Abstract
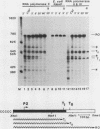
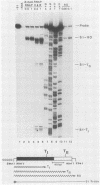
We have studied transcription elongation and termination in the human c-myc gene. Transcription of c-myc gene sequences with purified mammalian RNA polymerase II revealed several sites of transcription termination and pausing in the vicinity of the exon 1-intron 1 junction. This region previously has been shown to block transcription elongation in vivo by nuclear run-on analysis (D. Bentley and M. Groudine, Nature [London] 321:702-706, 1986). These sites were recognized by purified RNA polymerase II, and we therefore designated them intrinsic sites of termination and pausing. Two of these sites cause termination of RNA polymerase III transcription as well. RNA polymerase II terminated transcription in a cluster of seven consecutive T residues in the nontranscribed strand and paused during transcription at three additional sites in this region. The intrinsic sites of transcription termination and pausing described here correspond closely to the 3' ends of transcripts synthesized in Xenopus oocytes injected with plasmids containing the c-myc termination region (D. Bentley and M. Groudine, Cell 53:245-256, 1988). This correspondence suggests that the intrinsic recognition of these termination and pause sites by purified RNA polymerase II may play a role in the transcription elongation block observed in vivo.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bender T. P., Thompson C. B., Kuehl W. M. Differential expression of c-myb mRNA in murine B lymphomas by a block to transcription elongation. Science. 1987 Sep 18;237(4821):1473–1476. doi: 10.1126/science.3498214. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986 Jun 12;321(6071):702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. Sequence requirements for premature termination of transcription in the human c-myc gene. Cell. 1988 Apr 22;53(2):245–256. doi: 10.1016/0092-8674(88)90386-8. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Brown D. D. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981 Apr;24(1):261–270. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- Briat J. F., Bollag G., Kearney C. A., Molineux I., Chamberlin M. J. Tau factor from Escherichia coli mediates accurate and efficient termination of transcription at the bacteriophage T3 early termination site in vitro. J Mol Biol. 1987 Nov 5;198(1):43–49. doi: 10.1016/0022-2836(87)90456-6. [DOI] [PubMed] [Google Scholar]
- Brosius J. Toxicity of an overproduced foreign gene product in Escherichia coli and its use in plasmid vectors for the selection of transcription terminators. Gene. 1984 Feb;27(2):161–172. doi: 10.1016/0378-1119(84)90137-9. [DOI] [PubMed] [Google Scholar]
- Cesarman E., Dalla-Favera R., Bentley D., Groudine M. Mutations in the first exon are associated with altered transcription of c-myc in Burkitt lymphoma. Science. 1987 Nov 27;238(4831):1272–1275. doi: 10.1126/science.3685977. [DOI] [PubMed] [Google Scholar]
- Chamberlin M. J., Nierman W. C., Wiggs J., Neff N. A quantitative assay for bacterial RNA polymerases. J Biol Chem. 1979 Oct 25;254(20):10061–10069. [PubMed] [Google Scholar]
- Chung J., Sussman D. J., Zeller R., Leder P. The c-myc gene encodes superimposed RNA polymerase II and III promoters. Cell. 1987 Dec 24;51(6):1001–1008. doi: 10.1016/0092-8674(87)90586-1. [DOI] [PubMed] [Google Scholar]
- Cozzarelli N. R., Gerrard S. P., Schlissel M., Brown D. D., Bogenhagen D. F. Purified RNA polymerase III accurately and efficiently terminates transcription of 5S RNA genes. Cell. 1983 Oct;34(3):829–835. doi: 10.1016/0092-8674(83)90540-8. [DOI] [PubMed] [Google Scholar]
- Dedrick R. L., Chamberlin M. J. Studies on transcription of 3'-extended templates by mammalian RNA polymerase II. Parameters that affect the initiation and elongation reactions. Biochemistry. 1985 Apr 23;24(9):2245–2253. doi: 10.1021/bi00330a019. [DOI] [PubMed] [Google Scholar]
- Dedrick R. L., Kane C. M., Chamberlin M. J. Purified RNA polymerase II recognizes specific termination sites during transcription in vitro. J Biol Chem. 1987 Jul 5;262(19):9098–9108. [PubMed] [Google Scholar]
- Eick D., Bornkamm G. W. Transcriptional arrest within the first exon is a fast control mechanism in c-myc gene expression. Nucleic Acids Res. 1986 Nov 11;14(21):8331–8346. doi: 10.1093/nar/14.21.8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans R., Weber J., Ziff E., Darnell J. E. Premature termination during adenovirus transcription. Nature. 1979 Mar 22;278(5702):367–370. doi: 10.1038/278367a0. [DOI] [PubMed] [Google Scholar]
- Fort P., Rech J., Vie A., Piechaczyk M., Bonnieu A., Jeanteur P., Blanchard J. M. Regulation of c-fos gene expression in hamster fibroblasts: initiation and elongation of transcription and mRNA degradation. Nucleic Acids Res. 1987 Jul 24;15(14):5657–5667. doi: 10.1093/nar/15.14.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser N. W., Sehgal P. B., Darnell J. E., Jr Multiple discrete sites for premature RNA chain termination late in adenovirus-2 infection: enhancement by 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2571–2575. doi: 10.1073/pnas.76.6.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli G., Guise J. W., McDevitt M. A., Tucker P. W., Nevins J. R. Relative position and strengths of poly(A) sites as well as transcription termination are critical to membrane versus secreted mu-chain expression during B-cell development. Genes Dev. 1987 Jul;1(5):471–481. doi: 10.1101/gad.1.5.471. [DOI] [PubMed] [Google Scholar]
- Gilman M. Z., Chamberlin M. J. Developmental and genetic regulation of Bacillus subtilis genes transcribed by sigma 28-RNA polymerase. Cell. 1983 Nov;35(1):285–293. doi: 10.1016/0092-8674(83)90231-3. [DOI] [PubMed] [Google Scholar]
- Gilmour D. S., Lis J. T. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol Cell Biol. 1986 Nov;6(11):3984–3989. doi: 10.1128/mcb.6.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N., Wiggs J., Chamberlin M. J. A simple procedure for resolution of Escherichia coli RNA polymerase holoenzyme from core polymerase. Arch Biochem Biophys. 1977 Aug;182(2):404–408. doi: 10.1016/0003-9861(77)90521-5. [DOI] [PubMed] [Google Scholar]
- Hay N., Aloni Y. Attenuation in SV40 as a mechanism of transcription-termination by RNA polymerase B. Nucleic Acids Res. 1984 Feb 10;12(3):1401–1414. doi: 10.1093/nar/12.3.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N., Aloni Y. Attenuation of late simian virus 40 mRNA synthesis is enhanced by the agnoprotein and is temporally regulated in isolated nuclear systems. Mol Cell Biol. 1985 Jun;5(6):1327–1334. doi: 10.1128/mcb.5.6.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N., Skolnik-David H., Aloni Y. Attenuation in the control of SV40 gene expression. Cell. 1982 May;29(1):183–193. doi: 10.1016/0092-8674(82)90102-7. [DOI] [PubMed] [Google Scholar]
- Hess J., Perez-Stable C., Wu G. J., Weir B., Tinoco I., Jr, Shen C. K. End-to-end transcription of an Alu family repeat. A new type of polymerase-III-dependent terminator and its evolutionary implication. J Mol Biol. 1985 Jul 5;184(1):7–21. doi: 10.1016/0022-2836(85)90039-7. [DOI] [PubMed] [Google Scholar]
- Hodo H. G., 3rd, Blatti S. P. Purification using polyethylenimine precipitation and low molecular weight subunit analyses of calf thymus and wheat germ DNA-dependent RNA polymerase II. Biochemistry. 1977 May 31;16(11):2334–2343. doi: 10.1021/bi00630a005. [DOI] [PubMed] [Google Scholar]
- Kadesch T. R., Chamberlin M. J. Studies of in vitro transcription by calf thymus RNA polymerase II using a novel duplex DNA template. J Biol Chem. 1982 May 10;257(9):5286–5295. [PubMed] [Google Scholar]
- Kane C. M., Chamberlin M. J. Studies on transcription of 3'-extended DNA templates by mammalian RNA polymerase II. Partial purification and characterization of a factor from HeLa cells that facilitates renaturation of the DNA template. Biochemistry. 1985 Apr 23;24(9):2254–2262. doi: 10.1021/bi00330a020. [DOI] [PubMed] [Google Scholar]
- Kane C. M. Renaturase and ribonuclease H: a novel mechanism that influences transcript displacement by RNA polymerase II in vitro. Biochemistry. 1988 May 3;27(9):3187–3196. doi: 10.1021/bi00409a010. [DOI] [PubMed] [Google Scholar]
- Kao S. Y., Calman A. F., Luciw P. A., Peterlin B. M. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987 Dec 3;330(6147):489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- Levin J. R., Chamberlin M. J. Mapping and characterization of transcriptional pause sites in the early genetic region of bacteriophage T7. J Mol Biol. 1987 Jul 5;196(1):61–84. doi: 10.1016/0022-2836(87)90511-0. [DOI] [PubMed] [Google Scholar]
- Maderious A., Chen-Kiang S. Pausing and premature termination of human RNA polymerase II during transcription of adenovirus in vivo and in vitro. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5931–5935. doi: 10.1073/pnas.81.19.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather E. L., Nelson K. J., Haimovich J., Perry R. P. Mode of regulation of immunoglobulin mu- and delta-chain expression varies during B-lymphocyte maturation. Cell. 1984 Feb;36(2):329–338. doi: 10.1016/0092-8674(84)90226-5. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mazabraud A., Scherly D., Müller F., Rungger D., Clarkson S. G. Structure and transcription termination of a lysine tRNA gene from Xenopus laevis. J Mol Biol. 1987 Jun 20;195(4):835–845. doi: 10.1016/0022-2836(87)90488-8. [DOI] [PubMed] [Google Scholar]
- McCachren S. S., Jr, Salehi Z., Weinberg J. B., Niedel J. E. Transcription interruption may be a common mechanism of c-myc regulation during HL-60 differentiation. Biochem Biophys Res Commun. 1988 Feb 29;151(1):574–582. doi: 10.1016/0006-291x(88)90633-x. [DOI] [PubMed] [Google Scholar]
- Mok M., Maderious A., Chen-Kiang S. Premature termination by human RNA polymerase II occurs temporally in the adenovirus major late transcriptional unit. Mol Cell Biol. 1984 Oct;4(10):2031–2040. doi: 10.1128/mcb.4.10.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepveu A., Marcu K. B. Intragenic pausing and anti-sense transcription within the murine c-myc locus. EMBO J. 1986 Nov;5(11):2859–2865. doi: 10.1002/j.1460-2075.1986.tb04580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins J. R., Wilson M. C. Regulation of adenovirus-2 gene expression at the level of transcriptional termination and RNA processing. Nature. 1981 Mar 12;290(5802):113–118. doi: 10.1038/290113a0. [DOI] [PubMed] [Google Scholar]
- Pfeiffer P., Hay N., Pruzan R., Jakobovits E. B., Aloni Y. In vitro premature termination in SV40 late transcription. EMBO J. 1983;2(2):185–191. doi: 10.1002/j.1460-2075.1983.tb01403.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport J., Reinberg D., Zandomeni R., Weinmann R. Purification and functional characterization of transcription factor SII from calf thymus. Role in RNA polymerase II elongation. J Biol Chem. 1987 Apr 15;262(11):5227–5232. [PubMed] [Google Scholar]
- Reinberg D., Roeder R. G. Factors involved in specific transcription by mammalian RNA polymerase II. Transcription factor IIS stimulates elongation of RNA chains. J Biol Chem. 1987 Mar 5;262(7):3331–3337. [PubMed] [Google Scholar]
- Reines D., Wells D., Chamberlin M. J., Kane C. M. Identification of intrinsic termination sites in vitro for RNA polymerase II within eukaryotic gene sequences. J Mol Biol. 1987 Jul 20;196(2):299–312. doi: 10.1016/0022-2836(87)90691-7. [DOI] [PubMed] [Google Scholar]
- Schmidt M. C., Chamberlin M. J. nusA protein of Escherichia coli is an efficient transcription termination factor for certain terminator sites. J Mol Biol. 1987 Jun 20;195(4):809–818. doi: 10.1016/0022-2836(87)90486-4. [DOI] [PubMed] [Google Scholar]
- Shuman S., Broyles S. S., Moss B. Purification and characterization of a transcription termination factor from vaccinia virions. J Biol Chem. 1987 Sep 5;262(25):12372–12380. [PubMed] [Google Scholar]
- Thayer G. C., Brosius J. In vivo transcription from deletion mutations introduced near Escherichia coli ribosomal RNA promoter P2. Mol Gen Genet. 1985;199(1):55–58. doi: 10.1007/BF00327509. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Roeder R. G. Multiple proteins bind to VA RNA genes of adenovirus type 2. Mol Cell Biol. 1987 Mar;7(3):1021–1031. doi: 10.1128/mcb.7.3.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson J. B., Chandler D. W., Gralla J. D. Specific termination of in vitro transcription by calf thymus RNA polymerase III. Nucleic Acids Res. 1984 Jul 11;12(13):5369–5384. doi: 10.1093/nar/12.13.5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ar-Rushdi A., Nishikura K., Erikson J., Watt R., Rovera G., Croce C. M. Differential expression of the translocated and the untranslocated c-myc oncogene in Burkitt lymphoma. Science. 1983 Oct 28;222(4622):390–393. doi: 10.1126/science.6414084. [DOI] [PubMed] [Google Scholar]