Abstract
Sirtuins are NAD+-dependent protein deacetylases regulating metabolism, stress responses and ageing processes. Among the seven mammalian Sirtuins, Sirt1 is the physiologically best-studied isoform. It regulates nuclear functions such as chromatin remodelling and gene transcription, and it appears to mediate beneficial effects of a low calorie diet which can partly be mimicked by the Sirt1 activating polyphenol resveratrol. The molecular details of Sirt1 domain architecture and regulation, however, are little understood. It has a unique N-terminal domain and CTD (C-terminal domain) flanking a conserved Sirtuin catalytic core and these extensions are assumed to mediate Sirt1-specific features such as homo-oligomerization and activation by resveratrol. To analyse the architecture of human Sirt1 and functions of its N- and C-terminal extensions, we recombinantly produced Sirt1 and Sirt1 deletion constructs as well as the AROS (active regulator of Sirt1) protein. We then studied Sirt1 features such as molecular size, secondary structure and stimulation by small molecules and AROS. We find that Sirt1 is monomeric and has extended conformations in its flanking domains, likely disordered especially in the N-terminus, resulting in an increased hydrodynamic radius. Nevertheless, both termini increase Sirt1 deacetylase activity, indicating a regulatory function. We also find an unusual but defined conformation for AROS protein, which fails, however, to stimulate Sirt1. Resveratrol, in contrast, activates the Sirt1 catalytic core independent of the terminal domains, indicating a binding site within the catalytic core and suggesting that small molecule activators for other isoforms might also exist.
Keywords: activation, AROS, oligomerization, resveratrol, Sirtuin, structure
Abbreviations: AROS, active regulator of Sirt1; DTT, dithiothreitol; FdL, Fluor-de-Lys; PDE, phosphodiesterase; SEC, size-exclusion chromatography; STAC, Sirtuin-activating compound; SV, sedimentation velocity
INTRODUCTION
Sirtuins form an evolutionarily conserved family of protein deacetylases [1]. In contrast with other deacetylase families, they hydrolyse one NAD+ for each lysine residue they deacetylate, rendering them metabolic sensors [2,3]. Sirtuins contribute to metabolic regulation, stress responses and various ageing-related pathologies [4,5]. Of the seven mammalian Sirtuin isoforms, Sirt1 is the closest homologue to the family founding member, yeast Sir2. Sirt1 is mainly located in the nucleus and deacetylates various targets, such as histones and transcription factors [6]. Thereby, Sirt1 regulates, e.g., cellular responses to stress factors and nutrient availability, and it appears to mediate beneficial health effects of caloric restriction in higher organisms [3,4]. Sirt1 is thus considered an attractive drug target for metabolic and ageing-related diseases [7].
Sirtuins are defined by an evolutionarily conserved, ~250 residue large catalytic domain [1], which comprises a Rossmann fold subdomain and a smaller Zn2+-binding subdomain [8,9]. The acetyllysine-containing polypeptide substrate occupies a cleft between these two subdomains. The catalytic core comprises the whole enzyme for some Sirtuins, but is flanked by N- and C-terminal extensions in other family members [1,10]. For example, an extended N-terminal loop mediates a monomer–trimer equilibrium of yeast Hst2 in the low to mid micromolar range [11], and trimer formation was also suggested for Sirt1 based on SEC (size-exclusion chromatography) [12]. In human Sirt3, an ~120 residue N-terminal extension acts as autoinhibitory targeting sequence [10,13,14]. Most other human isoforms have smaller extensions, mostly of unknown function [2], whereas Sirt1 has even larger extensions of ~230 residues at the N-terminus and ~220 residues at the C-terminus, respectively. The Sirt1 N-terminus was reported to mediate recruitment of the substrate histone H1 [12], similar to Sirt6 recruitment to chromatin through its relatively short extensions [15]. Both, N- and C-terminus, increase Sirt1 catalytic activity, and a short amino acid stretch within the C-terminal part, which stably associates with the catalytic core, appears to be of particular importance for Sirt1 activity [16,17]. A variety of posttranslational modifications in the Sirt1 extensions have been reported, some of them with a confirmed effect on the enzyme's activity [18] indicating a function in dynamic regulation of Sirt1 activity for these regions. Consistently, the N-terminal extension was suggested to mediate the regulatory interaction with the protein AROS (active regulator of Sirt1) [19] and pharmacological activation of Sirt1. Mammalian Sirt1 and its yeast homologue Sir2 can be stimulated in vitro by resveratrol and other STACs (Sirtuin-activating compounds) [20,21], and their modulation appears to induce beneficial health and lifespan effects [22–24]. The Sirt1 N-terminal extension was reported to be essential for STAC-dependent activation [20] and resveratrol effects on Sirt1 appear to be substrate dependent [25,26], but further mechanistic insights are lacking. Since the compound affects a variety of other proteins [24,27], insights into its activation mechanism and the development of improved compounds are urgently required [9].
Here, we study the functions of the N- and C-terminal extensions in human Sirt1. We test their role in Sirt1 oligomerization, revealing that the protein is monomeric in solution and that its large hydrodynamic radius is likely caused by extended termini, especially at the N-terminus. We further analyse Sirt1 modulation by several putative Sirt1 regulators, showing that the regulator protein AROS has an unusual conformation and that the Sirt1 catalytic domain is sufficient for modulation by resveratrol, providing the basis for further mechanistic studies on Sirt1 regulation.
MATERIALS AND METHODS
Chemicals
All chemicals were obtained from Sigma if not stated differently. SRT1720 was from Cayman Europe, and peptides were synthesized by GL Biochem.
Cloning, recombinant expression and purification of human Sirt1 constructs
Full length and deletion constructs of Sirt1 were cloned between the NdeI and XhoI sites of pET15b vector (Merck) resulting in a construct with N-terminal His-tag and a thrombin protease cleavage site. Sirt1 constructs were expressed in Escherichia coli Rosetta 2 (DE3) cells (Merck) in LB (Luria–Bertani)/ampicillin/chloramphenicol medium at 37°C until the A600 had reached 0.5–0.7. Expression was induced with 0.1 mM IPTG (isopropyl β-D-thiogalactopyranoside), and cells were incubated overnight at 22°C. Cells were harvested by centrifugation, resuspended in lysis buffer (50 mM Tris, pH 7.5, 300 mM NaCl and 0.2 mM PMSF) and disrupted using a Microfluidizer (Microfluidics), and cell debris was removed by 45 min centrifugation at 18000 rev./min at 4°C in an Avanti J-26XP centrifuge with a JA-30.50 Ti rotor (Beckman Coulter). The supernatant was incubated with talon resin (Clontech) for 1 h at 4°C and the resin was then incubated for 30 min at 4°C with 20 μg/ml (bed volume) of bovine RNase A and 10 μg/ml of bovine DNase in 25 mM Tris, pH 7.5, 20 mM NaCl, 5 mM MgCl2 and 0.2 mM PMSF. The column was washed with 20 bed volumes of wash buffer (lysis buffer containing 10 mM imidazole) followed by elution with lysis buffer containing 150 mM imidazole. The eluted protein was concentrated and applied to a pre-equilibrated Superdex200 16/60 SEC column (GE Healthcare) and eluted in buffer A [25 mM HEPES, pH 7.5, 100 mM KCl, 2 mM DTT (dithiothreitol), 0.1 mM PMSF and 0.5 mM EDTA]. The protein was bound to a HiTrapQ anion exchange column (GE Healthcare) in buffer A and eluted in a gradient to buffer A supplemented with 400 mM KCl. Fractions were analysed by SDS/PAGE, and Sirt1 fractions were pooled and transferred into storage buffer (25 mM HEPES, pH 7.5, 100 mM KCl and 2 mM DTT) using a NAP column (GE Healthcare), aliquoted, snap-frozen in liquid nitrogen and stored at −80°C.
SEC
Analytical SEC runs of Sirt1 constructs were performed on a Superose 12 10/300 GL column (24 ml bed volume; GE Healthcare) pre-equilibrated in Sirt1 storage buffer. Size exclusion standards were purchased from Biorad and diluted in storage buffer before applying to the Superose 12 column.
Analytical ultracentrifugation
Experiments were carried out in a Beckman-Coulter XL-A analytical ultracentrifuge in 25 mM Hepes, pH 7.5, 0.1 M KCl and 0.5 mM DTT using an An-50 Ti rotor. Concentration profiles were measured at 280 or 230 nm depending on protein concentration using the UV/Vis scanning optics of the centrifuge. SV (sedimentation velocity) experiments were performed at a rotor speed of 40000 rev./min at 20°C using standard 3 or 12 mm centrepieces filled with 100 or 400 μl sample, respectively. The program package SEDFIT [28] was used for converting measured concentration profiles into diffusion-corrected differential sedimentation coefficient [c(s)] distributions and for determination of the molecular mass from diffusion broadening of the sedimenting boundary. Buffer density and viscosity and partial specific volumes of the proteins were determined with the program SEDNTERP [29] and used for correcting experimental sedimentation coefficients to s20,w. For non-associating systems, when a decrease of the sedimentation coefficient with increasing protein concentration c was observed, s20,w was corrected by linear extrapolation to c=0 [30]. Sedimentation equilibrium experiments were conducted at 4°C and rotor speeds of 6000, 9000, 12000 and 18000 rev./min as described previously [31], determination of molar mass was performed using a model of a single species [32]. Protein concentrations were determined spectrophotometrically using absorption coefficients at 280 nm calculated from amino acid composition [33].
EM (electron microscopy)
Purified Sirt1 was diluted as required in buffer (20 mM Tris/HCl, pH 7.8, 150 mM NaCl and 2 mM DTT) and adsorbed for 1 min to glow-discharged carbon-coated grids. The grids were washed and negatively stained with 0.07% uranyl formate (SPI Supplies) and imaged with a Jeol JEM1400 operated at 120 kV. Electron micrographs were recorded at a magnification of ×50000 on Kodak SO-163 films.
CD spectra
Protein samples were diluted in 2.5 mM HEPES, pH 7.5, 10 mM KCl and 0.2 mM DTT to a concentration of 9.5 μM (full-length Sirt1) or 10 μM (AROS), respectively. CD spectra were then measured at 15°C using a Jasco J600A CD spectrophotometer. Spectra were recorded in continuous scanning mode at 100 nm/min between 260 nm and 180 nm with 1 nm bandwidth, an accumulation of eight scans per spectrum and a response time of 1 s, and they were baseline corrected using the corresponding buffer spectrum.
Peptide deacetylation assays
Deacetylase activity of Sirt1 against the fluorogenic, p53-derived FdL-1 (Fluor-de-Lys-1) peptide RHK-acetylK-fluorophor was tested with a commercial Sirt1 assay kit (Enzo Life Sciences). Reactions were incubated at 37°C with 0.5 μg Sirt1, 25 μM fluorogenic peptide and 0.25 mM NAD+. Detection of a reaction product was performed according to the instructions of the manufacturer. For assays with a non-fluorogenic, non-modified substrate peptide, deacetylation of 100 μM of a p53-derived peptide (RHK-acetylK-LMFK) was monitored in a coupled, continuous enzymatic assay at 25°C and 1 mM NAD+ as described before [34,35]. In control reactions and blanks, the same amount of solvent used for its stock solution or compound analysed, respectively, was included. All assays were done in triplicate, and results shown are representatives of at least two independent replications.
Homology modelling
The catalytic core of human Sirt1 was homology modelled using the Phyre2 [36] server at http://www.sbg.bio.ic.ac.uk/phyre2.
RESULTS
Apparent size of Sirt1 and Sirt1 deletion constructs
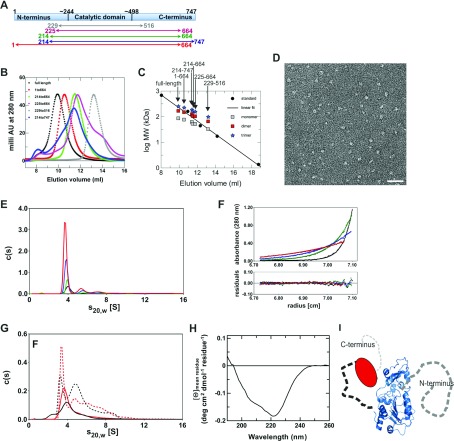
Sirt1 was suggested to form trimers [12], while Sirtuin catalytic domains are mainly monomeric (see the Discussion section), and protein oligomerization often contributes to regulation [37]. To analyse a potential regulatory function of trimer formation and the role of the N- and C-terminal extensions in Sirt1 oligomerization and regulation, we recombinantly produced full-length protein and deletion variants of human Sirt1 (Figure 1A). The apparent molecular masses were then analysed through SEC (Figure 1B) and compared with molecular mass standards (Figure 1C). This analysis indicated a size of ~230 kDa for full-length Sirt1, which would fit well to a trimer of the 84 kDa protein, consistent with the apparent formation of trimers detected by SEC, as reported before [12]. Truncation of the C-terminal region (Sirt1-1-664, calculated mass 75 kDa) resulted in elution comparable with a ~150 kDa protein, close to an apparent dimer, whereas truncation of the N-terminus (Sirt1-214-747, calculated mass 62 kDa) resulted in elution like a 90 kDa protein, between the monomer and dimer mass. Removing both extensions (Sirt1-214-664 and Sirt1-225-665, calculated masses 53 and 52 kDa, respectively) gave proteins eluting comparable with 70–80 kDa proteins, again a mass between monomer and dimer. A construct completely reduced to the catalytic core, Sirt1-229-516, eluted as expected for the 33 kDa monomer. SEC elution volumes depend not only on the molecular mass but also on the protein's shape [38], with deviation from spherical shape resulting in lower elution volumes corresponding to higher apparent molecular masses. An elution volume expected for a spherical monomer was observed for Sirt1 only when both terminal extensions were removed, indicating that its termini either mediate oligomerizations or cause an elongated, non-spherical shape.
Figure 1. Overall architecture of human Sirt1.
(A) Schematic view of full-length human Sirt1 with tentative borders indicated for the conserved Sirtuin catalytic domain. The Sirt1 deletion constructs used in our studies, which lack N- and/or C-terminal extensions to the catalytic core, are indicated. (B) Elution profiles of full-length human Sirt1 and Sirt1 deletion constructs in SEC experiments. (C) Comparison of the elution volumes of Sirt1 constructs to a molecular mass marker. For all constructs, the calculated sizes of a monomer, dimer and trimer are indicated at the respective elution volume. (D) EM image of negatively stained human Sirt1, showing defined, monomeric particles with a small, roughly spherical core. Scale bar, 40 nm. (E) SV analysis of 0.8 μM full-length Sirt1 monitored at 230 nm (black) and of 6.5 μM (green), 20 μM (blue) and 40 μM (red) full-length Sirt1 monitored at 280 nm. For better comparison all c(s) distributions have been converted to 12 mm path length. (F) Sedimentation equilibrium analysis of 6.5 μM full-length Sirt1. Concentration gradients were measured at 280 nm and 6000 rev./min (red), 9000 rev./min (blue), 12000 rev./min (green) and 18000 rev./min (black) with path length 12 mm. Global fitting using a single species model yielded a molecular mass of 93 kDa (solid curves). The lower panel represents differences between calculated and measured values. (G) SV analysis of N-terminally shortened variants Sirt1-214-747 (red) and Sirt1-214-664 (black) at 1.2 μM (solid lines, 230 nm) and 25 μM or 30 μM (broken lines, 280 nm), respectively. (H) CD spectrum of full-length human Sirt1. (I) Scheme for the architecture of human Sirt1: a conserved Sirtuin catalytic core (shown as a homology model) is flanked by a largely disordered N-terminal extension (right, dark grey) and a C-terminal region folded back on to the catalytic core (left, red) via an extended or disordered linker, possibly followed by a smaller disordered region (light grey).
Sirt1 is a monomeric protein with disordered regions or elongated shape
The observed SEC behaviour could be due to Sirt1 homo-oligomerization or be caused by an extended shape of the protein. To characterize its molecular shape, we negatively stained the full-length Sirt1 protein and analysed it by EM (Figure 1D). We observed small homogenous particles of spherical to elongated shape indicating that Sirt1 is indeed a monomer with a thin and elongated or largely disordered part.
To clarify whether full-length Sirt1 forms oligomers in solution, we analysed Sirt1 in SV experiments in the analytical ultracentrifuge in a concentration range from 0.8 to 40 μM. Calculation of c(s) distributions revealed that the main species of full-length Sirt1, which comprises approximately 80% of the total signal, sedimented with an s20,w of 4.0 S (Figure 1E). If this species were trimeric Sirt1, a frictional ratio of 3.7 would be obtained when compared with a spherical, unhydrated trimer. This value is much higher than expected for an at least partially folded protein [39]. Analysis of the diffusion broadening of the sedimenting boundary yielded a molecular mass of 76 kDa for the main Sirt1 full-length species. Since the calculated monomer mass is 83.9 kDa, full-length Sirt1 predominantly exists as a monomer in solution. A frictional ratio of 1.8 can be calculated from s20,W for this monomer. Since a frictional ratio of 1.1–1.2 would be expected for a hydrated, spherical protein [40], the Sirt1 behaviour deviates substantially from that of a sphere, most probably due to disordered regions and/or a highly elongated shape. The high frictional ratio of full-length Sirt1 is likely the reason why SEC analysis yielded an apparent molecular mass much higher than expected for a monomer. Comparison of c(s) distributions at different protein concentrations revealed that the s-value of the main species slightly decreases with increasing protein concentration (Figure 1E), which is typical for non-associating systems, especially if the protein shows some asymmetry [30]. The fraction of species in c(s) distributions with s-values higher than the main peak did not increase when increasing the protein concentration 50-fold (Figure 1E), indicating that these species represent impurities rather than higher oligomers of Sirt1. We performed further sedimentation equilibrium experiments with full-length Sirt1, which were analysed with a model of a single species and yielded a molecular mass of 93 kDa (Figure 1F). This value is in good agreement with the monomeric Sirt1 mass (83.9 kDa), with the slight deviation most probably being caused by the faster sedimenting impurities observed in SV analysis. We thus conclude that full-length Sirt1 is monomeric in solution and that its shape is non-spherical, likely due to disorder or an extended shape.
SV analysis of the C-terminally truncated Sirt1 construct Sirt1-1-664 in a concentration range from 0.9 to 39 μM yielded very similar results to the full-length protein (Supplementary Figure S1A available at http://www.bioscirep.org/bsr/033/bsr033e037add.htm). The s20,w-value of the main species was slightly reduced to 3.9 S and the frictional ratio, relative to an unhydrated, spherical monomer slightly decreased to 1.7. The C-terminal region thus contributes only slightly to Sirt1 asymmetry. From c(s) analysis a molecular mass of approximately 71 kDa was obtained (calculated mass 75.1 kDa) and a sedimentation equilibrium experiment analysed with a model of a single species yielded a molecular mass of 79 kDa (Supplementary Figure S1B). We therefore conclude that the C-terminally truncated version Sirt1-1-664, like the full-length protein, predominantly forms an elongated or partly unstructured monomer in solution. The N-terminally truncated proteins Sirt1-214-747 and Sirt1-214-664, however, showed a different behaviour. In c(s) distributions several peaks were visible and the weight average c(s) increased with increasing protein concentration (Figure 1G). This result indicates that the N-terminally truncated forms of Sirt1 can undergo a concentration-dependent oligomerization [41] and resembles the observations for yeast Hst2 [11]. An explanation for the different behaviour of N-terminally truncated Sirt1 compared with the full-length enzyme could be an interference of the highly negatively charged patches in the N-terminus with oligomer formation of full-length Sirt1. Such a scenario could have physiological relevance as a regulation mechanism for Sirt1 oligomerization, e.g., by interaction partners binding the Sirt1 N-terminus and relieving its shielding effect. However, the observation that N-terminally truncated Sirt1 forms a variety of different oligomeric species and that oligomer amounts are significant only at higher micromolar concentrations, which likely exceed physiological Sirt1 concentrations, strongly suggest that this oligomerization is non-physiological. Taken together, our results suggest that Sirt1 is monomeric in solution, and that its shape deviates from a sphere due to disorder or an extended shape especially in its N-terminal extension.
In order to determine the secondary structure content of Sirt1 and thereby to distinguish between disorder and an elongated shape for its extensions, we recorded a CD spectrum of the full-length protein (Figure 1H). The spectrum shows typical features for a protein with repetitive secondary structure, with a prominent minimum at 222 nm. However, the signal intensity is weak for an 82 kDa protein, and fitting the spectrum with reference spectra [42] indeed indicated only ~11% α-helical and ~23% β-sheet structure. Since sequence homology strongly indicates the generic Sirtuin catalytic core structure for the central part of Sirt1, we conclude that large parts of the N- and/or C-terminal Sirt1 extensions have no regular secondary structure or are disordered. Consistently, statistical analysis of the Sirt1 sequence using DisEMBL [43] indicates a disordered region covering, with small exceptions, altogether ~35 residues, the Sirt1 N-terminus up to position 175 (Supplementary Figure S2A available at http://www.bioscirep.org/bsr/033/bsr033e037add.htm). A mixture of ordered and disordered regions is predicted C-terminal from residue 530, after the ordered catalytic core. Analogous analysis using GlobPlot [44] resulted in comparable predictions, with disordered regions from 1 to 180 and 660 to 747 (Supplementary Figure S2B). Integrating with our results the recent finding that an ~20 residue stretch around position 640 appears to interact with and activate the catalytic core [17] indicates the following Sirt1 architecture (Figure 1I): a compact catalytic domain, which can be modelled reliably due to its homology to structurally characterized Sirtuins, has a C-terminal extension of a defined structure that folds back, via an extended or disordered linker, on the catalytic domain for activity regulation. The complete N-terminal extension, in contrast, is largely disordered in the absence of an interaction partner.
Testing Sirt1 regulation through AROS
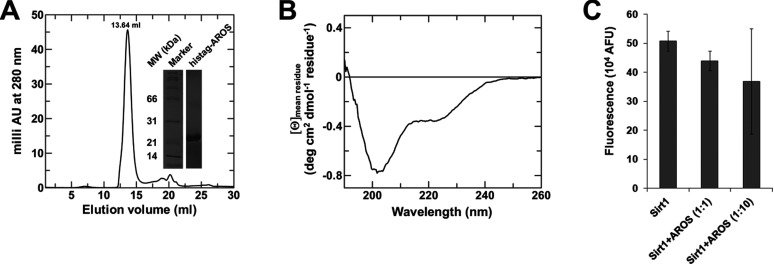
Besides oligomerization, additional Sirt1-specific features supposedly mediated by the N- and C-terminal extensions have been reported, such as regulation by resveratrol (see below), phosphorylation (see [18] for a review), and regulation by the proteins AROS and DBC1 (deleted in breast cancer 1) [19,45]. To test the role of the Sirt1 N-terminus as an AROS binding site (residues 114-217 [19]), which we speculated could stabilize this region and thus enable its structural characterization, we generated recombinant AROS protein with an N-terminal His-tag. The protein was efficiently overexpressed but insoluble. We therefore established a refolding protocol that yielded pure protein eluting as a single peak in SEC with an apparent molecular mass corresponding to a monomer (Figure 2A), indicating that the protein was folded in a globular shape. To test whether the protein has a defined structure, we analysed its secondary structure through CD spectroscopy. The AROS CD spectrum (Figure 2B) indeed shows a deviation from random conformation, yet partially an unusual protein secondary structure. The spectrum can be fitted with a combination of reference spectra [42] beyond 210 nm, indicating ~33% α-helical and ~12% β-sheet structure. The deep minimum at 203 nm and steep increase below 200 nm, however, does not fit typical spectra of α-helix, β-sheet and random conformations. However, this minimum closely resembles spectra of poly(Pro) conformations [46]. The CD spectrum thus suggests not only a folded AROS protein featuring regions in α-helix and β-sheet conformation but also a significant part with poly(Pro) conformation. Adding the recombinant AROS protein to aminomethylcoumarin-peptide based FdL assays [47] on Sirt1 deacetylation activity resulted in a very weak inhibitory effect, which was not statistically significant (Figure 2C), but an activating effect in these assays can be ruled out. However, we cannot exclude the possibility that our AROS protein is partially not folded, since no functional test is available and no natively purified AROS for comparison, although our data strongly suggest proper folding. Furthermore, the recombinant AROS might lack cofactors or further interacting proteins required for binding and/or regulating Sirt1.
Figure 2. Characterization of the Sirt1 activator protein AROS.
(A) Refolding overexpressed AROS resulted in pure protein, judged based on SDS/PAGE (inset), eluting in SEC experiments in the volume expected for a spherical monomer. (B) CD spectrum of refolded AROS protein, featuring minima at 222 nm and 203 nm. (C) FdL-based assay on Sirt1 activity in the presence and absence of AROS protein. Even a 10-fold molar excess of AROS protein did not result in a statistically significant change in Sirt1 activity.
The N-terminal Sirt1 domain is dispensable for modulation by resveratrol
The Sirt1-specific N-terminal domain was reported to be essential for the enzyme's activation by resveratrol [20], but other reports showed resveratrol-dependent activation also for mammalian Sirt5 [48] and yeast Sir2 [21], which have N-terminal domains unrelated to Sirt1. We therefore tested resveratrol effects on our Sirt1 constructs covering the catalytic domain with or without the N- and C-terminal extensions to identify the Sirt1 parts required for stimulation. Although the fluorophor of the FdL substrate was shown to influence activation, resveratrol effects have also been observed with non-modified substrates [48,49] and a better understanding of this mechanism is urgently needed [9]. For our tests on the domains involved we thus used the FdL assay, despite the non-physiological nature of the substrate, to keep our tests as comparable as possible to the previous study. The Sirt1 deletion constructs showed a significant reduction in basal activity (Figure 3A), consistent with previous reports [16,17]. However, resveratrol stimulated all constructs, and comparing stimulated activities to the respective basal activity in fact indicates that the strength of the stimulatory effect varies only slightly between constructs (Figure 3B). Even a minimal construct of the Sirt1 catalytic core (residues 229–516) was significantly activated, although the stimulatory effect was approximately 2-fold smaller than for full-length Sirt1. We further tested the effect of piceatannol, a resveratrol-related compound also reported to activate Sirt1 [21]. As for resveratrol, comparable relative increases of basal activity were observed for all constructs tested (Figure 3C). We thus conclude that resveratrol-dependent activation requires only the catalytic core of Sirt1, whereas its unique N-terminal domain is not necessary for the effect.
Figure 3. Regulation of Sirt1 activity by small molecules.
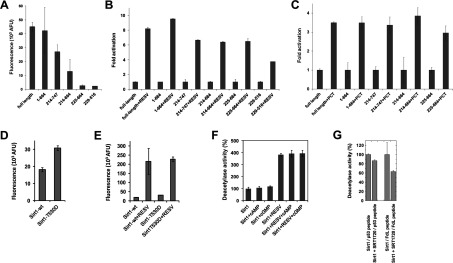
(A) Deacetylation activity of full-length Sirt1 and Sirt1 deletion constructs in the FdL assay. (B) Activation of Sirt1 and Sirt1 deletion constructs in the FdL assay by 100 μM resveratrol (RESV) shows only a weak decrease in the relative stimulation for shortened constructs. (C) Activity in FdL assays of full length Sirt1 and deletion constructs in the presence of 100 μM of the resveratrol-related compound piceatannol (PCT), relative to their basal activity. (D) FdL activity of the phosphorylation mimicking variant Sirt1-T530D compared with the activity of the same amount of wild-type (wt) enzyme. (E) Comparison of basal and resveratrol (RESV)-stimulated (100 μM) FdL activity of full-length human Sirt1-wt and Sirt1-T530D. (F) FdL deacetylation activity of full-length Sirt1 in the presence (100 μM) and the absence of the cyclic nucleotides cAMP and cGMP. The same set of experiments was done in the absence (left) and in the presence (right) of 100 μM resveratrol. The activity in the absence of any ligands was used as reference and set to 100%. (G) Effects of SRT1720 (1 μM) on the Sirt1-dependent deacetylation of FdL-peptide and of a p53-derived non-modified peptide.
A regulatory mechanism reported for the Sirt1 C-terminus is to act as a modification site for regulatory phosphorylations [18]. We indeed find that a Sirt1 mutation mimicking phosphorylation at residue 530 (T530D), immediately C-terminal to the catalytic core, increases basal activity (Figure 3D), consistent with a previous report [50]. Using the T530D variant, we then tested whether Sirt1 activation through Thr530 phosphorylation and by resveratrol exploit a common mechanism, such as a shared docking site or conformational change, or whether they are additive. Comparison of Sirt1-T530D activity in the presence and absence of resveratrol clearly showed a stimulatory effect of the compound (Figure 3E). We thus conclude that the regulatory Sirt1 phosphorylation at Thr530 and stimulation by resveratrol exploit two independent Sirt1 regulation mechanisms.
Testing Sirt1 modulation by cAMP, cGMP and SRT1720
Resveratrol was recently reported to be a competitive inhibitor of several PDEs (phosphodiesterases) [51], showing that the compound can occupy binding sites for the cyclic nucleotides, cAMP and cGMP. We thus speculated that the other way round, cAMP/cGMP could be physiological ligands for the resveratrol-binding site of Sirt1. However, testing Sirt1 alone and in the presence of cAMP or cGMP, respectively, yielded no significant changes in Sirt1 activity (Figure 3F), indicating that these cyclic nucleotides do not regulate Sirt1. Furthermore, adding cAMP or cGMP in the presence of resveratrol did not influence resveratrol-dependent Sirt1 activation, showing that the cyclic nucleotides do not bind to the Sirt1 site that mediates activation by resveratrol.
SRT1720 is a resveratrol-unrelated small molecule that stimulates Sirt1-dependent deacetylation of a substrate peptide [20], has resveratrol-like physiological effects [7,52] and was speculated to act as a PDE inhibitor [51]. In contrast with the previously reported activating effect in assays on an extended peptide, we find that SRT1720 has a weak inhibitory effect on Sirt1 in the FdL assay and against a non-modified p53 peptide (in a coupled enzymatic assay; Figure 3G), consistent with a previous study [53]. These different effects on Sirt1 activity against different substrates, as observed for resveratrol (see the Discussion section), might indicate the exciting possibility to develop substrate-specific modulators for Sirt1.
DISCUSSION
Sirt1 regulates central cellular processes and constitutes an attractive drug target for metabolic and ageing-related diseases [4,7]. However, its domain architecture and regulation are little understood on a molecular level. Sirt1 was suggested to form homo-trimers based on SEC experiments [12], and a monomer–trimer equilibrium mediated by N- and C-terminal extensions was observed for the yeast Sirtuin Hst2 [11]. Since Sirtuin catalytic domains have been described as monomers [8,54], the Sirt1 termini might mediate oligomerization. Our experiments show, however, that Sirt1 is a monomer that migrates fast in SEC due to an extended conformation, apparently mainly due to a disordered N-terminus, consistent with bioinformatics analyses reported here and previously [55]. The C-terminus also appears to harbour disordered regions, but partially seems to adopt a defined conformation that interacts with the catalytic core [17]. In fact, both termini increase the low activity of the catalytic core (our data and [16]), and we speculate two mechanisms could be involved: they might increase the stability of the catalytic core, which would be consistent with the low long-term stability we observed especially for short Sirt1 constructs, and they seem to mediate dedicated regulation mechanisms. Consistent with the latter idea, the Sirt3 N-terminus was also found to influence catalytic core activity [13,14], several regulatory phosphorylation sites are known in the Sirt1 termini [18], and the binding site for the Sirt1 activator AROS was located to the N-terminal domain [19]. In fact, the large apparent size of Sirt1 from natural sources can be explained by its disordered termini, but could also be caused by formation of larger complexes through interactions of the termini, similar to the recruitment of the substrate histone H1 through the N-terminus of Sirt6 [12]. Interestingly, full-length Sirt2 from natural sources also behaves like a trimer in SEC [56], which could indicate an extended conformation of the Sirt2 C-terminus (~120 residues) and/or formation of larger complexes. Bioinformatics analyses indeed indicate a high probability for disorder and coiled-coil formation in the ~60 Sirt2 C-terminal residues (Supplementary Figures S3A and S3B available at http://www.bioscirep.org/bsr/033/bsr033e037add.htm), which thus might be structurally related to the disordered/extended Sirt1 N-terminus despite a lack of significant sequence homology. Interestingly, for affinity purified recombinant Sirt1, a high absorption at 260 nm and treatment with DNase/RNase indicated that RNA is copurified (Supplementary Figures S4A and S4B available at http://www.bioscirep.org/bsr/033/bsr033e037add.htm). However, we could neither detect nucleic acid binding nor effects of nucleic acids on Sirt1 activity (results not shown). Since nucleic acids might also bind to the metal ions used for affinity purification, a nucleic acid-binding function for Sirt1 remains speculative.
For Sirt1 activation by resveratrol and other STACS, the N-terminus was reported to be essential, in particular residues 183–225 [20]. Depending on the substrate used, also Sirtuin inhibition has been observed in the presence of resveratrol [48,49], but a similar mechanism as for activation has to be assumed since no available evidence suggests otherwise. In contrast with the previous study on Sirt1 domains involved [20], we find that the entire N- and C-terminal Sirt1 extensions are dispensable for activation, which in fact is supported by data shown in [20] that activation is reduced but still detectable for the catalytic core. Since both termini also increase catalytic core activity (our work and [16]), shorter constructs yield lower absolute activities even when stimulated, which might have led to a misinterpretation that no significant stimulation occurs. However, we conclude that the N-terminus might indirectly affect resveratrol-dependent activation through its effect on catalytic core stability and/or activity, but the catalytic core contains all elements essential for stimulation by resveratrol. This activation mechanism thus appears to differ from activation by AROS, which binds to the Sirt1 N-terminus [19]. Since the catalytic core is evolutionarily conserved in all Sirtuins, it is tempting to speculate that small molecule activators might also exist for other family members, such as other mammalian Sirtuin isoforms. In fact, compounds activating non-specifically Sirt1, 2 and 3 were described [57], and the resveratrol-related compound piceatannol was shown to inhibit Sirt2 [58] and affect Sirt5 depending on the substrate used [48]. Interestingly, the activating effect of Sirt1-Thr530 phosphorylation is substrate-dependent and resveratrol can either activate or inhibit Sirt1 depending on the substrate used [25,49], which might indicate that resveratrol-like compounds could stimulate other Sirtuins if tested with the appropriate substrate. In fact, our failure to observe Sirt1 activation by AROS could also be due to a substrate-dependent effect. These substrate-dependent effects might indicate either a direct interaction between Sirt1-bound substrate and compound/modification, or an allosteric interaction between both sites, but further structural studies will be needed for fully understanding the modulation mechanism. Our results show that for such studies, the N- and C-terminal extensions of Sirt1 can be neglected, and thus provide the basis for further mechanistic studies on Sirt1 regulation.
AUTHOR CONTRIBUTIONS
Mahadevan Lakshminarasimhan performed and analysed protein expression, purification, gel filtration, CD and activity assay experiments. Ute Curth performed and analysed analytical ultracentrifugation experiments. Sebastien Moniot performed and analysed activity assay experiments. Shyamal Mosalaganti performed and analysed electron microscopy experiments. Stefan Raunser planned and analysed electron microscopy experiments. Clemens Steegborn designed the study and analysed biochemical results. Mahadevan Lakshminarasimhan, Ute Curth and Clemens Steegborn wrote the paper.
ACKNOWLEDGEMENTS
We thank Lidia Litz and Norbert Grillenbeck for excellent technical assistance and the members of the Steegborn laboratory for helpful discussions.
FUNDING
This work was supported by Deutsche Forschungsgemeinschaft [grant number RA1781/1], the Max Planck Society (to S.R.), the International Max Planck Research School Dortmund (to M.L. and S.M.), and Elite Network Bavaria (to M.L. and C.S.). This publication was funded by the German Research Foundation (DFG) and the University of Bayreuth in the funding program Open Access Publishing.
References
- 1.Sauve A. A., Wolberger C., Schramm V. L., Boeke J. D. The biochemistry of sirtuins. Annu. Rev. Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- 2.Michan S., Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem. J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guarente L., Picard F. Calorie restriction–the SIR2 connection. Cell. 2005;120:473–482. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 4.Haigis M. C., Sinclair D. A. Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lakshminarasimhan M., Steegborn C. Emerging mitochondrial signaling mechanisms in physiology, aging processes, and as drug targets. Exp. Gerontol. 2011;46:174–177. doi: 10.1016/j.exger.2010.08.024. [DOI] [PubMed] [Google Scholar]
- 6.Zhang T., Kraus W. L. SIRT1-dependent regulation of chromatin and transcription: Linking NAD(+) metabolism and signaling to the control of cellular functions. Biochim. Biophys. Acta. 2010;1804:1666–1675. doi: 10.1016/j.bbapap.2009.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milne J. C., Denu J. M. The Sirtuin family: therapeutic targets to treat diseases of aging. Curr. Opin. Chem. Biol. 2008;12:11–17. doi: 10.1016/j.cbpa.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 8.Sanders B. D., Jackson B., Marmorstein R. Structural basis for sirtuin function: what we know and what we don’t. Biochim. Biophys. Acta. 2010;1804:1604–1616. doi: 10.1016/j.bbapap.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moniot S., Weyand M., Steegborn C. Structures, substrates, and regulators of Mammalian sirtuins–opportunities and challenges for drug development. Front Pharmacol. 2012;3:16. doi: 10.3389/fphar.2012.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gertz M., Steegborn C. Function and regulation of the mitochondrial Sirtuin isoform Sirt5 in Mammalia. Biochim. Biophys. Acta. 2010;1804:1658–1665. doi: 10.1016/j.bbapap.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 11.Zhao K., Chai X., Clements A., Marmorstein R. Structure and autoregulation of the yeast Hst2 homolog of Sir2. Nat. Struct. Biol. 2003;10:864–871. doi: 10.1038/nsb978. [DOI] [PubMed] [Google Scholar]
- 12.Vaquero A., Scher M., Lee D., Erdjument-Bromage H., Tempst P., Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 13.Schwer B., North B. J., Frye R. A., Ott M., Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J. Cell Biol. 2002;158:647–657. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlicker C., Gertz M., Papatheodorou P., Kachholz B., Becker C. F., Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J. Mol. Biol. 2008;382:790–801. doi: 10.1016/j.jmb.2008.07.048. [DOI] [PubMed] [Google Scholar]
- 15.Tennen R. I., Berber E., Chua K. F. Functional dissection of SIRT6: identification of domains that regulate histone deacetylase activity and chromatin localization. Mech. Ageing Dev. 2010;131:185–192. doi: 10.1016/j.mad.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan M., Yuan H., Brent M., Ding E. C., Marmorstein R. SIRT1 contains N- and C-terminal regions that potentiate deacetylase activity. J. Biol. Chem. 2012;287:2468–2476. doi: 10.1074/jbc.M111.285031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang H., Suh J. Y., Jung Y. S., Jung J. W., Kim M. K., Chung J. H. Peptide switch is essential for sirt1 deacetylase activity. Mol. Cell. 2011;44:203–213. doi: 10.1016/j.molcel.2011.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flick F., Luscher B. Regulation of sirtuin function by posttranslational modifications. Front Pharmacol. 2012;3:29. doi: 10.3389/fphar.2012.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim E. J., Kho J. H., Kang M. R., Um S. J. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol. Cell. 2007;28:277–290. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 20.Milne J. C., Lambert P. D., Schenk S., Carney D. P., Smith J. J., Gagne D. J., Jin L., Boss O., Perni R. B., Vu C. B., et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howitz K. T., Bitterman K. J., Cohen H. Y., Lamming D. W., Lavu S., Wood J. G., Zipkin R. E., Chung P., Kisielewski A., Zhang L. L., et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 22.Barger J. L., Kayo T., Vann J. M., Arias E. B., Wang J., Hacker T. A., Wang Y., Raederstorff D., Morrow J. D., Leeuwenburgh C., et al. A low dose of dietary resveratrol partially mimics caloric restriction and retards aging parameters in mice. PLoS ONE. 2008;3:e2264. doi: 10.1371/journal.pone.0002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wood J. G., Rogina B., Lavu S., Howitz K., Helfand S. L., Tatar M., Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 24.Baur J. A., Sinclair D. A. Therapeutic potential of resveratrol: the in vivo evidence. Nat. Rev. Drug. Discov. 2006;5:493–506. doi: 10.1038/nrd2060. [DOI] [PubMed] [Google Scholar]
- 25.Kaeberlein M., McDonagh T., Heltweg B., Hixon J., Westman E. A., Caldwell S. D., Napper A., Curtis R., DiStefano P. S., Fields S., et al. Substrate-specific activation of sirtuins by resveratrol. J. Biol. Chem. 2005;280:17038–17045. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- 26.Borra M. T., Smith B. C., Denu J. M. Mechanism of human SIRT1 activation by resveratrol. J. Biol. Chem. 2005;280:17187–17195. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- 27.Pirola L., Frojdo S. Resveratrol: one molecule, many targets. IUBMB Life. 2008;60:323–332. doi: 10.1002/iub.47. [DOI] [PubMed] [Google Scholar]
- 28.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laue M. T., Shah B. D., Rigdeway T. M., Pelletier S. L. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding S. E., Rowe A. J., Horton J. C., editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Cambridge, U.K.: Royal Society of Chemistry; 1992. pp. 90–125. [Google Scholar]
- 30.Laue T. M., Stafford W. F., III Modern applications of analytical ultracentrifugation. Annu. Rev. Biophys. Biomol. Struct. 1999;28:75–100. doi: 10.1146/annurev.biophys.28.1.75. [DOI] [PubMed] [Google Scholar]
- 31.Wyszomirski K. H., Curth U., Alves J., Mackeldanz P., Moncke-Buchner E., Schutkowski M., Kruger D. H., Reuter M. Type III restriction endonuclease EcoP15I is a heterotrimeric complex containing one Res subunit with several DNA-binding regions and ATPase activity. Nucleic Acids Res. 2012;40:3610–3622. doi: 10.1093/nar/gkr1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Witte G., Urbanke C., Curth U. Single-stranded DNA-binding protein of Deinococcus radiodurans: a biophysical characterization. Nucleic Acids Res. 2005;33:1662–1670. doi: 10.1093/nar/gki310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith B. C., Hallows W. C., Denu J. M. A continuous microplate assay for sirtuins and nicotinamide-producing enzymes. Anal. Biochem. 2009;394:101–109. doi: 10.1016/j.ab.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fischer F., Gertz M., Suenkel B., Lakshminarasimhan M., Schutkowski M., Steegborn C. Sirt5 deacylation activities show differential sensitivities to nicotinamide inhibition. PLoS ONE. 2012;7:e45098. doi: 10.1371/journal.pone.0045098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kelley L. A., Sternberg M. J. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 37.Hashimoto K., Nishi H., Bryant S., Panchenko A. R. Caught in self-interaction: evolutionary and functional mechanisms of protein homooligomerization. Phys. Biol. 2011;8:035007. doi: 10.1088/1478-3975/8/3/035007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Batas B., Jones H. R., Chaudhuri J. B. Studies of the hydrodynamic volume changes that occur during refolding of lysozyme using size-exclusion chromatography. J. Chromatogr. A. 1997;766:109–119. doi: 10.1016/s0021-9673(96)01020-5. [DOI] [PubMed] [Google Scholar]
- 39.Salvay A. G., Communie G., Ebel C. Sedimentation velocity analytical ultracentrifugation for intrinsically disordered proteins. Methods Mol. Biol. 2012;896:91–105. doi: 10.1007/978-1-4614-3704-8_6. [DOI] [PubMed] [Google Scholar]
- 40.Lebowitz J., Lewis M. S., Schuck P. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 2002;11:2067–2079. doi: 10.1110/ps.0207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown P. H., Balbo A., Schuck P. Characterizing protein–protein interactions by sedimentation velocity analytical ultracentrifugation. Curr. Protoc. Immunol. 2008 doi: 10.1002/0471142735.im1815s81. Chapter 18, Unit 18 15. [DOI] [PubMed] [Google Scholar]
- 42.Louis-Jeune C., Andrade-Navarro M. A., Perez-Iratxeta C. Prediction of protein secondary structure from circular dichroism using theoretically derived spectra. Proteins. 2011;80:374–381. doi: 10.1002/prot.23188. [DOI] [PubMed] [Google Scholar]
- 43.Linding R., Jensen L. J., Diella F., Bork P., Gibson T. J., Russell R. B. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 44.Linding R., Russell R. B., Neduva V., Gibson T. J. GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31:3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim J. E., Chen J., Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 46.Woody R. W. Circular dichroism spectrum of peptides in the poly(Pro)II conformation. J. Am. Chem. Soc. 2009;131:8234–8245. doi: 10.1021/ja901218m. [DOI] [PubMed] [Google Scholar]
- 47.Heltweg B., Trapp J., Jung M. In vitro assays for the determination of histone deacetylase activity. Methods. 2005;36:332–337. doi: 10.1016/j.ymeth.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 48.Gertz M., Nguyen G. T. T., Fischer F., Suenkel B., Schlicker C., Fränzel B., Tomaschewski J., Aladini F., Becker C., Wolters D., Steegborn C. A Molecular Mechanism for Direct Sirtuin Activation by Resveratrol. PLoS ONE. 2012;7:e49761. doi: 10.1371/journal.pone.0049761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dai H., Kustigian L., Carney D., Case A., Considine T., Hubbard B. P., Perni R. B., Riera T. V., Szczepankiewicz B., Vlasuk G. P., Stein R. L. SIRT1 activation by small molecules: kinetic and biophysical evidence for direct interaction of enzyme and activator. J. Biol. Chem. 2010;285:32695–32703. doi: 10.1074/jbc.M110.133892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nasrin N., Kaushik V. K., Fortier E., Wall D., Pearson K. J., de Cabo R., Bordone L. JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PLoS ONE. 2009;4:e8414. doi: 10.1371/journal.pone.0008414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park S. J., Ahmad F., Philp A., Baar K., Williams T., Luo H., Ke H., Rehmann H., Taussig R., Brown A. L., et al. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell. 2012;148:421–433. doi: 10.1016/j.cell.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feige J. N., Lagouge M., Canto C., Strehle A., Houten S. M., Milne J. C., Lambert P. D., Mataki C., Elliott P. J., Auwerx J. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8:347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 53.Pacholec M., Bleasdale J. E., Chrunyk B., Cunningham D., Flynn D., Garofalo R. S., Griffith D., Griffor M., Loulakis P., Pabst B., et al. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J. Biol. Chem. 2010;285:8340–8351. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Finnin M. S., Donigian J. R., Pavletich N. P. Structure of the histone deacetylase SIRT2. Nat. Struct. Biol. 2001;8:621–625. doi: 10.1038/89668. [DOI] [PubMed] [Google Scholar]
- 55.Autiero I., Costantini S., Colonna G. Human sirt-1: molecular modeling and structure-function relationships of an unordered protein. PLoS ONE. 2009;4:e7350. doi: 10.1371/journal.pone.0007350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vaquero A., Scher M. B., Lee D. H., Sutton A., Cheng H. L., Alt F. W., Serrano L., Sternglanz R., Reinberg D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mai A., Valente S., Meade S., Carafa V., Tardugno M., Nebbioso A., Galmozzi A., Mitro N., De Fabiani E., Altucci L., Kazantsev A. Study of 1,4–dihydropyridine structural scaffold: discovery of novel sirtuin activators and inhibitors. J. Med. Chem. 2009;52:5496–5504. doi: 10.1021/jm9008289. [DOI] [PubMed] [Google Scholar]
- 58.Trapp J., Jochum A., Meier R., Saunders L., Marshall B., Kunick C., Verdin E., Goekjian P., Sippl W., Jung M. Adenosine mimetics as inhibitors of NAD+-dependent histone deacetylases, from kinase to sirtuin inhibition. J. Med. Chem. 2006;49:7307–7316. doi: 10.1021/jm060118b. [DOI] [PubMed] [Google Scholar]