2009. Regulation of small RNA accumulation in the maize shoot apex. Plos Genetics 5, e1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira FTS, Madi S, Chitwood DH, Juarez MT, Timmermans MCP. 2007. Two small regulatory RNAs establish opposing fates of a developmental axis. Genes & Development 21, 750–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongaro V, Bainbridge K, Williamson L, Leyser O. 2008. Interactions between axillary branches of Arabidopsis . Molecular Plant 1, 388–400. [DOI] [PubMed] [Google Scholar]
- Ouyang S, Buell CR. 2004. The TIGR Plant Repeat Databases: a collective resource for the identification of repetitive sequences in plants. Nucleic Acids Research 32, D360––D363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik JF, Wollmann H, Schommer C, et al. 2007. Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Developmental Cell 13, 115–125. [DOI] [PubMed] [Google Scholar]
- Pang MX, Woodward AW, Agarwal V, Guan XY, Ha M, Ramachandran V, Chen XM, Triplett BA, Stelly DM, Chen ZJ. 2009. Genome-wide analysis reveals rapid and dynamic changes in miRNA and siRNA sequence and expression during ovule and fiber development in allotetraploid cotton (Gossypium hirsutum L.). Genome Biology 10, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piriyapongsa J, Jordan IK. 2008. Dual coding of siRNAs and miRNAs by plant transposable elements. RNA 14, 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. 2006. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana . Genes & Development 20, 3407–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Kuhlemeier C. 2002. Plant architecture. EMBO Reports 3, 846–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JL, Chua NH. 2007. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. The Plant Journal 49, 592–606. [DOI] [PubMed] [Google Scholar]
- Schmitz G, Theres K. 2005. Shoot and inflorescence branching. Current Opinion in Plant Biology 8, 506–511. [DOI] [PubMed] [Google Scholar]
- Schwarz S, Grande A, Bujdoso N, Saedler H, Huijser P. 2008. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis . Plant Molecular Biology 67, 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D, Wang S, Chen H, Zhu QH, Helliwell C, Fan LJ. 2009. Molecular phylogeny of miR390-guided trans-acting siRNA genes (TAS3) in the grass family. Plant Systematics and Evolution 283, 125–132. [Google Scholar]
- Shimizu-Sato S, Mori H. 2001. Control of outgrowth and dormancy in axillary buds. Plant Physiology 127, 1405–1413. [PMC free article] [PubMed] [Google Scholar]
- Stafstrom JP, Ripley BD, Devitt ML, Drake B. 1998. Dormancy-associated gene expression in pea axillary buds. Planta 205, 547–552. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24, 1596–1599. [DOI] [PubMed] [Google Scholar]
- Thiebaut F, Grativol C, Carnavale-Bottino M, Rojas CA, Tanurdzic M, Farinelli L, Martienssen RA, Hemerly AS, Ferreira PCG. 2012. Computational identification and analysis of novel sugarcane microRNAs. BMC Genomics 13, 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Wu R, Wood M, Walton E, Hellens R. 2007. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H. 2006. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes & Development 759–771. [DOI] [PubMed] [Google Scholar]
- Vazquez F. 2006. Arabidopsis endogenous small RNAs: highways and byways. Trends in Plant Science 11, 460–468. [DOI] [PubMed] [Google Scholar]
- Voinnet O. 2009. Origin, biogenesis, and activity of plant microRNAs. Cell 136, 669–687. [DOI] [PubMed] [Google Scholar]
- Wang L, Mai YX, Zhang YC, Luo QA, Yang HQ. 2010. MicroRNA171c-targeted SCL6-II, SCL6-III, and SCL6-IV genes regulate shoot branching in Arabidopsis . Molecular Plant 3, 794–806. [DOI] [PubMed] [Google Scholar]
- Wang XF, Elling AA, Li XY, Li N, Peng ZY, He GM, Sun H, Qi YJ, Liu XS, Deng XW. 2009. Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 21, 1053–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH, Li JY. 2008. Molecular basis of plant architecture. Annual Review of Plant Biology 59, 253–279. [DOI] [PubMed] [Google Scholar]
- Wei B, Cai T, Zhang RZ, et al. 2009. Novel microRNAs uncovered by deep sequencing of small RNA transcriptomes in bread wheat (Triticum aestivum L.) and Brachypodium distachyon (L.) Beauv. Functional & Integrative Genomics 9, 499–511. [DOI] [PubMed] [Google Scholar]
- Yan YS, Zhang YM, Yang K, Sun ZX, Fu YP, Chen XY, Fang RX. 2011. Small RNAs from MITE-derived stem–loop precursors regulate abscisic acid signaling and abiotic stress responses in rice. The Plant Journal 65, 820–828. [DOI] [PubMed] [Google Scholar]
- Zanca AS, Vicentini R, Ortiz-Morea FA, Del Bem LEV, da Silva MJ, Vincentz M, Nogueira FTS. 2010. Identification and expression analysis of microRNAs and targets in the biofuel crop sugarcane. BMC Plant Biology 10, 260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai JX, Jeong DH, De Paoli E, et al. 2011. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes & Development 25, 2540–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Spriggs A, Matthew L, Fan L, Kennedy G, Gubler F, Helliwell C. 2008. A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Research 18, 1456–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.