Abstract
SKD1 (suppressor of K+ transport growth defect 1) is an AAA-type ATPase that functions as a molecular motor. It was previously shown that SKD1 accumulates in epidermal bladder cells of the halophyte Mesembryanthemum crystallinum. SKD1 knock-down Arabidopsis mutants showed an imbalanced Na+/K+ ratio under salt stress. Two enzymes involved in protein post-translational modifications that physically interacted with McSKD1 were identified. McCPN1 (copine 1), a RING-type ubiquitin ligase, has an N-terminal myristoylation site that links to the plasma membrane, a central copine domain that interacts with McSKD1, and a C-terminal RING domain that catalyses protein ubiquitination. In vitro ubiquitination assay demonstrated that McCPN1 was capable of mediating ubiquitination of McSKD1. McSnRK1 (sucrose non-fermenting 1-related protein kinase) is a Ser/Thr protein kinase that contains an N-terminal STKc catalytic domain to phosphorylate McSKD1, and C-terminal UBA and KA1 domains to interact with McSKD1. The transcript and protein levels of McSnRK1 increased as NaCl concentrations increased. The formation of an SKD1–SnRK1–CPN1 ternary complex was demonstrated by yeast three-hybrid and bimolecular fluorescence complementation. It was found that McSKD1 preferentially interacts with McSnRK1 in the cytosol, and salt induced the re-distribution of McSKD1 and McSnRK1 towards the plasma membrane via the microtubule cytoskeleton and subsequently interacted with RING-type E3 McCPN1. The potential effects of ubiquitination and phosphorylation on McSKD1, such as changes in the ATPase activity and cellular localization, and how they relate to the functions of SKD1 in the maintenance of Na+/K+ homeostasis under salt stress, are discussed.
Key words: AAA-type ATPase, protein kinase, protein ubiquitination, RING-type ubiquitin ligase, salt stress, SKD1, SnRK1.
Introduction
The halophyte ice plant Mesembryanthemum crystallinum L. has unique features for tolerating high salinity environments. A critical feature of the ice plant is its ability to sequester Na+ in the enlarged vacuoles of epidermal bladder cells (EBCs), enabling the avoidance of sodium toxicity (Adams et al., 1998). EBCs are prominent cells that cover the surface of the above-ground portion of ice plants, and EBC-less mutants show reduced growth and reproduction under 400mM NaCl (Agarie et al., 2007). EBCs are not merely a water and salt reservoir in the ice plant but they are also metabolically active and function in maintaining ion homeostasis (Jou et al., 2007). Previously, a salt-induced gene SKD1 (suppressor of K+ transport growth defect; also known as vacuolar protein sorting 4, VPS4), was found to be expressed at high levels in EBCs. Ice plant SKD1 (McSKD1) can complement the K+ uptake defect phenotype of yeast K+ uptake mutants, suggesting that SKD1 is involved in facilitating K+ transport (Jou et al., 2004). Glycophytes display a typical K+-deficient symptom and a high Na+/K+ ratio in high salt conditions because excessive external Na+ competes with the uptake of the essential nutrient potassium (Watad et al., 1983). Knock-down SKD1 mutants were found to show abnormal root morphology and an imbalanced Na+/K+ ratio under salt stress (Ho et al., 2010), indicating that SKD1 is involved in the salt-tolerant mechanism in higher plants.
SKD1 proteins have a variable N-terminal region containing a microtubule-interacting and trafficking (MIT) domain, followed by a highly conserved AAA (ATPase associated with various cellular activities)–ATPase cassette and a C-terminal oligomerization domain (Babst et al., 1998). The ATP hydrolysis activity of SKD1 has been demonstrated in yeast (Babst et al., 1998), ice plant (Jou et al., 2006), and Arabidopsis (Haas et al., 2007; Shahriari et al., 2010). The AAA-type ATPases are molecular motors, usually assembled into ring-shaped homo-oligomers, participating in energy-acquiring processes such as disassembly of protein complexes (Hanson and Whiteheart, 2005). The best studied function of SKD1/VPS4 is in participation in protein sorting in multivescular bodies (MVBs), a versatile organelle where exocytic and endocytic pathways meet (Babst et al., 2002). Membrane-bound proteins targeted for vacuolar degradation are first conjugated to one ubiquitin molecule (Katzmann et al., 2001), sorted in MVBs, and subsequently delivered to vacuoles. The sorting complex on the MVB membrane consists of four ESCRT (endosomal sorting complex required for transport) complexes, ESCRT-0, -I, -II, and -III (reviewed in Hurley and Hanson, 2010). The N-terminal MIT domain of SKD1 interacts with ESCRT-III components VPS2, 20, and 24, and catalyses the dissociation of ESCRT complexes after cargo proteins are sorted (Obita et al., 2007; Shim et al., 2007; Stuchell-Brereton et al., 2007). The C-terminal regions of SKD1 interact with each other to form homo-oligomers with the help of Vta1/LIP5 and other proteins to stabilize the structure and stimulate ATPase activity (Azmi et al., 2006). In the model proposed by Shim et al. (2008) after sequential assembly of ESCRT-III components, SKD1/VPS4 and Vta1/LIP5 form an organized intertwined complex which leads to completion of the sorting process on the MVB membrane.
Many proteins are sorted by the ESCRT machinery on the MVB membrane, including proteins targeted to lysosomes/vacuoles, secretary proteins, and membrane receptors. The vacuolar hydrolase carboxypeptidase S (CPS) was used as a marker for characterization of components of ESCRT complexes in yeast (Babst et al., 2002). In mammalian cells, the ESCRT machinery is involved in the down-regulation of several activated growth factor receptors (Rodahl et al., 2009). In higher plants, the potential cargo proteins of MVB sorting pathways include arabinogalactan-rich glycoproteins (Herman and Lamb, 1992), regulator of cell specification DEK1 and CR4 (Tian et al., 2007), and auxin carriers PIN1, PIN2, and AUX1 (Spitzer et al., 2009). All of these proteins are localized in the plasma membrane and are also detected in the MVB. DEK1 is involved in cell–cell communication in epidermal cell fate specification (Lid et al., 2002) while PIN1, PIN2, and AUX1 are carriers of polar auxin transport (Kleine-Vehn and Friml, 2008), showing that the ESCRT machinery participates in the growth and development of higher plants.
The ESCRT machinery is also involved in cellular responses to environmental stress (Huang et al., 2007; Logg et al., 2008; Ruotolo et al., 2008). In a screen for altered response to cadmium and nickel, the vps4 mutant showed cadmium sensitivity but nickel resistance (Ruotolo et al., 2008). This suggested that the ESCRT machinery is specifically involved in down-regulation of plasma membrane-bound cadmium transporters as well as participating in targeting of vacuolar proteins responsible for cadmium detoxification. A potential ESCRT cargo protein, P-type sodium effluxer ENA1, is the main component of the sodium homeostasis system in yeast. Mutants defective in the ESCRT-II component VPS22 or ESCRT-III components VPS20 and SNF7 (sucrose non-fermenting 7) exhibit mild salt sensitivity, and overexpression of ENA1 substantially reduced the salt sensitivity in these mutants (Logg et al., 2008). However, overexpression of ENA1 did not suppress salt sensitivity in mutants defective in VPS4, causing Logg and co-workers (2008) to conclude that VPS4 may have additional ESCRT-independent functions.
To determine the potential functions of the McSKD1-related pathway in the salt-tolerant mechanism of the ice plant, yeast two-hybrid (Y2H) screening was performed to identify its interacting partners using a library constructed from the salt-stressed ice plant. Several candidates were identified including a VPS protein. The focus of this study is on the characterization of two enzymes involved in the post-translational modifications of proteins. One is a RING-type E3 ligase that catalyses the addition of ubiquitin molecules on target proteins and the other is a serine/threonine protein kinase that phosphorylates target proteins. The McSKD1-interacting RING-type E3 ligase has high homology to Arabidopsis RGLG1/RGLG2 (RING domain ligase1 and 2). Mutants defective in both RGLG1 and RGLG2 show loss of apical dominance, alteration of leaf phyllotaxy, and a decrease in the abundance of the auxin carrier PIN1 (Yin et al., 2007). Cheng et al. (2012) showed that RGLG2 down-regulates the drought response through targeting a drought-induced transcription factor AtERF53 for degradation. Another homologue found in pepper, CaRFP1 (RING-finger protein 1), acts as an early regulator in the pathogen defence response and osmotic stress tolerance (Hong et al., 2007). Therefore, this specific group of RING-type E3 ligases play critical roles in development and stress responses of higher plants. The McSKD1-interacting protein kinase is SnRK1 (sucrose non-fermenting 1-related kinase 1). Plant SnRK1s play important roles in protecting cells against nutritional deprivation and environmental stresses (Halford and Hardie, 1998; Halford et al., 2003). Two other SnRK families, SnRK2 and SnRK3, are unique to plants, and certain members are involved in abscisic acid (ABA)-mediated signalling and response to high salinity (Hrabak et al., 2003). As for high salinity stress, the most extensively studied SnRK is SnRK3.11 (Liu et al., 2000), also known as salt overly sensitive 2 (SOS2) or CBL-interacting protein kinase 24 (CIPK24). The members of the SnRK family participating in salt tolerance have been summarized (Coello et al., 2011). The unique roles of plant SnRKs in interconnecting metabolic and stress signalling led Coello et al. (2011) to suggest them as potential candidates for genetic manipulations that could improve crop yield under stressful environments.
In this report, these two SKD1-interacting proteins were first characterized, then the domains responsible for the protein–protein interaction were identified, and finally possible modulation of SKD1 functions through ubiquitination and phosphorylation was examined.
Materials and methods
Plant materials
The growth conditions for ice plants (M. crystallinum L.) and callus were the same as used in Jou et al. (2006). Six-week-old plants were treated with 200mM NaCl for 3 d and root RNAs were collected for construction of a cDNA library used in the Y2H screen. One-week-old callus was transferred to medium containing 0–200mM NaCl and cultured for another week. Samples were collected for microsome isolation, reverse transcription–PCR (RT–PCR), and immunoblotting. Three-day-old dark-grown seedlings were used for immunolabelling experiments.
Yeast two-hybrid assays
An ice plant cDNA library was constructed from poly(A)+ RNA isolated from salt-stressed roots using a BD Matchmaker™ library Construction & Screening Kit (BD Biosciences, Clontech, USA) into yeast strain AH109. Library cells were mated with the yeast strain Y187 bearing a GAL4-binding domain (BD) fusion of McSKD1 and selected on high stringency medium (SD/–Leu/–Trp/–His/–Ade). Putative positive colonies were used for plasmid DNA extraction and β-galactosidase assays. The full-length or partial sequences of McSKD1, McCPN1, and McSnRK1 were cloned into pGADT7 or pGBKT7 vectors. Different combinations of plasmids were co-transformed into strain AH109 using the lithium acetate method (Gietz et al. 1995). The transformants that emerged from the SD/–Trp/–Leu medium were subsequently serially diluted onto SD/–Trp/–Leu/–His and SD/–Trp/–Leu/–His/–Ade selection plates. Combinations able to grow in SD/–Trp/–Leu/–His/–Ade selection medium were further tested for β-galactosidase activity.
Yeast three-hybrid assay
Plasmid pGADT7-McCPN1 carrying full-length McCPN1 used in the Y2H assay was applied again in the yeast three-hybrid (Y3H) assay. Plasmid pBridge (Clontech) was used for the expression of McSKD1 and the UK [the region between the ubiquitin-associated domain (UBA) and the kinase-associated 1 (KA1) domains] or the KA1 domain of McSnRK1. pBridge plasmids carrying only BD-McSKD1 or UK were used as negative controls, and those carrying pGADT7 and pBridge were empty vector controls. For the growth assay, transformants were serially diluted onto selection media SD/–Trp/–Leu/–His/–Met or SD/–Trp/–Leu/–His/–Ade/–Met and grown at 30 °C for 7 d.
Rapid amplification of cDNA ends (RACE)
A BD SMART™ RACE cDNA Amplification Kit (Clontech) was used in both 5’-terminal and 3’-terminal RACE. Touchdown PCR and nested PCR were performed using the GeneAmp® PCR system 2700 (Applied Biosystems, USA). The PCR products were cloned into the pGEM-T easy vector (Promega, USA) and subjected to DNA sequencing.
Protein overexpression, purification and antibody production
Recombinant proteins were expressed in Escherichia coli, affinity-purified, and subsequently used for enzyme assays and antibody production. The open reading frames (ORFs) of McCPN1 and McSnRK1 were cloned into pGEX 4T-1 vectors (GE Healthcare, USA) to give a glutathione S-transferase (GST) fusion at the N-terminus and expressed in E. coli BL21 strain. Isopropyl-β-d-thiogalactopyranoside (IPTG)-induced protein extracts were affinity-purified by Glutathione Sepharose™ 4 Fast Flow resin on an ÄKTAprime™plus system (GE Healthcare, USA). Purified protein was used directly for assaying enzyme activity or cut with thrombin protease to remove the GST tag for antibody production in either BALB/c mice or New Zealand white rabbits. The expression and purification of McSKD1-(His)6 and McSnRK1-(His)6 were essentially as described by Jou et al. (2006). The BL21-AI™ strain (Invitrogen™, USA) harbouring pDEST-AtUBC8 or pDEST-CIP8, used for the in vitro ubiquitination assay, was induced by 0.2% (w/v) l-arabinose and purified by BD TALON™ Metal Affinity Resin (BD Bioscience, USA) or a glutathione column, respectively.
In vitro ubiquitination assay and in vitro kinase assay
In vitro ubiquitination assay was performed according to Stone et al. (2005). One reaction contains 55ng of E1 (human recombinant, Boston Biochem, USA), 250ng of E2 (His)6-AtUBC8, 300ng of E3 GST–CIP8 (positive control) or GST–McCPN1, and 2 μg of ubiquitin (Sigma, USA). Protein–ubiquitin conjugates were analysed by 8% SDS–PAGE and detected by immunoblotting with either anti-ubiquitin (Sigma, USA) or anti-McCPN1 antiserum. In testing the potential substrates of McCPN1, 50–1000ng of McSKD1-(His)6 and/or McSnRK1-(His)6 were added to the reaction mixtures and detected by anti-McSKD1 or anti-McSnRK1 antiserum. In vitro kinase assay was performed based on Fujii and Zhu (2009) with some modifications. Reaction mixtures containing 20mM TRIS-HCl, pH 7.2, 10mM MgCl2, 0.5mM CaCl2, 0.01mM ATP, 2mM dithiothreitol, 5 μCi of [γ-32P]ATP (specific activity ~220 TBq mmol–1; Amersham, UK), and 3 μg of purified GST–McSnRK1 were incubated at 30 °C for 1h. After separation by 10% SDS–PAGE, gels were transferred to PVDF (polyvinylidene fluoride) and radioactive signals were visualized using a Fujifilm BAS-2500 PhosphorImager (Fuji Medical Systems, USA). One constitutively active mutant (GST-McSnRK1 T172D) and one inactive mutant (GST-McSnRK1 T172A) were generated by site-directed mutagenesis using a QuikChange™ Site-Directed Mutagenesis kit (Stratagene, USA). The expression, purification, and assaying of these two mutant proteins were the same as those of the wild-type protein.
Co-immunoprecipitation and pull-down assay
In vitro transcription and translation reactions were carried out using a TNT T7-Coupled Reticulocyte Lysate System (Promega, USA). Equal amounts of [35S]methionine-labelled cMyc-McSKD1 and [35S]methionine-labelled haemagglutinin (HA)-McCPN1, -copine, -McSnRK1, or -UK were immunoprecipitated with either cMyc or HA antibody using a Matchmaker™ Co-IP kit (Clontech). Products were separated by 10% SDS–PAGE, and visualized on a PhosphorImager. Pull-down assay was performed using protein extracts isolated from ice plant callus treated with 200mM NaCl for 6h. Crude extract was separated into microsomal and soluble fractions according to Jou et al. (2006). Protein A–agarose beads (GE Healthcare, USA) were first conjugated with anti-SKD1 and then incubated with either microsomal or soluble ice plant protein overnight. Antibody–protein conjugates were eluted from agarose beads, separated by 10% SDS–PAGE, and detected by immunoblotting with anti-SKD1, anti-CPN1, and anti-SnRK1 antiserum.
Bimolecular fluorescence complementation (BiFC) and immunofluorescence double labelling
The Gateway cloning system was used to generate constructs for the BiFC protein interaction assay (Invitrogen). Full-length McSKD1, McSnRK1, and McCPN1 were transferred from their respective entry clones into the destination vectors pE3130, 3132, 3134, and 3136. Plasmids containing the eYFPC (enhanced yellow fluorescent protein C-terminal) fusion and the eYFPN fusion were co-transfected into Arabidopsis protoplasts. Protoplast isolation and transfection were performed according to Yoo et al. (2007). After 16h incubation at room temperature, the YFP signal was detected by confocal laser scanning microscopy.
Isolated ice plant cells were obtained from root tips of 3-day-old aseptically grown seedlings. Seedlings were fixed in 4% paraformaldehyde in PME solution (10mM PIPES, pH 6.9, 2mM MgSO4·7H2O, 10mM EGTA) for 45min at room temperature. After treatment with 1% cellulase for 30min, the squashed root tips were fixed on polylysine-coated slides. Immunostaining and detection were performed according to Jou et al. (2006). Anti-SKD1, -CPN1, and -SnRK1 were used as the primary antibodies. Secondary antibodies were conjugated with either Alexa Fluor 488 (Invitrogen, USA) or Cy3 (Jackson ImmunoResaerch, USA). The cells were observed under a confocal microscope (Olympus FV-1000). The confocal scanner used an excitation multi-argon laser and the beam splitter DM405/488/543. Filters SDM560 and a mirror collected the emitted fluorescence. From 10 to 20 slides were scanned for each treatment. The images were collected sequentially from the same optical section, analysed by FV10-ASW software, and quantified by Adobe Photoshop software. The red (R), green (G), and yellow (Y) pixel areas in single sections were measured, and the percentage of McSKD1 co-localized with McCPN1 or McSnRK1 was defined as Y/R, while the percentage of McCPN1 or McSnRK1 co-localized with McSKD1 was defined as Y/G.
RT–PCR and western blotting
Total RNA extracted from cultured ice plant cells was reverse transcribed into cDNA using ReverTraAce (Toyobo, Japan). The reverse transcription products were then PCR amplified using gene-specific primers of McCPN1, McSnRK1, and FNR1 (see Supplementary Table S2 at JXB online). Total protein extracted from cultured ice plant cells was separated by 12% SDS–PAGE. The membrane was probed with anti-McCPN1 or anti-McSnRK1 antiserum followed by peroxidase-conjugated AffinitiPure Goat anti-mouse IgG and detected by chemiluminescence (Millipore, USA).
Sequence data can be found in GenBank under the following accession numbers: McSnRK1 (JN558778), VPS13 (JN558779), and McCPN1 (JN558780).
Results
Identification of McSKD1-interacting proteins by yeast two-hybrid screen
To explore the interactive network of McSKD1 in response to high salinity, full-length McSKD1 was used as bait to screen an ice plant cDNA library. The library was constructed from roots of 6-week-old ice plants that had been salt treated for 3 d, conditions known to induce McSKD1 expression (Jou et al., 2004). Several positive clones were identified from a repeated screening process. Six McSKD1-interacting candidates that have potential roles in protein trafficking are listed in Supplementary Table S1 available at JXB online.
According to this list, the sequence of one candidate matched the C-terminal vacuolar protein sorting-associated domain MRS6 of the VPS13 protein. 5’-RACE was performed nine times to identify the full-length ice plant VPS13. The ice plant VPS13 encodes a large 416kDa protein with 60% identity to Arabidopsis At4g17140. Redding et al. (1996) previously proposed a role for VPS13 in protein trafficking between the trans-Golgi network and MVBs; however, the exact molecular mechanism of action of VPS13 in vacuolar protein sorting remains unclear. Five enzymes, namely three hydrolytic enzymes, polygalacturonase, aspartic proteinase, and epoxide hydrolase, and two protein-modifying enzymes, RING-type E3 ligase and SnRK1, were identified as potential McSKD1-interacting proteins. RING-type E3 ligase catalyses the attachment of ubiquitin to the substrate proteins, and SnRK1 catalyses the addition of phosphate groups to the substrate proteins. Both enzymes participate in protein post-translational modifications. Growth on SD/–Trp/–Leu/–His/–Ade selection medium indicated that McSKD1undergoes protein–protein interactions with certain regions of these two protein-modifying enzymes (Supplementary Fig. S1 at JXB online).
It is worth mentioning that the cDNAs of the ice plant Y2H library represent the transcripts expressed under salt treatment. In screening of this library, known SKD1-interacting proteins, all of which are involved in the operation of the ESCRT machinery in yeast, were not identified. It is possible that the gene expression level of such ESCRT components might be low under salt stress and thus the associated cDNAs are not sufficiently enriched to be detectable under the screening conditions that were used. As currently no report on post-translational modification of SKD1 exists in the literature, E3 ligase and protein kinase were chosen to explore the possible effects of ubiquitination and phosphorylation on SKD1 functions.
Characterization of RING-type ubiquitin ligase McCPN1
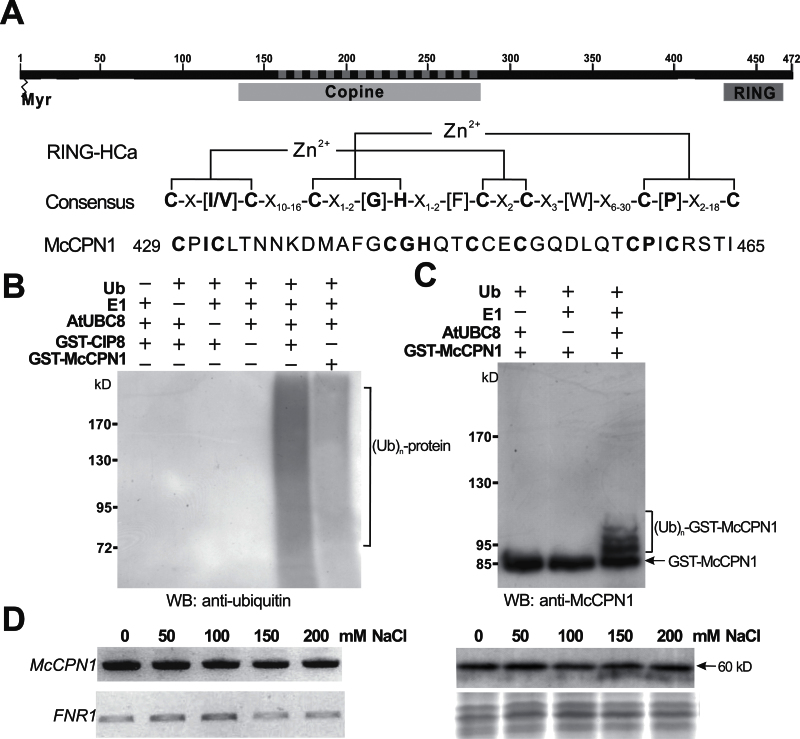
The Y2H screen identified that the McSKD1-interacting region of the putative E3 ligase was the central copine (or von Willebrand factor type A; vWA) domain, a coiled-coil domain capable of interacting with a variety of proteins that also contain a coiled-coil structure (Tomsig et al., 2003), leading it to be named McCPN1. The full-length cDNA of McCPN1 was obtained using 5’- and 3’-RACE. It was ~1770bp in length with an ORF of 1419bp corresponding to 472 amino acids. Sequence analysis revealed that the copine domain spans the region from position 134 to 280 and that a RING finger domain exists from position 429 to 465, close to the C-terminus (Fig. 1A). According to the classification by Stone and colleagues (2005), the RING domain of McCPN1 belongs to the HCa subgroup (Fig. 1A). A predicted N-myristoylation site was found at the N-terminus, indicating that McCPN1 may be linked to membranes through fatty acid linkage. The deduced amino acid sequence of McCPN1 has 72% and 75% identity to Arabidopsis RGLG1 and RGLG2, respectively. The results showed that McCPN1 is a RING-type copine, a subgroup of plant-specific copine-related proteins, composed of an N-myristoylation site at the N-terminus, a copine domain in the centre, and a RING-finger domain at the C-terminal end.
Fig. 1.
Characterization of RING-type E3 ligase McCPN1. (A) Predicted domain structures and RING-HCa consensus sequence of McCPN1. Myr indicates the potential N-myristoylation site, Copine indicates the copine domain, RING indicates the RING domain, and the dashed line indicates the sequences identified from the Y2H screen. Conserved metal ligand binding positions including eight consensus cysteine and histidine residues and zinc (Zn2+) coordinating amino acid pairs are shown in bold. (B) Western blotting using anti-ubiquitin detects protein–ubiquitin conjugates of the in vitro ubiquitination product. In the presence of ubiquitin, E1, and E2 (AtUBC8), both GST–CIP8 and GST–McCPN1 display ubiquitin E3 ligase activity. (C) Western blotting using anti-McCPN1 detects the McCPN1–ubiquitin conjugates, and shifted bands indicate the attachment of 1–3 ubiquitin molecules. The arrow indicates the positions of GST–McCPN1 (85kDa). (D) Gene expression and protein accumulation of McCPN1 in cultured ice plant cells treated with different concentrations of NaCl for 1 week. Left panel: the expression of McCPN1 was analysed by RT–PCR. The expression of FNR1 (ferredoxin-NADP + reductase 1) was used as an internal control. Right panel: the accumulation of McCPN1 was analysed by western blotting using anti-McCPN1 antiserum (top). Coomassie blue-stained total protein was used as a loading control (bottom). The arrow indicates the position of McCPN1 (60kDa).
In vitro ubiquitination assays were performed to determine if McCPN1 has the capacity for E2-dependent protein ubiquitination. The assay contained ubiquitin, human E1, His-tagged AtUBC8 as E2, and GST-fused McCPN1 or CIP8 (COP1-interacting protein 8). CIP8, a known Arabidopsis RING-type E3, was used as a positive control. Western blot analysis using anti-ubiquitin antibodies showed a smear in the high molecular weight region indicating that protein ubiquitination had occurred. Omission of ubiquitin, E1, E2, or E3 from the reaction mixture resulted in a loss of protein ubiquitination (Fig. 1B). Western blotting using anti-McCPN1 detected four polypeptides of 85, 94, 101, and 108kDa in the complete reaction (Fig. 1C). The 85kDa polypeptide was GST–McCPN1, whereas the other three polypeptides were its ubiquitin-conjugated forms. In vitro assay showed that the RING domain-containing McCPN1 functions as an E3 ubiquitin ligase.
The expression of McSKD1 is salt induced in cultured ice plant cells (Jou et al., 2004). As a potential interacting partner of McSKD1, gene expression and protein accumulation of McCPN1 under salt stress were analysed (Fig. 1D). The result showed that McCPN1 was constitutively expressed independently of the addition of NaCl. At the protein level, addition of NaCl did not affect the accumulation of 60kDa McCPN1. It is concluded that McCPN1 is a constitutively expressed protein in the presence of NaCl.
Characterization of serine/threonine protein kinase McSnRK1
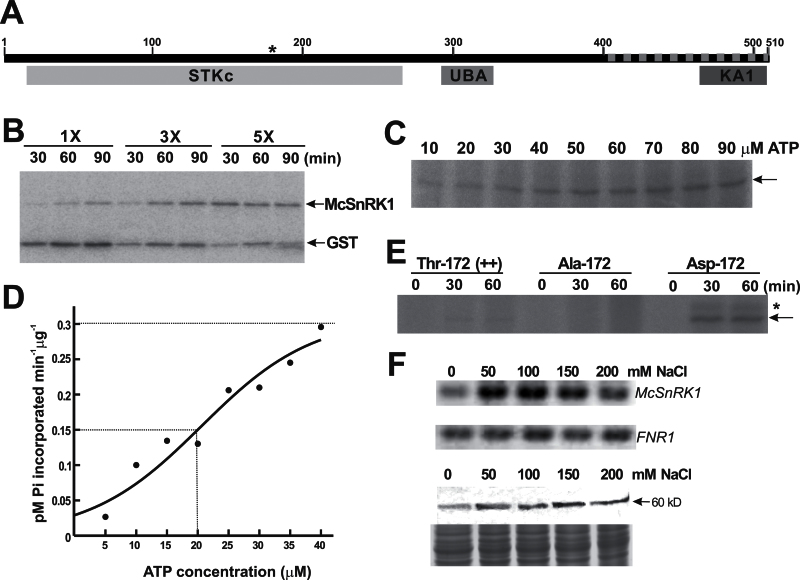
The Y2H screen identified a clone covering the C-terminal regulatory domain of SnRK1 (Fig. 2A). It was therefore named McSnRK1. The full-length McSnRK1 was obtained by 5’- and 3’-RACE. The cDNA contained 2356bp, whereby the length of the ORF was 1533bp, corresponding to 510 amino acids. McSnRK1 contains three conserved domains (Fig. 2A), namely a Ser/Thr protein kinase catalytic domain (STKc; position 16–268), a UBA domain (position 291–327), and a KA1 domain (position 463–509). A conserved threonine residue in the activation loop of the STKc domain, critical for the full activation of SnRKs (Sugden et al., 1999a), is located at position 172 in McSnRK1 (Fig. 2A). The region indentified from the Y2H screen contained an entire KA1 domain and regions between UBA and KA1; hereafter, this will be referred to as the UK region. McSnRK1 has 87–96% similarity with Arabidopsis, tomato, potato, rice, and Cucumis sativus, indicating that McSnRK1 belongs to this highly conserved protein kinase family.
Fig. 2.
Characterization of SNF1-related protein kinase McSnRK1. (A) Predicted domain structures and conserved activation loop sequence of McSnRK1. STKc indicates the Ser/Thr protein kinase catalytic domain, UBA indicates the ubiquitin-associated domain, KA1 indicates the kinase-associated domain 1, and the dashed line indicates the sequences identified from the yeast two-hybrid screen. The position of Thr172 responsible for the catalytic activity is marked by an asterisk. (B) Analysis of autophosphorylation and substrate phosphorylation activity of McSnRK1 by in vitro kinase assay. Different amounts (1×, 0.6 μg; 3×, 1.8 μg; 5×, 3 μg) of purified GST–McSnRK1 were incubated with 10 μM ATP in the presence of [γ-32P]ATP. The reaction mixture was treated with thrombin protease before separation by SDS–PAGE. The signals were detected by autoradiography. (C) The effects of ATP on GST–McSnRK1 phosphorylation. ATP concentrations from 10 μM to 90 μM were tested. The arrow indicates the position of GST–McSnRK1 (88kDa). The autoradiogram shown is representative of three independent experiments with similar results. (D) ATP dependence of protein phosphorylation activity of McSnRK1. The ATPase activity was measured at the range of 0–40 μM ATP. The value of K 0.5 is equivalent to the ATP concentration at which V o is one-half V max (dashed lines). The experiments were repeated three times and data were calculated from one representative experiment. (E) Protein phosphorylation activities of wild-type McSnRK1 (Thr172), inactive McSnRK1 (Ala172), and constitutive active McSnRK1 (Asp172). Purified GST–McSnRK1 (Thr172), Ala172, and Asp172 were subjected to kinase assay under an excess concentration of ATP (0.1mM) for 30min and 60min. The arrow indicates the position of GST–McSnRK1, and the asterisk indicates a 100kDa phosphorylated polypeptide appearing in the Asp172 mutant of McSnRK1. (F) Gene expression and protein accumulation of McSnRK1 in cultured ice plant cells treated with different concentrations of NaCl for 1 week. Top: the expression of McSnRK1 was analysed by RT–PCR. The expression of FNR1 was used as an internal control. Bottom: the accumulation of McSnRK1 was analysed by western blotting using anti-McSnRK1 antiserum. Coomassie blue-stained total protein was used as a loading control.
A homology search also revealed that the catalytic STKc domain is highly conserved in all eukaryotes, while the greatest variation occurs in the regulatory domain at the C-terminus. The STKc domain of McSnRK1 had 81% similarity to yeast Snf1 (Supplementary Fig. S2 at JXB online). Yeast Snf1 is a key regulator in the process called glucose repression (Carlson, 1999). Yeast mutants defective in Snf1 are unable to utilize sucrose, galactose, maltose, or non-fermentable carbon sources such as glycerol and ethanol. Alderson and co-workers (1991) demonstrated that plant SnRK1was able to complement the snf1 phenotype in yeast. A similar complementation assay was performed by expressing McSnRK1 in a yeast snf1 mutant, and it was found that McSnRK1 was able partially to complement the snf1 mutation, proving that McSnRK1 is a functional SnRK1 (Supplementary Fig. S2).
Several plant SnRKs have been shown to exhibit both autophosphorylation and substrate phosphorylation activity in vitro (Guo et al., 2001; Belin et al., 2006; Jossier et al., 2009). McSnRK1 was fused with GST and the phosphorylation activity of McSnRK1 was tested in vitro. After incubation with [γ-32P]ATP, GST–McSnRK1 was separated into GST and McSnRK1 by cutting with protease. As shown in Fig. 2B, under a low protein concentration (1×) and a short incubation time (30min), phosphorylation sites were primarily found on the GST portion of the fusion protein. During prolonged incubation, McSnRK1 became self-phosphorylated. A 5-fold increase in protein concentration efficiently promoted self-phosphorylation of McSnRK1. The results indicated that, at least in vitro, McSnRK1 exhibits autophosphorylation and substrate phosphorylation activity.
The effects of ATP were further analysed under assay conditions favourable for autophosphorylation. The degree of protein phosphorylation increased as the ATP concentration increased, approaching saturation at 50 μM ATP (Fig. 2C). The kinetic property of GST–McSnRK1 phosphorylation was then determined to lie in the range of 0–40 μM ATP (Fig. 2D). The data were fitted into sigmoid kinetics, with V max approaching 0.3 pmol μg–1 min–1 and K 0.5 [ATP] ~20 μM. The sigmoid curve indicates that ATP may also serve as a stimulatory modulator of this reaction. The kinetic properties showed that the V max was substantially lower while K 0.5 was higher than for other plant SnRKs (Toroser et al., 2000; Guo et al., 2001; Gong et al., 2002a, b ). The low rate of McSnRK1-catalysed phosphorylation may result from the enzyme being in a less activated state. Therefore, constitutively active and inactive forms of McSnRK1 were generated by substituting the conserved Thr172 in the activation loop of the STKc domain with aspartate and alanine, respectively. As shown in Fig. 2E, the degree of phosphorylation of the wild-type GST–McSnRK1 (Thr172) increased during 1h of incubation. Substituting Thr172 for Ala172 completely abolished the kinase activity, while replacement by Asp172 significantly boosted the kinase activity by a minimum of 20-fold (Fig. 2E).
The expression of McSnRK1 and accumulation of McSnRK1 were examined under salt stress in ice plants (Fig. 2F). McSnRK1 was expressed in cells maintained in salt-free culture medium. When the culture medium contained even a low concentration (50mM NaCl) of salt, the transcript level of McSnRK1 increased. A salt-induced accumulation of 60kDa McSnRK1 was also observed; however, the degree of induction was lower than that of the transcript level. These results showed that McSnRK1, like its interacting partner McSKD1, is responsive to NaCl. The overall results showed that McSnRK1 is a salt-inducible protein kinase that possesses protein phosphorylation activity.
Interaction between McSKD1, McCPN1, and McSnRK1 through distinct domains
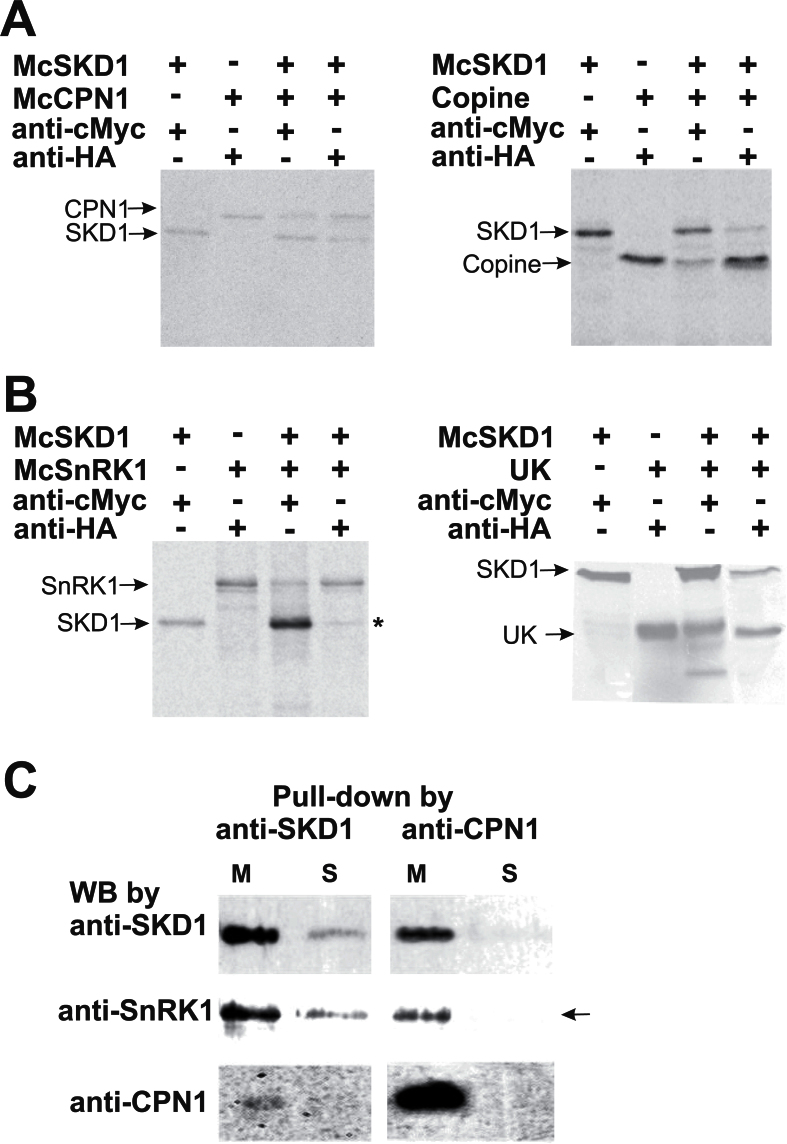
The interactions between McSKD1 and its two partners were examined by co-immunoprecipitation (Co-IP) using [35S]methionine-labelled in vitro synthesized proteins. McSKD1 was translated into a fusion protein with a c-Myc epitope tag. McCPN1 and McSnRK1, either the full-length cDNA or sequences identified from the Y2H screen, were fused with an HA epitope tag. Anti-cMyc antibody immunoprecipitated McSKD1-cMyc and co-precipitated McCPN1; reciprocal Co-IP using anti-HA was also able to co-precipitate a quantity of McSKD1 (Fig. 3A, left panel). Replacement of full-length McCPN1 by the copine domain of McCPN1 yielded similar results (Fig. 3A, right panel). As for McSnRK1, anti-cMyc antibody immunoprecipitated McSKD1-cMyc and co-precipitated McSnRK1; however, reciprocal Co-IP using anti-HA antibody showed a strong McSnRK1-HA band and a faint McSKD1 band (Fig. 3B, left panel). When the UK region of McSnRK1 was used in the Co-IP assay, a greater amount of McSKD1 co-precipitated with the UK fragment (Fig. 3B, right panel), indicating that the UK region alone has a higher affinity for McSKD1.
Fig. 3.
Confirmation of McSKD1 interacting with McCPN1 and McSnRK1 by co-immunoprecipitation (Co-IP) and pull-down assay. (A) [35S]Methionine-labelled cMyc-McSKD1, HA-McCPN1, and HA-copine domain were immunoprecipitated with either anti-cMyc or anti-HA antibodies as indicated on the top. The resulting gels were autoradiographed using a PhosphorImager. The positions of the corresponding protein bands are indicated on the left. Left panel: cMyc-McSKD1 co-immunoprecipitated with HA-McCPN1; right panel: cMyc-McSKD1 co-immunoprecipitated with HA-copine domain of McCPN1. The experiments were repeated twice yielding similar results. (B) Co-IP of [35S]methionine-labelled cMyc-McSKD1 and HA-McSnRK1 (left panel) or cMyc-McSKD1 and the HA-UK region of McSnRK1 (right panel). The asterisk indicates a weak McSKD1 band. The experiments were repeated twice yielding similar results. (C) Anti-SKD1 or anti-CPN1 antibodies were first immobilized on the protein A–agarose beads and incubated with the microsomal (M) or soluble (S) protein fractions isolated from salt-stressed ice plant callus. The pull-down protein complex eluted from protein A beads was separated by 10% SDS–PAGE, and western blotting against anti-SKD1, anti-SnRK1, or anti-CPN1 antibodies was performed as indicated. The arrow indicates that McSnRK1 was detected in the pull-down sample using anti-CPN1 in the microsomal fraction but not in the soluble fraction (negative control).
Polypeptides generated by in vitro transcription and translation systems may be folded improperly and lack post-translational modifications which can cause the failure of McSKD1 to co-precipitate with its full-length partners. Therefore, a pull-down assay was performed using protein extract obtained from salt-stressed ice plant cells. A crude extract was separated into microsomal and soluble fractions following the discovery by Jou et al. (2006) that McSKD1 is located primarily in membrane-bound compartments. When anti-SKD1 antibody was used in the pull-down assay, greater amounts of McSKD1 were distributed in the microsomal fraction. When the same blot was re-probed with either anti-SnRK1 or anti-CPN1, it was possible to detect McSnRK1 in both microsomal and soluble fractions, while McCPN1 was detected solely in the microsomal fraction (Fig. 3C). This result showed that McSKD1 is able to interact with the intact forms of McSnRK1 and McCPN1 in ice plant. Furthermore, McCPN1 was found to be enriched in the microsomal fraction, possibly through N-terminal linked lipids. Anti-CPN1 antibody was then used to pull down proteins in the microsomal fraction and it was found that McSKD1 was bound to McCPN1, and, interestingly, McSnRK1 was also detected in the pull-down products (Fig. 3C). This indicates that McSKD1, McCPN1, and McSnRK1 could form a complex on the membrane of ice plant cells.
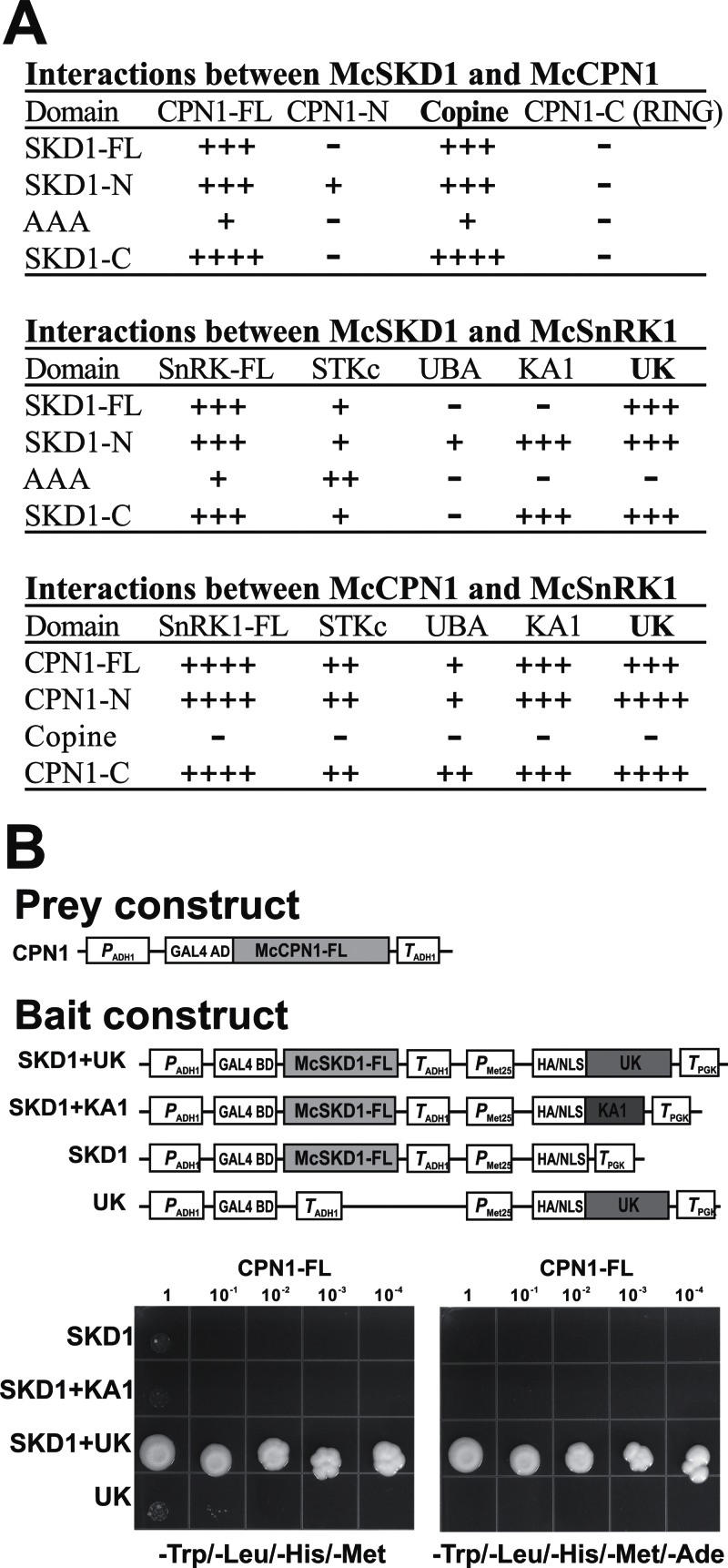
A pairwise Y2H interaction assay was then used to identify domains responsible for protein–protein interaction. All three proteins were divided into three parts according to their assigned domains (Supplementary Fig. S3A at JXB online). A UK construct containing a sequence from the UBA to the KA1 domain of McSnRK1 was also included. The interaction assay was performed by plating transformants in high stringent medium (–Trp/–Leu/–His/–Ade). The activity of β-galactosidase was measured in all combinations capable of growing in a high stringent medium. The growth of yeast is shown in Supplementary Fig. S3 and the results are summarized in Fig. 4A. The results showed that McCPN1 and McSnRK1 interacted with McSKD1, primarily via the copine domain and UK region, respectively, confirming the result obtained from the initial Y2H screen. The UK region of McSnRK1 also interacted with McCPN1. The copine domain of McCPN1 that exhibited interaction with McSKD1 did not interact with McSnRK1, suggesting that McCPN1 interacts with McSKD1 and McSnRK1 through different domains.
Fig. 4.
Identification of the UK region of McSnRK1 is necessary for interaction with McSKD1 and McCPN1. (A) Full-length or domain–domain interactions between McSKD1, McCPN1, and McSnRK1. Results were combined from yeast growth assays and β-galactosidase activity assays shown in Supplementary Fig. S3 at JXB online. –, No interaction (yeast cells grown only on SD-2); +, low degree of interaction (yeast cells grown slowly on SD-3); ++, medium degree of interaction (yeast cells grown on SD-3); +++, high degree of interaction (yeast cells grown on SD-4); ++++, very high degree of interaction (yeast cells grown on SD-4 with β-galactosidase >15 Miller units). (B) Investigation of ternary protein complexes containing McSKD1, McCPN1, and the UK region of McSnRK1 by yeast three-hybrid assay. Schematic representation of the constructs used in the Y3H assay is shown on the top. Grey boxes indicate the sequences cloned into pBridge vector (bait constructs) or pGADT7 vector (prey construct). The growth assay on –Trp/–Leu/–His/–Met medium (left panel) and –Trp/–Leu/–His/–Met/–Ade medium (right panel) is shown on the bottom. Yeast AH109 transformants carrying pBridge-based bait plasmids and pGADT7-based full-length McCPN1 were serially diluted, spotted on selection plates, and incubated at 30 °C for 7 d. Bait plasmids carrying only full-length McSKD1 (SKD1) or a UK region of McSnRK1 (UK) are used as negative controls showing no growth in high stringent medium. Two independent experiments were performed, yielding similar results.
The pairwise interaction assay showed that McSKD1, McCPN1, and McSnRK1 can interact with one another. In fact, McSnRK1 can interact with McSKD1 as well as McCPN1 directly via the C-terminal UK region (Fig. 4A). A Y3H assay was established to evaluate whether these three proteins could form a stable ternary complex. The full-length McSKD1 and UK region of McSnRK1 were cloned into a pBridge vector and full-length McCPN1 was cloned into a GAL4 AD vector (Fig. 4B). pBridge vectors containing McSKD1 or a UK region alone were used as negative controls. The effect of the KA1 domain in the formation of the ternary complex was also tested because the KA1 domain within the UK region showed interaction with certain domains of McSKD1 and McCPN1. Based on the results, cells carrying McSKD1 and UK (SKD1+UK) interacted significantly with McCPN1 (Fig. 4B). Low levels of interactions were observed in constructs carrying SKD1+KA1, SKD1, or UK alone in –Trp/–Leu/–His/–Met, medium and no interactions were observed in these combinations under high stringent (–Trp/–Leu/–His/–Met/–Ade) medium. The Y3H results suggest that these three proteins are able to form a protein complex through the UK region of McSnRK1 forming a ‘bridge’ to stabilize the interaction between McSKD1 and McCPN1.
McSKD1 is a potential substrate for McSnRK1 and McCPN1
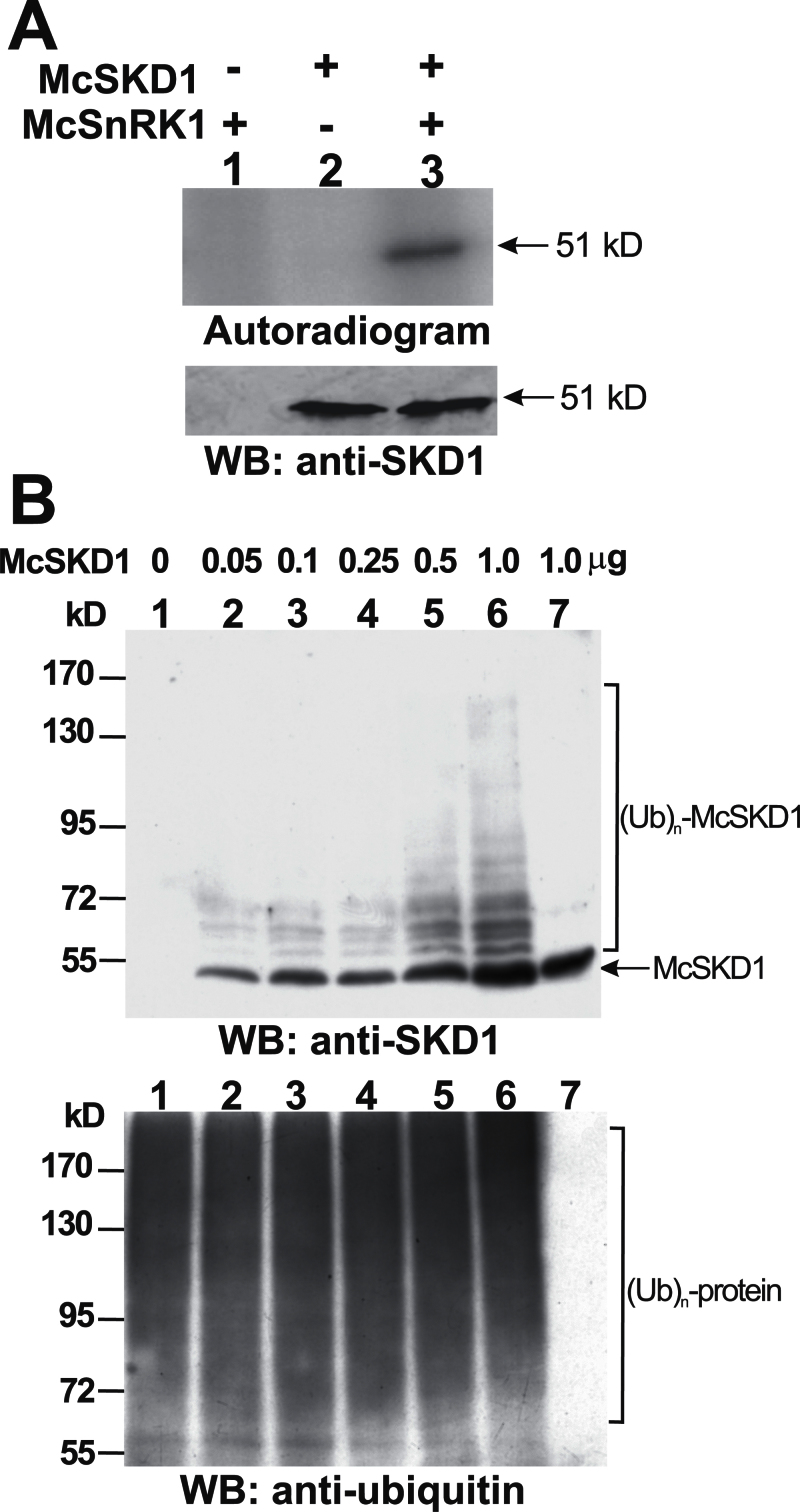
It has been shown that McCPN1 possesses E3 ligase activity while McSnRK1 possesses protein kinase activity and that these two proteins interact with McSKD1 through distinct domains. It was therefore tested whether McSKD1 could serve as substrate for these two enzymes. Purified McSKD1 were added to the reaction mixture of the kinase assay in the presence of N-ethylmaleimide (NEM), an inhibitor of AAA-type ATPases, to prevent McSKD1-catalysed hydrolysis of ATP. When McSnRK1 was added to the kinase reaction mixture, a strong 51kDa signal was detected, indicating that McSKD1 was phosphorylated (Fig. 5A). No signal was detected by adding McSKD1 alone. This result shows that McSKD1 is a potential substrate for McSnRK1.
Fig. 5.
In vitro analysis of McSKD1 phosphorylation by McSnRK1 and ubiquitination by McCPN1. (A) The substrate phosphorylation reaction by McSnRK1 was performed using McSKD1 as the substrate in the presence of 10mM N-ethylmaleimide (NEM) (lanes 1–3). A 1 μg aliquot of purified McSKD1-(His)6 was added alone (negative control) or with 1 μg of purified GST–McSnRK1 to the in vitro kinase reaction. After separation by 10% SDS–PAGE, the resulting gels were transferred to a PVDF membrane and autoradiographed using a PhosphorImager. Subsequently, the same membrane was analysed by western blotting using anti-McSKD1 antiserum. The arrow indicates the position of McSKD1. (B) In vitro ubiquitination reaction by McCPN1 was performed using McSKD1 as the substrate. Various concentrations of purified McSKD1-(His)6, up to 1 μg, were added to the complete in vitro ubiquitination mixture containing McCPN1 as E3 (lanes 1–6). Lane 7 is a reaction without E3 as a negative control showing that no ubiquitination occurred. Western blotting using anti-SKD1 antiserum was used to detect McSKD1 and its ubiquitin-conjugated forms (top), and anti-ubiquitin antibody was used to detect protein–ubiquitin conjugates (bottom). The arrow indicates the position of McSKD1 (51kDa) and the laddered bands indicate ubiquitin–McSKD1 conjugates.
The in vitro ubiquitination assay shown in Fig 1B was performed in the presence of McSKD1. By adding McSKD1 to the reaction mixture following detection by anti-SKD1 antibody, a typical laddered pattern of protein ubiquitination was detected (Fig. 5B, top). A 51kDa major band was identified as McSKD1-(His)6, while others with greater molecular weights were identified as McSKD1-(His)6–ubiquitin conjugates such that the degree of McSKD1 ubiquitination increased as the amount of McSKD1 in the reaction mixture increased. When the same blot was probed by an anti-ubiquitin antibody, smear bands were detected in all lanes, confirming that protein ubiquitination had occurred in all reactions (Fig. 5B, bottom). Results from thein vitro assay indicate that McSKD1 is a potential substrate of E3 McCPN1.
Because McSnRK1 also interacted with McCPN1, the McCPN1-mediated ubiquitination of McSnRK1 was tested but only the 60kDa McSnRK1 band was found (Supplementary Fig. S4 at JXB online). Lack of band shift indicated that McSnRK1 may not be a suitable substrate for McCPN1. Whether McSnRK1 plays any role in McCPN1-mediated McSKD1 ubiquitination was next examined. In a complete in vitro ubiquitination reaction using McSKD1 as the substrate, purified McSnRK1 was added to the assay. The result showed that McSKD1 can be ubiquitinated in the absence or presence of McSnRK1 (Supplementary Fig. S5 at JXB online). No dramatic changes in the patterns of McSKD1 ubiquitination indicate that McSnRK1 has no direct enhancing or inhibiting effect on McSKD1 ubiquitination in vitro.
McCPN1 and McSnRK1 co-localize with McSKD1
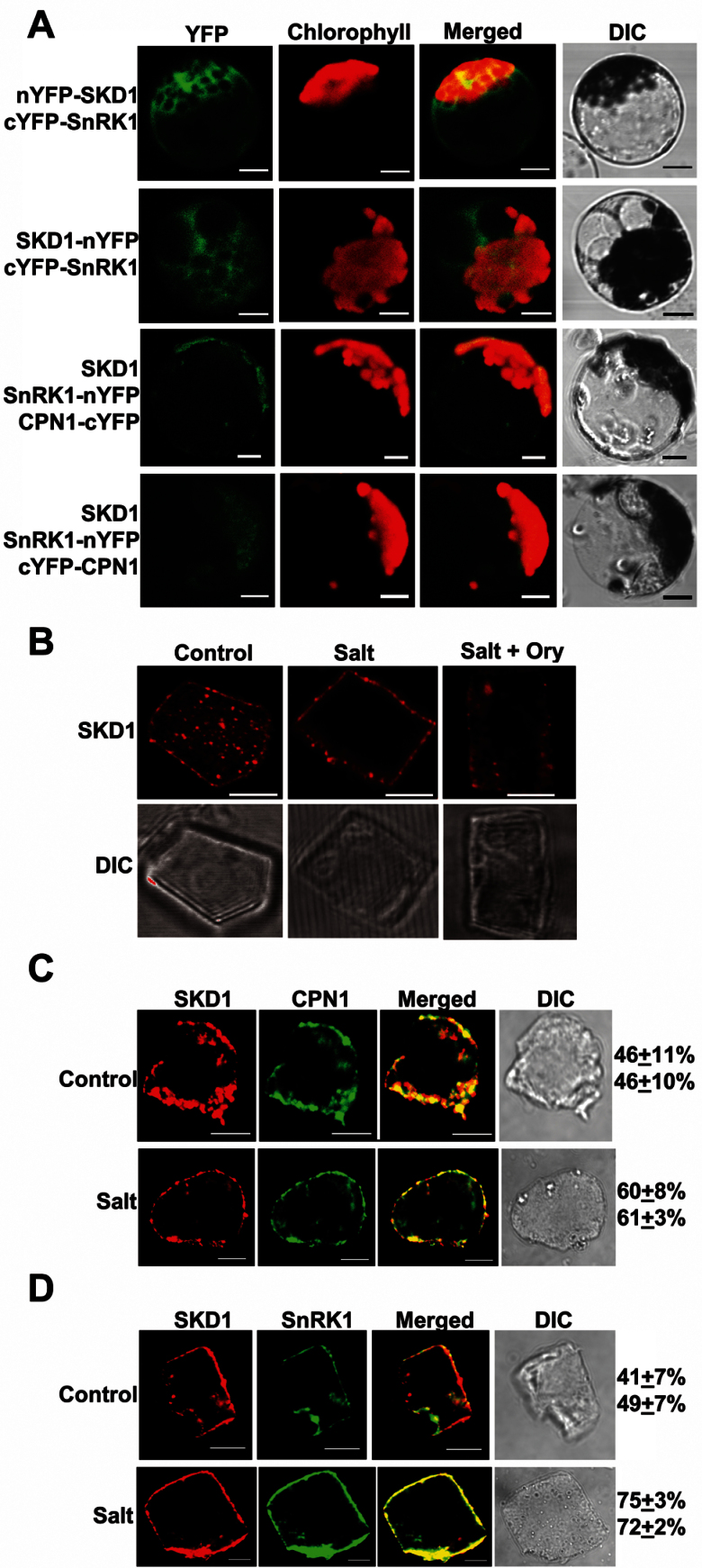
BiFC was performed using Arabidopsis protoplasts to examine the protein interactions in vivo. When plasmid carrying N-terminal YFP fused with McSKD1 (nYFP–McSKD1) was co-transfected with C-terminal YFP fused with McSnRK1 (cYFP–McSnRK1), strong fluorescence appeared throughout the cytoplasm, showing that these two proteins interact in vivo (Fig. 6A, top panel). YFP signal was also detected in the McSKD1–nYFP and cYFP–McSnRK1 combination (Fig. 6A, second panel). No fluorescence was detected in the split YFP fusion of McSKD1 or McSnRK1 co-transfected with complementary empty split YFP vector (data not shown). However, BiFC signal between McSKD1 and McCPN1 could not be detected in any of the eight combinations (data not shown).
Fig. 6.
BiFC and immunofluorescence detecting cellular localization of McSKD1, McCPN1, and McSnRK1. (A) Visualization of the protein complex using BiFC in Arabidopsis protoplasts. Protoplasts were co-transfected with plasmids encoding McSKD1, McSnRK1, or McCPN1 fused with nYFP (N-terminal residues 1–175) or cYFP (C-terminal residues 175–end) as indicated on the left. Column 1 shows signals from eYFP; column 2 shows chlorophyll autofluorescence; column 3 shows merged images of column 1 and 2; column 4 shows bright field images. Bar=10 μm. (B) Root cells isolated from 3-day-old ice plant seedlings (control) or treated either with 200mM NaCl for 6h (salt) or with 30 μM oryzalin for 30min prior to addition of 200mM NaCl (salt+ory) were immunolabelled by anti-SKD1. The image of McSKD1 localization is shown by red fluorescence (top panel). The lower panel shows the cell image under Nomarski optics (differential interference contrast). At least 15–20 cells were examined in each treatment. Root cells isolated from 3-day-old seedlings (control; top panel) or treated with 200mM NaCl for 6h (salt; lower panel) were dual labelled by anti-SKD1 and anti-CPN1 (C) or with anti-SKD1 and anti-SnRK1 (D). Confocal images represent single images of Cy3/Alexa 488 dual labelling of a representative cell. Red fluorescence indicates the McSKD1 image while McCPN1 or McSnRK1 is shown by green fluorescence. The presence of yellow in the merged image indicates the co-localization area of the two proteins. Numbers on the right indicate the percentage of overlapping area (top, yellow/red; bottom, yellow/green). At least 10 cells were calculated in each treatment. All bars=5 μm.
The Y3H results showed that McSnRK1 stabilizes the interaction between McSKD1 and McCPN1 (Fig. 4B). Three constructs, SKD1, McSnRK1–nYFP, and McCPN1–cYFP, were co-transfected into Arabidopsis protoplasts. Weak fluorescence was detected in the plasma membrane (Fig. 6A, third panel). Yin et al. (2007) showed that RGLG2, the McCPN1 homologue, is localized in the plasma membrane through N-terminal myristoylation modification. Signal arising from the plasma membrane was also detected in the construct carrying McCPN1 fused with full-length YFP (Supplementary Fig. S6 at JXB online). It is very likely that spatial separation of plasma membrane-localized McCPN1 limits its interaction with cytosolic-localized McSKD1 or McSnRK1. Therefore, McCPN1–cYFP (membrane bound) was replaced by cYFP–McCPN1 (cytosolic form) and fluorescence arising from the cytosol was found (Fig. 6A, bottom panel), proving that these three proteins interact in vivo if they are present in the same compartment.
Previously it was shown that McSKD1 is enriched in microsomal fractions and forms punctate spots in the cytosol in cultured ice plant cells (Jou et al., 2006). Using the same immunofluorescence labelling method, a similar distribution of McSKD1 in the plasma membrane and in the cytosol of root tip cells was observed (Fig. 6B, control; Supplementary Fig. S7 at JXB online). When seedlings were treated with 200mM NaCl for 6h, the distribution of McSKD1 changed such that >80% of red fluorescence appeared around the plasma membrane (Fig. 6B, salt). Since the N-terminus of McSKD1 contains an MIT domain that can interact with microtubules, it is probable that microtubules are involved in the re-distribution of McSKD1. To test this possibility, ice plant seedlings were then stressed in the presence of oryzalin, a herbicide that disrupts the structure of microtubules. The result was a reduction in the previously observed salt-induced re-distribution of McSKD1 (Fig. 6B, salt+ory), showing that the movement of McSKD1 is probably mediated by the microtubule cytoskeleton.
Because protoplasts are sensitive to prolonged osmotic treatments, immunofluorescence double labelling was used to examine the endogenous distribution of McSKD1, McCPN1, and McSnRK1 in the ice plant and to estimate the change of protein distribution under salt stress. Root cells were fixed before and after salt treatment and stained with the corresponding antibodies. The red fluorescence indicated that McSKD1 was detected throughout the plasma membrane and in punctate spots in the cytosol; McCPN1 could be seen to have a similar cellular distribution as indicated by green fluorescence (Fig. 6C, control). Yin et al. (2007) observed that RGLG2 is occasionally present at the membrane of dynamically formed cytoplasm strands. The root tip cells used here contain a large central nucleus and conspicuous cytoplasmic strands; therefore, it is the reason why a higher portion of McCPN1 was detected in the cytoplasm of these actively dividing cells. By merging images, a greater degree of co-localization around the plasma membrane (yellow colour) and a lesser degree of co-localization in the cytosol was visualized, leading to the overall approximation of 50% co-localization. Upon salt stress, most of the McSKD1 protein was localized in the plasma membrane, and the percentage co-localization increased to an estimated 60% (Fig. 6C, salt).
The distribution of McSnRK1 also showed a similar pattern to that of McSKD1. Under control treatment, the degree of co-localization was <50%, increasing to 75% 6h after salt treatment (Fig. 6D). This can be explained by the fact that the majority of McSnRK1 was now located on the cell surface. Although the degrees of co-localization were higher under conditions of salt stress, some distinct spots appeared only for McSKD1 or McSnRK1 in the cytosol (Fig. 6D, salt). The difference in temporal distribution suggests that these two McSKD1-interacting proteins may interact with McSKD1 at distinct stages in the salt stress response.
Combining all results, the following model is proposed (Supplementary Fig. S8 at JXB online): under external stimuli, activated McSnRK1 interacts with McSKD1, the phosphorylated McSKD1–McSnRK1 complex then trafficks to the plasma membrane via the microtubule cytoskeleton, where McCPN1 ubiquitinates McSKD1. The UBA domain of McSnRK1 recognizes the ubiquitin molecules on McCPN1 or McSKD1 and reinforces the ternary complex. Modified McSKD1 probably executes its molecular motor action on the remodelling of membrane components such as ion transporters to avoid ion toxicity.
Discussion
Interactions with specific E3 ligase and protein kinase resemble enzyme–substrate binding activity
Many proteins are known to interact with VPS4/SKD1, and to date they are all functionally related to the ESCRT machinery (Shestakova et al., 2010). In yeast, Vta1/LIP5 interacts with SKD1 through the C-terminal β-domain to form a stable oligomer, and ESCRT-III components VPS2, VPS24, SNF7, and Did2 subsequently interact with the SKD1 oligomer through the N-terminal MIT domain to facilitate ESCRT-III disassembly (Yeo et al., 2003; Vajjhala et al., 2006, 2007). In this study, two post-translational modification enzymes, McCPN1 and McSnRK1, known to be involved in stress responses but with no apparent association with the ESCRT machinery, were identified. The interactions between McSKD1 and these two enzymes are more likely to resemble the relationship between substrate and enzyme, leading to changes in ATPase activity or/and cellular localization of SKD1. The regions interacting with SKD1, namely the copine domain of McCPN1 and the UK region of McSnRK1 (Fig. 4), are expected to be the substrate recognition sites that are distinct from the catalytic domains of both enzymes.
One explanation for why no known SKD1-interacting ESCRT-related proteins were identified in the initial Y2H screen is that these genes were under-represented in the salt-stressed ice plant Y2H library. Shahriari and co-workers (2011) recently established the Arabidopsis interactome of ESCRT-I, -II, and -III complexes using systematic pairwise Y2H assay of proteins showing homology to the yeast ESCRT core complex. This same strategy could be employed to identify the interactions between SKD1 and ESCRT-III components in ice plant.
Post-translational modifications of McSKD1 may be stress induced and ESCRT independent
McSKD1 was originally identified from a salt-enriched cDNA library of ice plant (Yen et al., 2000). It was highly expressed in EBCs, root cortex, pollen sac, and seed coat (Jou et al., 2004, 2006). AtSKD1, the McSKD1 homologue in Arabidopsis, is an essential gene that participates in MVB protein sorting (Haas et al., 2007) and vacuolar maintenance (Shahriari et al., 2010). In addition, SKD1 is also involved in stress responses. Impaired or reduced SKD1 expression results in the salt-sensitive phenotype in yeast (Logg et al., 2008) and Arabidopsis (Ho et al., 2010). It is well known that salt stress induces the accumulation of various proteinases. A high amount of cysteine proteinase accumulation was found in vacuoles of salt-stressed EBCs (Jou et al., 2007). In addition to two protein modification enzymes, three SKD1-interacting hydrolytic enzymes, polygalacturonase, aspartic proteinase, and epoxide hydrolase, were also identified in the Y2H screen (Supplementary Table S1 at JXB online), indicating that the Y2H library is enriched in salt-induced cDNAs. The prey proteins obtained here may represent salt-induced responses.
In addition to the function of SKD1 in the ESCRT machinery, several reports have suggested that SKD1 could mediate ESCRT-independent functions in yeast and mammalian cells. Du and co-workers (2013) found that ESCRT-III components are not required for the SKD1-mediated cholesterol transport in HeLa cells. Logg et al. (2008) found functional separation of SKD1 from ESCRT-III by analysis of salt tolerance in yeast. The fact that the SKD1 loss-of-function mutant is lethal in higher plants but not in yeast suggests that plant SKD1 may have other roles in addition to ESCRT-mediated MVB sorting. Further studies are necessary to elucidate whether SKD1–CPN1–SnRK1 interactions are salt stress specific and/or ESCRT independent.
CPN1-mediated SKD1 recruitment to the plasma membrane and regulation of ATPase activity
The E3 ligase McCPN1 interacts with McSKD1 via the copine domain and catalyses the ubiquitination of McSKD1 via the RING domain. Tomsig et al. (2003) showed that the copine domains of three human copine proteins have the capacity to recognize a set of proteins containing coiled-coil motifs. The McSKD1 N-terminus also contains a coiled-coil motif, and deletion of this motif greatly reduced K+ transport ability in a yeast complementation assay (Jou et al., 2004). The domain–domain interaction assay confirms that the N-terminal region of McSKD1 interacts with the copine domain of McCPN1 (Fig. 4). The interaction of SKD1 with McCPN1, a protein with N-myristoylation modification, may result in recruitment of McSKD1 to membrane surfaces and the regulation of the ATPase activity of McSKD1 by ubiquitination.
Ubiquitinated proteins are subjected to proteosomal degradation or preceded by other cellular events, such as changes in cellular localization or activity (Welchman et al., 2005). There are two reasons why it is suggested here that rather than regulating the abundance of McSKD1, ubiquitinated McSKD1 may change the ATPase activity of McSKD1. First, the accumulation of McSKD1 increased during salt stress (Jou et al., 2006), making it unlikely to be targeted for degradation. Secondly, it was found that the ATPase activity of McSKD1 is reduced by binding to ADP, suggesting that the ATPase activity of McSKD1 is regulated through the ATP- and ADP-binding states (Jou et al., 2006). Ubiquitination of McSKD1 may favour McSKD1 towards the active ATP-binding state or it may affect its participation in other functions. Further investigation is necessary to establish the effect of ubiquitination of McSKD1.
Arabidopsis RING-type copines, RGLG1 and RGLG2, participate in hormone-mediated plant development. In the rglg1 rglg2 mutant, the polarized distribution of the auxin effluxer PIN1 on the plasma membrane was greatly reduced, resulting in decreased auxin transport capacity (Yin et al., 2007). Spitzer et al. (2009) identified that auxin carriers PIN1, PIN2, and AUX1 are potential ESCRT cargo proteins of the MVB sorting pathway. Previous work showed that transgenic Arabidopsis with reduced SKD1 expression had altered root architecture due to reduced auxin transport capacity (Ho et al., 2010). Because SKD1 is a potential substrate of McCPN1, it is suggested that RING-type copine modulates ESCRT-mediated auxin transport via ubiquitination of SKD1 during normal plant growth and development.
The dynamic cellular localization of RING-type copine under stress is an interesting phenomenon. Cheng and co-workers (2012) observed a salt stress-induced translocation of RGLG2 from the plasma membrane into the nucleus to promote degradation of a drought-related transcription factor. Translocation of a lipid-linked protein from the plasma membrane to the nucleus requires specific vesicle trafficking machinery. Nevertheless, additional proof is necessary to show whether SKD1 is involved in the stress-induced protein trafficking process of McCPN1. Under salt stress, the interaction between McSKD1 and McCPN1 could be mutually beneficial in that McCPN1 promotes changes in activity of McSKD1 by ubiquitination, and subsequently the ubiquitinated McSKD1 facilitates the translocation of McCPN1.
Phosphorylation of SKD1 by SnRK1 promotes the re-distribution of SKD1
Yeast Snf1 plays critical roles in regulation of carbon metabolism and in response to stress (Sanz, 2003). Yeast Snf1 is the catalytic subunit of a heterotrimeric complex and is activated by upstream kinases in response to high salinity. Activated Snf1 controls the expression of ENA1, the major sodium pump in yeast, through a pathway that is distinct from the regulation of glucose metabolism (Ye et al., 2006, 2008). Plant SnRK1s also form heterotrimeric complexes resulting in transcriptional regulation of many genes in response to metabolic or stress signals (Ferrando et al., 2001) as well as post-translational modifications of certain enzymes. Purified 58kDa spinach SnRK1s phosphorylate 3-hydroxy-3-methylglutaryl-coenzyme A reductase, nitrate reductase, and sucrose phosphate synthase in vitro (Sugden et al., 1999b). It was found here that McSnRK1 exhibited kinase activity and phosphorylated McSKD1 in vitro, but with low catalytic efficiency. The idea that full activation of McSnRK1 may require an upstream kinase or the formation of an enzyme complex requires further investigation.
The C-terminal region of the SnRK1 family contains UBA and KA1 domains. The KA1 domain acts as an autoinhibitory domain of the N-terminal catalytic domain and is involved in protein localization. Mammalian MARKs (microtubule affinity-regulating kinases) are present in both the cytosol and the plasma membrane, and require the KA1 domain for plasma membrane localization (Göransson et al., 2006). Changes in subcellular localization were also observed in the pEg3 protein when the C-terminal KA1 domain was deleted (Chartrain et al., 2006). The fact that the KA1 domain of McSnRK1 interacts with domains of McSKD1 (Fig. 4A) and that these two proteins show strong interaction in vivo (Fig. 6A) leads to the suggestion that salt-induced re-distribution to the plasma membrane is mediated by the KA1 domain of McSnRK1 and is transported through microtubules via the MIT domain of McSKD1.
The UBA domain is commonly found in proteins involved in ubiquitination (Hofmann and Bucher, 1996), and implicated in binding to mono- or polyubiquitin (Raasi et al., 2004; Swanson et al., 2006). The present results showed that both McSKD1 and McCPN1 interacted more strongly with the UK (UBA+KA1) region than with the KA1 domain alone. In addition, only the complete C-terminal half of McSnRK1 (i.e. the UK region), but not the KA1 domain, exhibited strong interaction in the Y3H experiment (Fig. 4). These results indicate that the UBA domain of McSnRK1 facilitates the interaction with its protein partners and is necessary for the formation of the McSKD1–McCPN1–McSnRK1 ternary complex.
In conclusion, this article describes the interaction of the salt-induced protein McSKD1 with two enzymes, McCPN1 and McSnRK1, and potential modifications by ubiquitination and phosphorylation. These three proteins can further form a ternary complex through distinct domains. McSKD1 co-localizes with McCPN1 and McSnRK1 in the plasma membrane and cytosol, and salt induces re-distribution of McSKD1 towards the plasma membrane. This study points to two new lines of connection to the existing complex network of protein interactions for salt tolerance in higher plants.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. In vivo interaction between McSKD1 and partial sequences of RING-type E3 ligase (McCPN1) and SNF1-related protein kinase (McSnRK1).
Figure S2. Domain alignments and complementation assay of McSnRK1 to yeast Snf1 and related protein kinases.
Figure S3. Pairwise yeast two-hybrid analysis of domain–domain interaction between McSKD1, McCPN1, and McSnRK1.
Figure S4. In vitro ubiquitination reaction by McCPN1 using McSnRK1 as a substrate.
Figure S5. The effect of McSnRK1 on McSKD1 ubiquitination.
Figure S6. Localization of McCPN1–YFP in Arabidopsis protoplasts.
Figure S7. Co-localization of McSKD1 with the plasma membrane marker HKT1 using double labelling immunofluorescence in ice plant root tip cells.
Figure S8. Proposed model for the formation of a McSKD1–McSnRK1–McCPN1 ternary complex under high salinity.
Table S1. Putative McSKD1-interacting proteins identified from the salt-stressed ice plant cDNA library.
Table S2. Primers used in 5’-RACE, 3’-RACE, and RT–PCR of McCPN1 and McSnRK1.
Acknowledgements
We thank Dr Judy Callis, University of California, Davis for providing (His)6-AtUBC8 and GST–CIP8 constructs, Drs Wan-Sheng Sunny Lo and Long-Chi Kelvin Wang, Academic Sinica, Taiwan for providing the Δsnf1 yeast mutant and yeast expression vector, and Drs Ming-Hsiun Hsieh and Ju-Jiun Chen, Academic Sinica, Taiwan for their technical assistance with the in vitro kinase assay. We appreciate the help of Yung-Chung Wang with RACE, I-Chun Kuo for yeast complementation, Jia-Fang Ho for tissue culture, and Ting-Ting Yang for graphic illustration. We are grateful to the Centers of Biotechnology and Nanoscience and Nanotechnology, NCHU, Taiwan for assistance in the DNA sequencing and confocal microscopy. This work was supported by the National Science Council, Taiwan grant 98-2311-B-005-003-MY3 to HEY.
Glossary
Abbreviations:
- BiFC
bimolecular fluorescence complementation
- EBC
epidermal bladder cell
- Co-IP
co-immunoprecipitation
- NEM
N-ethylmaleimide
- Y2(3)H
yeast two- (three-) hybrid.
References
- Adams P, Nelson DE, Yamada S, Chmara W, Jensen RG, Bohnert HJ, Griffiths H. 1998. Growth and development of Mesembryanthemum crystallinum (Aizoaceae). New Phytologist 138, 171–190. [DOI] [PubMed] [Google Scholar]
- Agarie S, Shimoda T, Shimizu Y, Baumann K, Sunagawa H, Kondo A, Ueno O, Nakahara T, Nose A, Cushman JC. 2007. Salt tolerance, salt accumulation, and ionic homeostasis in an epidermal bladder-cell-less mutant of the common ice plant Mesembryanthemum crystallinum . Journal of Experimental Botany 58, 1957–1967. [DOI] [PubMed] [Google Scholar]
- Alderson A, Sabellit PA, Dickinsont JR, Coleo D, Richardson M, Kreis M, Shewryt PR, Halford NG. 1991. Complementation of snfl, a mutation affecting global regulation of carbon metabolism in yeast, by a plant protein kinase cDNA. Proceedings of the National Academy of Sciences, USA 88, 8602–8605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi I, Davies B, Dimaano C, Payne J, Eckert D, Babst M, Katzmann DJ. 2006. Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1. Journal of Cell Biology 172, 705–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Katzmann DJ, Estepa EJ, Meerloo T, Emr SD. 2002. ESCRT-III: an endosome-associated heterooligomeric protein complex required for MVB sorting. Developmental Cell 3, 271–282. [DOI] [PubMed] [Google Scholar]
- Babst M, Wendland B, Estepa EJ, Emr SD. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO Journal 17, 2982–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin C, de Franco PO, Bourbousse C, Chaignepain S, Schmitter JM, Vavasseur A, Giraudat J, Barbier-Brygoo H, Thomine S. 2006. Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant Physiology 141, 1316–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M. 1999. Glucose repression in yeast. Current Opinion in Microbiology 2, 202–207. [DOI] [PubMed] [Google Scholar]
- Chartrain I, Couturier A, Tassan JP. 2006. Cell cycle dependent cortical localization of pEg3 protein kinase in Xenopus and human cells. Biology of the Cell 98, 253–263. [DOI] [PubMed] [Google Scholar]
- Cheng MC, Hsieh EJ, Chen JH, Chen HY, Lin TP. 2012. Arabidopsis RGLG2, functioning as a RING E3 ligase, interacts with AtERF53 and negatively regulates the plant drought stress response. Plant Physiology 158, 363–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coello P, Hey SJ, Halford NG. 2011. The sucrose non-fermenting-1-related (SnRK) family of protein kinases: potential for manipulation to improve stress tolerance and increase yield. Journal of Experimental Botany 62, 883–893. [DOI] [PubMed] [Google Scholar]
- Du X, Kazim AS, Dawes IW, Brown AJ, Yang H. 2013. The AAA ATPase VPS4/SKD1 regulates endosomal cholesterol trafficking independently of ESCRT-III. Traffic 14, 107–119. [DOI] [PubMed] [Google Scholar]
- Ferrando A, Koncz-Kalman Z, Farrás R, Tiburcio A, Schell J, Koncz C. 2001. Detection of in vivo protein interactions between Snf1-related kinase subunits with intron-tagged epitope-labelling in plant cells. Nucleic Acids Research 29, 3685–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Zhu JK. 2009. An autophosphorylation site of the protein kinase SOS2 is important for salt tolerance in Arabidopsis. Molecular Plant 2, 183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11, 355–360. [DOI] [PubMed] [Google Scholar]
- Gong D, Gong Z, Guo Y, Zhu JK. 2002a. Expression, activation, and biochemical properties of a novel Arabidopsis protein kinase. Plant Physiology 129, 225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong D, Guo Y, Jagendorf AT, Zhu JK. 2002b. Biochemical characterization of the Arabidopsis protein kinase SOS2 that functions in salt tolerance. Plant Physiology 130, 256–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göransson O, Deak M, Wullschleger S, Morrice NA, Prescott AR, Alessi DR. 2006. Regulation of the polarity kinases PAR-1/MARK by 14-3-3 interaction and phosphorylation. Journal of Cell Science 119, 4059–4070. [DOI] [PubMed] [Google Scholar]
- Guo Y, Halfter U, Ishitani M, Zhu JK. 2001. Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. The Plant Cell 13, 1383–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas TJ, Sliwinski MK, Martínez DE, Preuss M, Ebine K, Ueda T, Nielsen E, Odorizzi G, Oteguia MS. 2007. The Arabidopsis AAA ATPase SKD1 is involved in multivesicular endosome function and interacts with its positive regulator LYST-INTERACTING PROTEIN5. The Plant Cell 19, 1295–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford NG, Hardie DG. 1998. SNF1-related protein kinases: global regulators of carbon metabolism in plants? Plant Molecular Biolology 37, 735–748. [DOI] [PubMed] [Google Scholar]
- Halford NG, Hey S, Jhurreea D, Laurie S, McKibbin RS, Paul M, Zhang YJ. 2003. Metabolic signalling and carbon partitioning: role of Snf1-related (SnRK1) protein kinase. Journal of Experimental Botany 54, 467–475. [DOI] [PubMed] [Google Scholar]
- Hanson PL, Whiteheart SW. 2005. AAA+ proteins: have engine, will work. Nature Reviews Molecular Cell Biology 6, 519–529. [DOI] [PubMed] [Google Scholar]
- Herman EM, Lamb CJ. 1992. Arabinogalactan-rich glycoproteins are localized on the cell surface and in intravacuolar multivesicular bodies. Plant Physiology 98, 264–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho LW, Yang TT, Shieh SS, Edwards GE, Yen HE. 2010. Reduced expression of a vesicle trafficking-related ATPase SKD1 decreases salt tolerance in Arabidopsis. Functional Plant Biology 37, 962–973. [Google Scholar]
- Hofmann K, Bucher P. 1996. The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends in Biochemical Sciences 21, 172–173. [PubMed] [Google Scholar]
- Hong JK, Choi HW, Hwang IS, Hwang BK. 2007. Role of a novel pathogen-induced pepper C3-H-C4 type RING-finger protein gene, CaRFP1, in disease susceptibility and osmotic stress tolerance. Plant Molecular Biology 63, 571–588. [DOI] [PubMed] [Google Scholar]
- Hrabak EM, Chan CWM, Gribskov M, et al. 2003. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiology 132, 666–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Reggiori F, Klionsky DJ. 2007. The transmembrane domain of acid trehalase mediates ubiquitin-independent multivesicular body pathway sorting. Molecular Biology of the Cell 18, 2511–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Hanson PI. 2010. Membrane budding and scission by the ESCRT machinery: it’s all in the neck. Nature Reviews Molecular Cell Biology 8, 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jossier M, Bouly JP, Meimoun P, Arjmand A, Lessard P, Hawley S, Hardie DG, Thomas M. 2009. SnRK1 (SNF1-related kinase 1) has a central role in sugar and ABA signaling in Arabidopsis thaliana . The Plant Journal 59, 316–328. [DOI] [PubMed] [Google Scholar]
- Jou Y, Chiang CP, Jauh GY, Yen HE. 2006. Functional characterization of ice plant SKD1, an AAA-type ATPase associated with the endoplasmic reticulum–Golgi network, and its role in adaptation to salt stress. Plant Physiology 141, 135–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jou Y, Chou PH, He M, Hung Y, Yen HE. 2004. Tissue-specific expression and functional complementation of a yeast potassium-uptake mutant by a salt-induced ice plant gene McSKD1 . Plant Molecular Biology 54, 881–893. [DOI] [PubMed] [Google Scholar]
- Jou Y, Wang YL, Yen HE. 2007. Vacuolar acidity, protein profile, and crystal composition of epidermal bladder cells of the halophyte Mesembryanthemum crystallinum . Functional Plant Biology 34, 353–359. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Babst M, Emr SD. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106, 145–155. [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J, Friml J. 2008. Polar targeting and endocytic recycling in auxin-dependent plant development. Annual Review of Cell and Developmental Biology 24, 447–473. [DOI] [PubMed] [Google Scholar]
- Lid SE, Gruis D, Jung R, Lorentzen JA, Ananiev E, Chamberlin M, Niu X, Meeley R, Nichols SE, Olsen OA. 2002. The defective kernel 1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proceedings of the National Academy of Sciences, USA 99, 5460–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ishitani M, Halfter U, Kim CS, Zhu JK. 2000. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proceedings of the National Academy of Sciences, USA 97, 3730–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logg K, Warringer J, Hashemi SH, Käll M, Blomberg A. 2008. The sodium pump Ena1p provides mechanistic insight into the salt sensitivity of vacuolar protein sorting mutants. Biochimica et Biophysica Acta 1783, 974–984. [DOI] [PubMed] [Google Scholar]
- Obita T, Saksena S, Ghazi-Tabatabai S, Gill DJ, Perisic O, Emr SD, Williams RL. 2007. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature 449, 735–739. [DOI] [PubMed] [Google Scholar]
- Raasi S, Orlov I, Fleming KG, Pickart CM. 2004. Binding of polyubiquitin chains to ubiquitin-associated (UBA) domains of HHR23A. Journal of Molecular Biology 341, 1367–1379. [DOI] [PubMed] [Google Scholar]
- Redding K, Brickner JH, Marschall LG, Nichols JW, Fuller RS. 1996. Allele-specific suppression of a defective trans-Golgi network (TGN) localization signal in Kex2p identifies three genes involved in localization of TGN transmembrane proteins. Molecular and Cellular Biology 16, 6208–6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodahl LM, Stuffers S, Lobert VH, Stenmark H. 2009. The role of ESCRT proteins in attenuation of cell signalling. Biochemical Society Transactions 37, 137–142. [DOI] [PubMed] [Google Scholar]
- Ruotolo R, Marchini G, Ottonello S. 2008. Membrane transporters and protein traffic networks differentially affecting metal tolerance: a genomic phenotyping study in yeast. Genome Biology 9, R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz P. 2003. Snf1 protein kinase: a key player in the response to cellular stress in yeast. Biochemical Society Transactions 31, 178–181. [DOI] [PubMed] [Google Scholar]
- Shahriari M, Keshavaiah C, Scheuring D, Sabovljevic A, Pimpl P, Häusler RE, Hülskamp M, Schellmann S. 2010. The AAA-type ATPase AtSKD1 contributes to vacuolar maintenance of Arabidopsis thaliana . The Plant Journal 64, 71–85. [DOI] [PubMed] [Google Scholar]
- Shahriari M, Richter K, Keshavaiah C, Sabovljevic A, Huelskamp M, Schellmann S. 2011. The Arabidopsis ESCRT protein–protein interaction network. Plant Molecular Biology 76, 85–96. [DOI] [PubMed] [Google Scholar]
- Shestakova A, Hanono A, Drosner S, Curtiss M, Davies BA, Katzmann DJ, Babst M. 2010. Assembly of the AAA ATPase Vps4 on ESCRT-III. Molecular Biology of the Cell 21, 1059–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S, Kimpler LA, Hanson PI. 2007. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic 8, 1068–1079. [DOI] [PubMed] [Google Scholar]
- Shim S, Merrill SA, Hanson PI. 2008. Novel interactions of ESCRT-III with LIP5 and VPS4 and their implications for ESCRT-III disassembly. Molecular Biology of the Cell 19, 2661–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer C, Reyes FC, Buono R, Sliwinski MK, Haas TJ, Otegui MS. 2009. The ESCRT-related CHMP1A and B proteins mediate multivesicular body sorting of auxin carriers in Arabidopsis and are required for plant development. The Plant Cell 21, 749–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Hauksdóttir H, Troy A, Herschleb J, Kraft E, Callis J. 2005. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiology 137, 13–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuchell-Brereton MD, Skalicky JJ, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI. 2007. ESCRT-III recognition by VPS4 ATPases. Nature 449, 740–744. [DOI] [PubMed] [Google Scholar]
- Sugden C, Crawford RM, Halford NG, Hardie DG. 1999a. Regulation of spinach SNF1-related (SnRK1) kinases by protein kinases and phosphatases is associated with phosphorylation of the T loop and is regulated by 5’-AMP. The Plant Journal 19, 433–439. [DOI] [PubMed] [Google Scholar]
- Sugden C, Donaghy PG, Halford NG, Hardie DG. 1999b. Two SNF1-related protein kinases from spinach leaf phosphorylate and inactivate 3-hydroxy-3-methylglutaryl-coenzyme A reductase, nitrate reductase, and sucrose phosphate synthase in vitro . Plant Physiology 120, 257–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson K, Hicke L, Radhakrishnan I. 2006. Structural basis for monoubiquitin recognition by the Ede1 UBA domain. Journal of Molecular Biology 358, 713–724. [DOI] [PubMed] [Google Scholar]
- Tian Q, Olsen L, Sun B, et al. 2007. Subcellular localization and functional domain studies of DEFECTIVE KERNEL1 in maize and Arabidopsis suggest a model for aleurone cell fate specification involving CRINKLY4 and SUPERNUMERARY ALEURONE LAYER1. The Plant Cell 19, 3127–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomsig JL, Snyder SL, Creutz CE. 2003. Identification of targets for calcium signaling through the copine family of proteins. Characterization of a coiled-coil copine-binding motif. Journal of Biological Chemistry 278, 10048–10054. [DOI] [PubMed] [Google Scholar]
- Toroser D, Plaut Z, Huber SC. 2000. Regulation of a plant SNF1-related protein kinase by glucose 6-phosphate. Plant Physiology 123, 403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajjhala PR, Catchpoole E, Nguyen CH, Kistler C, Munn AL. 2007. Vps4 regulates a subset of protein interactions at the multivesicular endosome. FEBS Journal 274, 1894–1907. [DOI] [PubMed] [Google Scholar]
- Vajjhala PR, Wong JS, To HY, Munn AL. 2006. The beta domain is required for Vps4p oligomerization into a functionally active ATPase. FEBS Journal 273, 2357–2373. [DOI] [PubMed] [Google Scholar]
- Watad A-EA, Reinhold L, Lerner HR. 1983. Comparison between a stable NaCl-selected Nicotiana cell line and the wild type: K+, Na+, and proline pools as a function of salinity. Plant Physiology 73, 624–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchman RL, Gordon C, Mayer RJ. 2005. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nature Reviews Molecular Cell Biology 6, 599–609. [DOI] [PubMed] [Google Scholar]
- Ye T, Elbing K, Hohmann S. 2008. The pathway by which the yeast protein kinase Snf1p controls acquisition of sodium tolerance is different from that mediating glucose regulation. Microbiology 154, 2814–2826. [DOI] [PubMed] [Google Scholar]
- Ye T, García-Salcedo R, Ramos J, Hohmann S. 2006. Gis4, a new component of the ion homeostasis system in the yeast Saccharomyces cerevisiae . Eukaryotic Cell 5, 1611–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen HE, Wu SM, Hong YH, Yen SK. 2000. Isolation of 3 salt-induced low-abundance cDNAs from light-grown calli of Mesembryanthemum crystallinum by suppression subtractive hybridization. Physiologia Plantarum 110, 402–409. [Google Scholar]
- Yeo SC, Xu L, Ren J, Boulton VJ, Wagle MD, Liu C, Ren G, Wong P, Zahn R, Sasajala P. 2003. Vps20p and Vta1p interact with Vps4p and function in multivesicular body sorting and endosomal transport in Saccharomyces cerevisiae . Journal of Cell Science 116, 3957–3970. [DOI] [PubMed] [Google Scholar]
- Yin XJ, Volk S, Ljung K, et al. 2007. Ubiquitin lysine 63 chain forming ligases regulate apical dominance in Arabidopsis. The Plant Cell 19, 1898–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.