Abstract
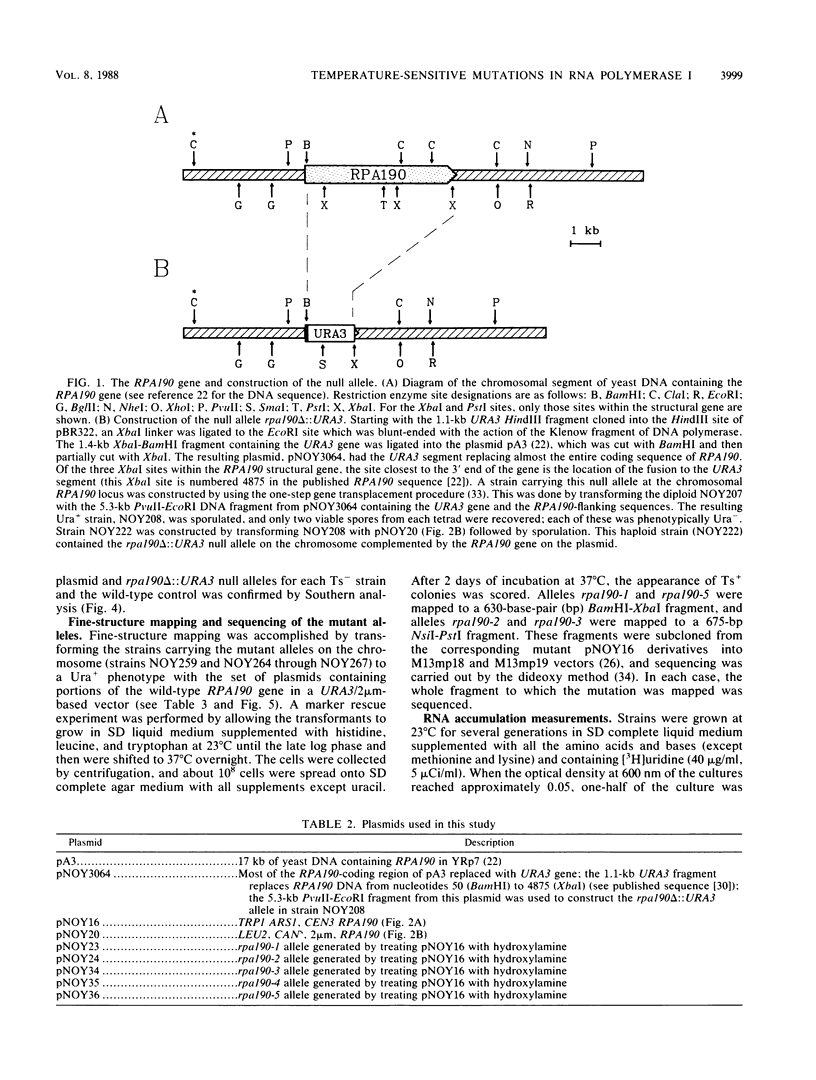
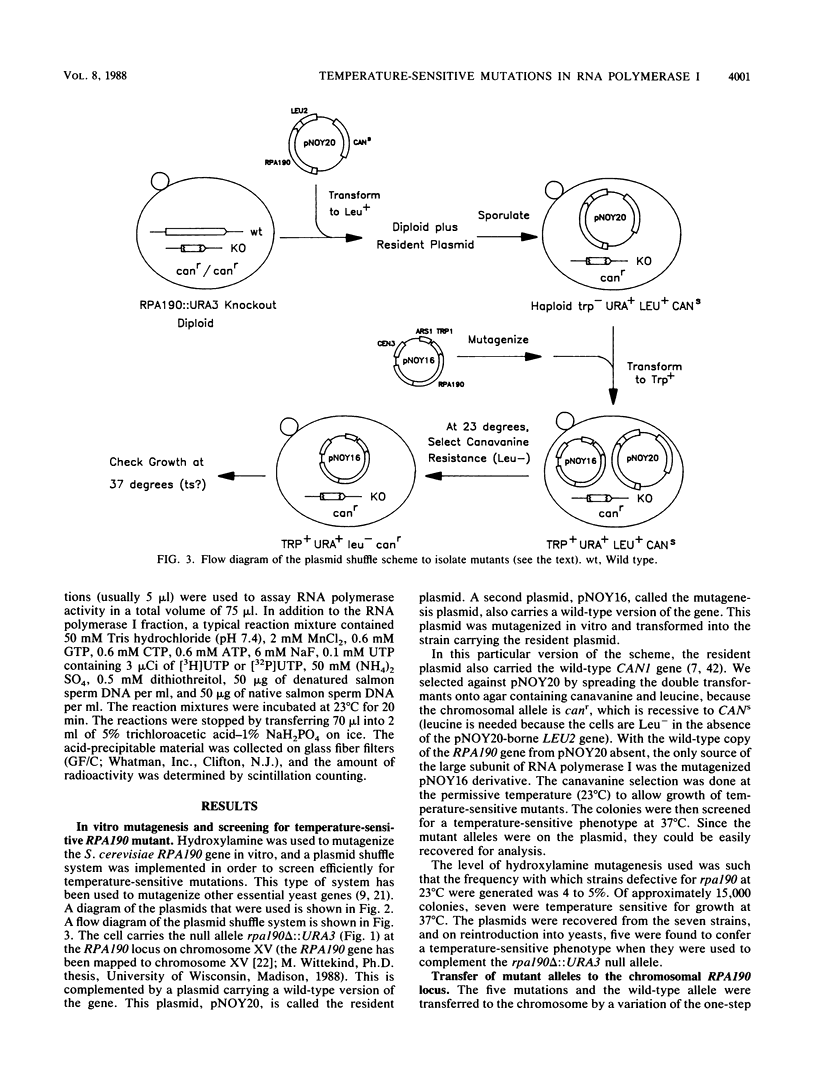
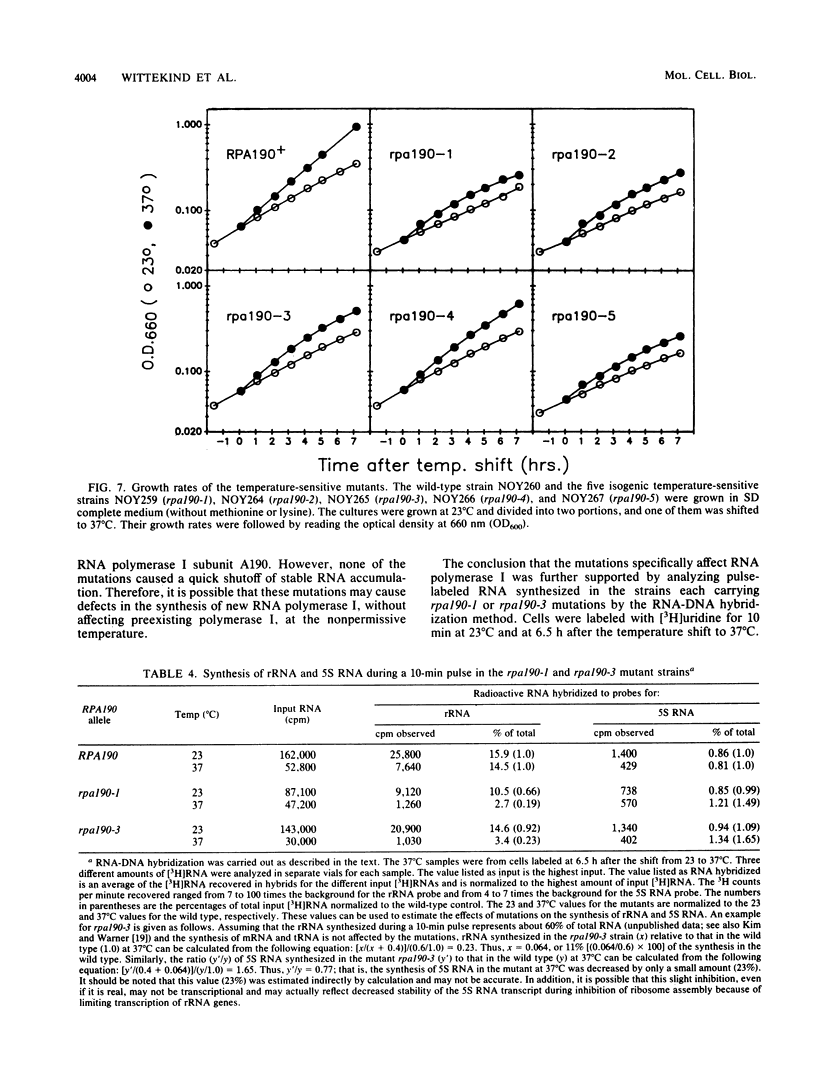
The isolation and characterization of temperature-sensitive mutations in RNA polymerase I from Saccharomyces cerevisiae are described. A plasmid carrying RPA190, the gene encoding the largest subunit of the enzyme, was subjected to in vitro mutagenesis with hydroxylamine. Using a plasmid shuffle screening system, five different plasmids were isolated which conferred a temperature-sensitive phenotype in haploid yeast strains carrying the disrupted chromosomal RPA190 gene. These temperature-sensitive alleles were transferred to the chromosomal RPA190 locus for mapping and physiology experiments. Accumulation of RNA was found to be defective in all mutant strains at the nonpermissive temperature. In addition, analysis of pulse-labeled RNA from two mutant strains at 37 degrees C showed that the transcription of rRNA genes was decreased, while that of 5S RNA was relatively unaffected. RNA polymerase I was partially purified from several of the mutant strains grown at the nonpermissive temperature and was shown to be deficient when assayed in vitro. Fine-structure mapping and sequencing of the mutant alleles demonstrated that all five mutations were unique. The rpa190-1 and rpa190-5 mutations are tightly clustered in region I (S.S. Broyles and B. Moss, Proc. Natl. Acad. Sci. USA 83:3141-3145, 1986), the putative zinc-binding region that is common to all eucaryotic RNA polymerase large subunits. The rpa190-3 mutation is located between regions III and IV, and a strain carrying it behaves as a mutant that is defective in the synthesis of the enzyme. This mutation lies within a previously unidentified segment of highly conserved amino acid sequence homology that is shared among the largest subunits of eucaryotic nuclear RNA polymerases. Another temperature-sensitive mutation, rpa190-2, creates a UGA nonsense codon.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahearn J. M., Jr, Bartolomei M. S., West M. L., Cisek L. J., Corden J. L. Cloning and sequence analysis of the mouse genomic locus encoding the largest subunit of RNA polymerase II. J Biol Chem. 1987 Aug 5;262(22):10695–10705. [PubMed] [Google Scholar]
- Allison L. A., Moyle M., Shales M., Ingles C. J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985 Sep;42(2):599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- Allison L. A., Wong J. K., Fitzpatrick V. D., Moyle M., Ingles C. J. The C-terminal domain of the largest subunit of RNA polymerase II of Saccharomyces cerevisiae, Drosophila melanogaster, and mammals: a conserved structure with an essential function. Mol Cell Biol. 1988 Jan;8(1):321–329. doi: 10.1128/mcb.8.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg J. M. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986 Apr 25;232(4749):485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- Biggs J., Searles L. L., Greenleaf A. L. Structure of the eukaryotic transcription apparatus: features of the gene for the largest subunit of Drosophila RNA polymerase II. Cell. 1985 Sep;42(2):611–621. doi: 10.1016/0092-8674(85)90118-7. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Broyles S. S., Moss B. Homology between RNA polymerases of poxviruses, prokaryotes, and eukaryotes: nucleotide sequence and transcriptional analysis of vaccinia virus genes encoding 147-kDa and 22-kDa subunits. Proc Natl Acad Sci U S A. 1986 May;83(10):3141–3145. doi: 10.1073/pnas.83.10.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd M., Campbell J. L. Temperature-sensitive mutations in the yeast DNA polymerase I gene. Proc Natl Acad Sci U S A. 1987 May;84(9):2838–2842. doi: 10.1073/pnas.84.9.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans R. M., Hollenberg S. M. Zinc fingers: gilt by association. Cell. 1988 Jan 15;52(1):1–3. doi: 10.1016/0092-8674(88)90522-3. [DOI] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Himmelfarb H. J., Simpson E. M., Friesen J. D. Isolation and characterization of temperature-sensitive RNA polymerase II mutants of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Jun;7(6):2155–2164. doi: 10.1128/mcb.7.6.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honzatko R. B., Crawford J. L., Monaco H. L., Ladner J. E., Ewards B. F., Evans D. R., Warren S. G., Wiley D. C., Ladner R. C., Lipscomb W. N. Crystal and molecular structures of native and CTP-liganded aspartate carbamoyltransferase from Escherichia coli. J Mol Biol. 1982 Sep 15;160(2):219–263. doi: 10.1016/0022-2836(82)90175-9. [DOI] [PubMed] [Google Scholar]
- Huet J., Sentenac A., Fromageot P. Spot-immunodetection of conserved determinants in eukaryotic RNA polymerases. Study with antibodies to yeast RNA polymerases subunits. J Biol Chem. 1982 Mar 10;257(5):2613–2618. [PubMed] [Google Scholar]
- Ingles C. J., Himmelfarb H. J., Shales M., Greenleaf A. L., Friesen J. D. Identification, molecular cloning, and mutagenesis of Saccharomyces cerevisiae RNA polymerase genes. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2157–2161. doi: 10.1073/pnas.81.7.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Wittekind M., Nomura M., Shiba K., Yura T., Miura A., Nashimoto H. A temperature-sensitive mutant of E. coli exhibiting slow processing of exported proteins. Cell. 1983 Mar;32(3):789–797. doi: 10.1016/0092-8674(83)90065-x. [DOI] [PubMed] [Google Scholar]
- Jinks-Robertson S., Gourse R. L., Nomura M. Expression of rRNA and tRNA genes in Escherichia coli: evidence for feedback regulation by products of rRNA operons. Cell. 1983 Jul;33(3):865–876. doi: 10.1016/0092-8674(83)90029-6. [DOI] [PubMed] [Google Scholar]
- Kim C. H., Warner J. R. Mild temperature shock alters the transcription of a discrete class of Saccharomyces cerevisiae genes. Mol Cell Biol. 1983 Mar;3(3):457–465. doi: 10.1128/mcb.3.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann C., Buhler J. M., Treich I., Sentenac A. RPC40, a unique gene for a subunit shared between yeast RNA polymerases A and C. Cell. 1987 Feb 27;48(4):627–637. doi: 10.1016/0092-8674(87)90241-8. [DOI] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mémet S., Gouy M., Marck C., Sentenac A., Buhler J. M. RPA190, the gene coding for the largest subunit of yeast RNA polymerase A. J Biol Chem. 1988 Feb 25;263(6):2830–2839. [PubMed] [Google Scholar]
- Nonet M., Scafe C., Sexton J., Young R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol. 1987 May;7(5):1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M., Sweetser D., Young R. A. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell. 1987 Sep 11;50(6):909–915. doi: 10.1016/0092-8674(87)90517-4. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Parent S. A., Fenimore C. M., Bostian K. A. Vector systems for the expression, analysis and cloning of DNA sequences in S. cerevisiae. Yeast. 1985 Dec;1(2):83–138. doi: 10.1002/yea.320010202. [DOI] [PubMed] [Google Scholar]
- Petes T. D., Botstein D. Simple Mendelian inheritance of the reiterated ribosomal DNA of yeast. Proc Natl Acad Sci U S A. 1977 Nov;74(11):5091–5095. doi: 10.1073/pnas.74.11.5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva M., Buhler J. M., Sentenac A., Fromageot P., Hawthorne D. C. Natural variation in yeast RNA polymerase A. Formation of a mosaic RNA polymerase A in a meiotic segregant from an interspecific hybrid. J Biol Chem. 1982 Apr 25;257(8):4570–4577. [PubMed] [Google Scholar]
- Riva M., Memet S., Micouin J. Y., Huet J., Treich I., Dassa J., Young R., Buhler J. M., Sentenac A., Fromageot P. Isolation of structural genes for yeast RNA polymerases by immunological screening. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1554–1558. doi: 10.1073/pnas.83.6.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Fink G. R. KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell. 1987 Mar 27;48(6):1047–1060. doi: 10.1016/0092-8674(87)90712-4. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian J., Takano I., Halvorson H. O. Independent regulation of the nuclear RNA polymerases I and II during the yeast cell cycle. Proc Natl Acad Sci U S A. 1974 Mar;71(3):769–773. doi: 10.1073/pnas.71.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentenac A. Eukaryotic RNA polymerases. CRC Crit Rev Biochem. 1985;18(1):31–90. doi: 10.3109/10409238509082539. [DOI] [PubMed] [Google Scholar]
- Shaw K. J., Olson M. V. Effects of altered 5'-flanking sequences on the in vivo expression of a Saccharomyces cerevisiae tRNATyr gene. Mol Cell Biol. 1984 Apr;4(4):657–665. doi: 10.1128/mcb.4.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B., Kuśmierek J. T. Chemical mutagenesis. Annu Rev Biochem. 1982;51:655–693. doi: 10.1146/annurev.bi.51.070182.003255. [DOI] [PubMed] [Google Scholar]
- Sweetser D., Nonet M., Young R. A. Prokaryotic and eukaryotic RNA polymerases have homologous core subunits. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1192–1196. doi: 10.1073/pnas.84.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan W. L., Gocke E., Manney T. R. The CAN1 locus of Saccharomyces cerevisiae: fine-structure analysis and forward mutation rates. Genetics. 1979 Jan;91(1):35–51. doi: 10.1093/genetics/91.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Yeast RNA polymerase II genes: isolation with antibody probes. Science. 1983 Nov 18;222(4625):778–782. doi: 10.1126/science.6356359. [DOI] [PubMed] [Google Scholar]



