Abstract
Efficient and faithful replication of telomeric DNA is critical for maintaining genome integrity. The G-quadruplex (G4) structure arising in the repetitive TTAGGG sequence is thought to stall replication forks, impairing efficient telomere replication and leading to telomere instabilities. However, pathways modulating telomeric G4 are poorly understood, and it is unclear whether defects in these pathways contribute to genome instabilities in vivo. Here, we report that mammalian DNA2 helicase/nuclease recognizes and cleaves telomeric G4 in vitro. Consistent with DNA2’s role in removing G4, DNA2 deficiency in mouse cells leads to telomere replication defects, elevating the levels of fragile telomeres (FTs) and sister telomere associations (STAs). Such telomere defects are enhanced by stabilizers of G4. Moreover, DNA2 deficiency induces telomere DNA damage and chromosome segregation errors, resulting in tetraploidy and aneuploidy. Consequently, DNA2-deficient mice develop aneuploidy-associated cancers containing dysfunctional telomeres. Collectively, our genetic, cytological, and biochemical results suggest that mammalian DNA2 reduces replication stress at telomeres, thereby preserving genome stability and suppressing cancer development, and that this may involve, at least in part, nucleolytic processing of telomeric G4.
Keywords: aneuploidy, cancer, DNA2 nuclease, G4, telomere
Introduction
Telomeres are specialized DNA–protein structures that protect chromosome ends from inappropriate degradation and fusion (Blackburn et al, 2006; O'Sullivan & Karlseder, 2010). Maintaining the integrity of telomeres is essential for genome stability and prevention of cancer development (Artandi et al, 2000; Davoli and de Lange, 2012; Gunes and Rudolph, 2013). Mammalian telomeres consist of hundreds to thousands copies of tandem TTAGGG repeats and terminate in short 3′-TTAGGG single-stranded overhangs (Blackburn et al, 2006). Telomerase maintains the length of telomeres and counteracts telomere shortening during each cell division (Blackburn et al, 2006). Telomeres also rely on proteins binding to telomeric DNA to provide the protective functions (de Lange, 2005; Palm and de Lange, 2008). A protein complex known as shelterin, consisting of TRF1, TRF2, POT1, TPP1, TIN2 and RAP1, binds to telomeric DNA. These proteins regulate telomere length homoeostasis and prevent inappropriate activation of DNA damage response and repair (de Lange, 2005; Palm and de Lange, 2008).
Telomere maintenance requires efficient and faithful replication of telomeric DNA, which is fundamental for preventing rapid loss of telomeres. However, in mammalian cells, replication of telomeric DNA sequences is problematic and replication forks stall frequently within the telomere repeats (Martinez et al, 2009; Sfeir et al, 2009). Stalled forks, if not restarted promptly, may collapse and prematurely terminate telomere replication, resulting in rapid telomere loss. Therefore, the conventional replication machinery needs assistance from other factors to successfully duplicate telomere repeats. Several studies suggest that efficient replication of telomere DNA is promoted in a variety of ways. For example, TRF1 has been suggested to recruit the BLM helicase to telomeres (Martinez et al, 2009; Sfeir et al, 2009), probably to resolve late-replicating intermediate structures and decatenated DNA structures at telomeres (Barefield and Karlseder, 2012). TRF2 and Apollo, working together with topoisomerase IIα, may relieve topological stress during telomere replication (Ye et al, 2010). The WRN helicase likely unwinds the G-quadruplex (G4) structure, promoting lagging strand telomere synthesis (Crabbe et al, 2004). The Ctc1/Stn1/Ten1 complex is required for efficient replication of lagging strand telomeres, perhaps by facilitating efficient DNA priming (Nakaoka et al, 2012; Gu et al, 2012; Huang et al, 2012; Stewart et al, 2012). The RECQL4 helicase unwinds telomeric D-loop structures with the help of shelterin proteins (Ghosh et al, 2012). Rqh1, the Schizosaccharomyces pombe RecQ ortholog, is thought to have a deleterious role on stalled telomere forks that is regulated by sumoylation (Rog et al, 2009). Sumoylation of Rqh1 helicase controls the fate of dysfunctional telomeres (Rog et al, 2009). Another helicase, RTEL1, disassembles T-loops and also counteracts the effect of ligands that stabilize G4 structures (Vannier et al, 2012). The tumour suppressor BRCA2 acts as the loader of RAD51 recombinase at telomeres during the S and G2 phases and facilitates telomere replication, although the mechanism remains to be defined (Badie et al, 2010). The requirement for diverse ways to promote efficient telomere replication perhaps lies within the unique nature of telomeres, including the highly repetitive and G-rich sequences that may form special structures, as well as abundant DNA-binding proteins which may constitute barriers to the replication fork (Gilson and Geli, 2007).
There is growing acceptance, although the evidence is not yet definitive, that telomeric T2AG3 repeats, as well as internal G-rich sequences containing runs of Gs, form G4 structures in vivo (Paeschke et al, 2005; Paeschke et al, 2011; Smith et al, 2011; Biffi et al, 2013). First, structure-specific antibodies or proteins recognizing G4 have detected G4 in vivo at ciliate telomeric T4G4 repeats as well as at human telomeres (Schaffitzel et al, 2001; Paeschke et al, 2005; Paeschke et al, 2008; Biffi et al, 2013). G4 structures form in a cell cycle-specific manner and are increased in S phase in human cells (Biffi et al, 2013). In ciliates G4 structure formation requires chaperones such as TEBP-α and -β (Paeschke et al, 2008; Juranek and Paeschke, 2012). These chaperones must be removed for resolution of the G4 DNA. A number of functions, such as transcriptional activation, stimulation of recombination in meiosis, chromatin formation, and rudimentary telomere capping, have been proposed for G4 structures (for review see Bochman et al, 2012). Second, ligands that stabilize G4 structures lead to telomere dysfunction. These molecules appear to inhibit telomere DNA replication, suggesting G4 DNA is also pathological (Tauchi et al, 2006; Rizzo et al, 2009). Consistent with this, previous biochemical studies demonstrated that G4 in DNA templates could inhibit DNA polymerase activity in vitro (Weitzmann et al, 1996; Han et al, 1999). Third, in addition to the proteins described above, numerous additional helicases have been shown to unwind G4 DNA, and several nucleases have been shown to bind, unwind, and cleave G4 (Liu and Gilbert, 1994; Ghosal and Muniyappa, 2005; Masuda-Sasa et al, 2008). In most cases, the in vivo roles of G4 at telomeres have not been determined. The mechanistic pathways for modulating putative telomeric G4 are particularly poorly defined. The key questions include: how do nucleases/helicases resolve telomeric G4, is G4 unwinding the only way to resolve G4, are these nucleases/helicases redundant or complementary, how do they interplay with shelterins and other telomere-associated proteins to maintain telomere stability? More importantly, it is still unclear whether defects in these G4-modulating pathways contribute to telomere and genome instabilities in vivo.
The DNA2 nuclease/helicase is a prime candidate for promoting replication through telomeres, perhaps by resolving telomeric G4 structure. DNA2 was initially identified and has, until recently, been most thoroughly studied in yeast (Budd and Campbell, 1995). It is an essential factor for Okazaki fragment processing in DNA replication (Budd and Campbell, 1997). DNA2 is thought to act only at a subset of difficult-to-replicate genomic regions, by cleaving the 5′ RNA/DNA primer within long RPA-bound single-stranded DNA (ssDNA) flaps, removing a portion of the flap (Bae et al, 2001). The resulting short flap structure is then cleaved by FEN1 nuclease to generate a DNA end suitable for ligation, which functions at the majority of Okazaki fragments that have short flaps and do not require DNA2 pre-processing (Zheng and Shen, 2011). In addition to its nuclease activity, DNA2 possesses a 5′ to 3′ helicase activity that can unwind DNA duplex and facilitate the production of the 5′ flap structure (Budd and Campbell, 1995; Bae et al, 2002; Kao et al, 2004). Interestingly, yeast DNA2 is also required for stable mitochondrial DNA maintenance (Budd et al, 2006). More recently, yeast DNA2 was shown to function in the DNA double-strand break (DSB) repair pathway (Zhu et al, 2008; Cejka et al, 2010; Niu et al, 2010; Nimonkar et al, 2011). Yeast DNA2 and RecQ helicase Sgs1 form a complex that processes 5′ DNA ends to generate 3′ ssDNA overhangs, which initiate the homologous recombination repair (Zhu et al, 2008; Cejka et al, 2010; Niu et al, 2010; Nimonkar et al, 2011). DNA2 is also a component of the S phase checkpoint (Kumar and Burgers, 2013; Lee et al, 2013). Most relevant to our studies, yeast DNA2 localizes preferentially to telomeres and does so in a cell cycle-specific manner (Choe et al, 2002). Strikingly, in G1 and G2 phase of the cell cycle DNA2 is found at telomeres, whereas during S phase DNA2 leaves telomeres and is found throughout the replicating DNA (Choe et al, 2002). Most important, genetic studies revealed directly that DNA2 is required for both telomerase-dependent and telomerase-independent (recombinational) telomere elongation in both Saccharomyces cerevisiae and Schizosaccharomyces pombe (Choe et al, 2002; Tomita et al, 2004). In addition to playing a role in telomere replication, yeast DNA2 also participates in resection of telomeres to produce the ubiquitous 3′ G-rich overhangs (Bonetti et al, 2009). Yeast and human DNA2 were found to bind to both inter-molecular and intra-molecular yeast, ciliate, or human telomeric G4 structures within ssDNA and to cleave DNA adjacent to these G4 structures. In the presence of the cognate RPA, DNA2 also cleaves within the G4 structure (Masuda-Sasa et al, 2008). In addition, both yeast and human DNA2 were found to unwind intermolecular G4 structures (Masuda-Sasa et al, 2008).
While the biochemical properties of mammalian DNA2 are similar to those of yeast DNA2 (Masuda-Sasa et al, 2006; Masuda-Sasa et al, 2008), the extent to which the biological functions of DNA2 are conserved in mammalian cells remains to be clarified. We and others previously showed that mammalian DNA2 is predominantly detected in the mitochondrion (Zheng et al, 2008; Duxin et al, 2009) and lacks classic nuclear localization signals. We showed that DNA2 plays a role in long-patch base excision repair (Zheng et al, 2008), and we recently demonstrated that three human progressive myopathies are linked to mutations in DNA2 that inactivate DNA2 helicase and nuclease activities and cause mitochondrial DNA instability (Ronchi et al, 2013). Although only a small fraction of human DNA2 is detected in the nucleus by immunofluorescence (IF; Duxin et al, 2009), mammalian DNA2 is apparently present in the nucleus at levels equivalent to other replication proteins, as evidenced by extensive analysis of co-immunoprecipitation between DNA2 and multiple replication proteins: pol δ, PCNA, And1, Mcm10 and RPA (Duxin et al, 2012; Peng et al, 2012). Cells lacking DNA2 show induction of the S phase checkpoint, numerous internuclear chromosome bridges, and increased aneuploidy, indicative of defects in DNA replication (Karanja et al, 2012; Peng et al, 2012). The role of DNA2 in chromosomal DSB resection is also conserved in humans (Nimonkar et al, 2011; Karanja et al, 2012; Peng et al, 2012). Interestingly, human DNA2 is overexpressed in human cancer cells, where it reduces replication stress and supports hyper-DNA replication (Peng et al, 2012). However, besides its role in repairing replication-linked chromosomal DSBs, the precise mechanistic roles of mammalian DNA2 in maintaining genome stability during S phase remain poorly understood. In particular, if and how mammalian DNA2 is involved in telomeric DNA replication has not yet been investigated.
In this study, we establish a knockout mouse model for the DNA2 gene and analyse the impact of DNA2 deficiency in both cells and mice. Homozygous DNA2 knockout is embryonic lethal, indicating that DNA2 is an essential gene. To determine if mammalian DNA2, like yeast DNA2, is important in telomere DNA metabolism, using cells derived from heterozygous DNA2 mice, we demonstrate that DNA2 deficiency results in fragile telomeres (FTs), an indication of telomere replication defect. We reveal that a subpopulation of mouse nuclear DNA2 is localized to telomeres and that DNA2 interacts with telomeric proteins TRF1 and TRF2, suggesting a potential role in telomere maintenance. We then purified DNA2 and have found that DNA2 nuclease cleaves intra-strand G4 formed by telomeric T2AG3 repeats. Our results suggest that DNA2 may utilize its nuclease activity to remove template G4 structures that may hinder fork progression at telomeric regions. Consistent with the biochemical results, small molecules stabilizing G4 elevates telomere replication stress in DNA2-deficient cells. Taken together, our data demonstrate that mammalian DNA2 alleviates telomere replication stress and maintains genome stability, perhaps through resolution of G4 structures.
Results
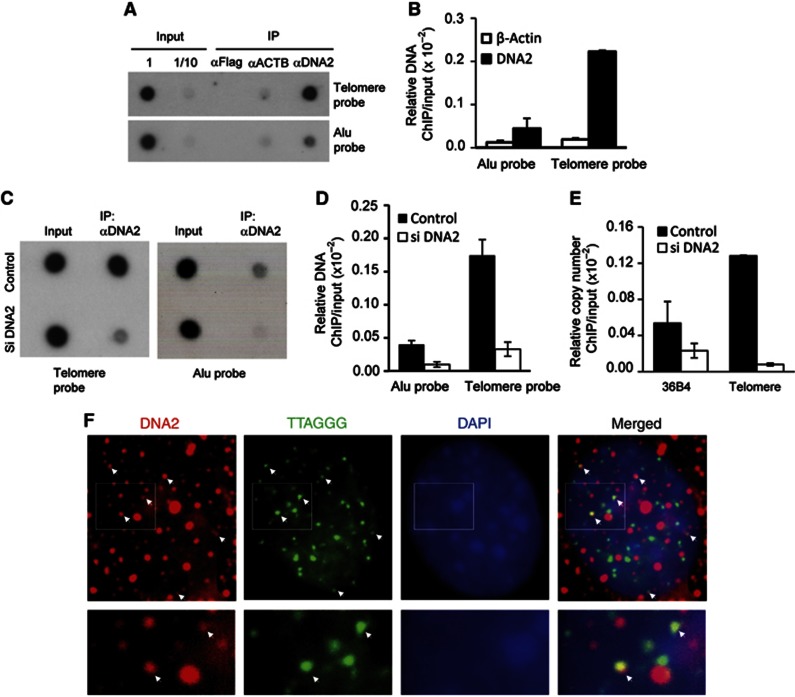
Mammalian DNA2 is localized at telomeres
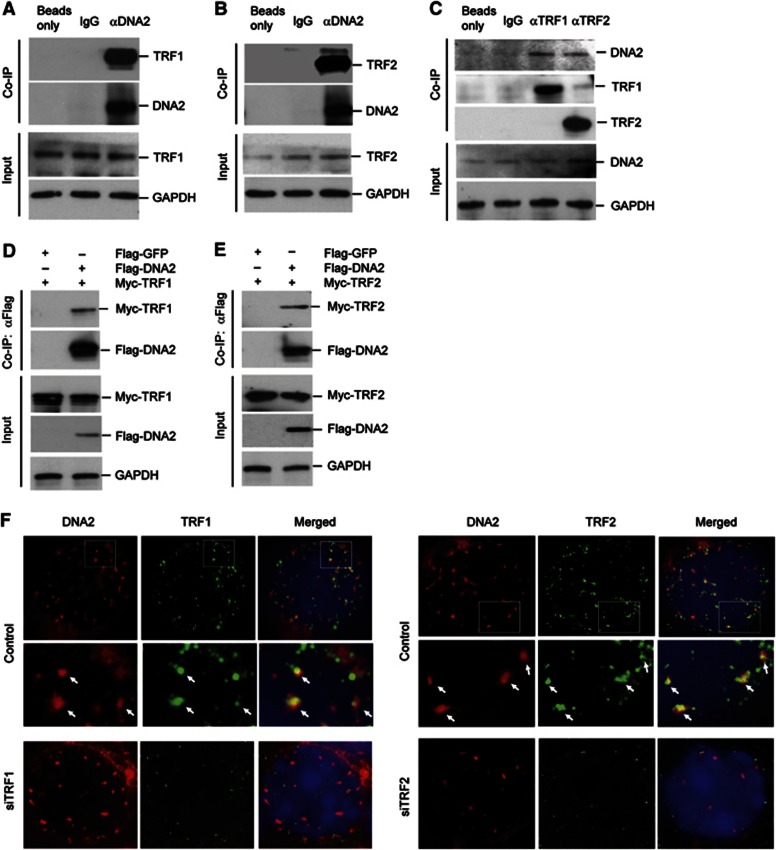
To determine if DNA2 played a role in maintaining telomere integrity, we sought to determine if DNA2 was associated with telomeres by conducting chromatin immunoprecipitation (ChIP) assays and IF-FISH assays on primary mouse embryo fibroblast (MEF) cells. ChIP results indicated that DNA2 was enriched at telomeres (telomere probe) by ∼10-fold compared with the internal regions of the nuclear genome (Alu probe) (Figure 1A and B). Furthermore, knockdown of mouse DNA2 in MEF cells abolished precipitation of telomere DNA by the anti-DNA2 antibody (Figure 1C–E), suggesting specific association of DNA2 with telomeres. IF-FISH confirmed that DNA2 was localized at telomeres (Figure 1F and Supplementary Figure S1). Not surprisingly, DNA2 also showed non-telomeric localization, in keeping with evidence that it has additional nuclear functions in maintaining genome integrity. Next, we performed co-immunoprecipitation to determine if DNA2 interacted with shelterin proteins such as TRF1 and TRF2 (de Lange, 2005). TRF1 and TRF2 were pulled down with DNA2 by the antibody against mouse DNA2 (Figure 2A and B), and DNA2 was also pulled down with TRF1 or TRF2 by the antibody against mouse TRF1 or TRF2 (Figure 2C). To further validate these observations, we then co-expressed Flag-tagged mouse DNA2 and Myc-tagged mouse TRF1 or TRF2. We pulled-down Flag-DNA2 with the anti-Flag and found that Myc-TRF1 and TRF2 were co-immunoprecipitated with Flag-DNA2 only from cells overexpressing Flag-DNA2 (Figure 2D and E). Consistent with these results, we observed that DNA2 nuclear foci co-localized with TRF1 or TRF2 in MEFs cells (Figure 2F). Taken together, these results suggested that mammalian DNA2 localized to telomeres and was in a complex with TRF1 and TRF2.
Figure 1.
DNA2 localizes to telomeres (A) ChIP assays on MEF cells. DNA was precipitated by a polyclonal antibody against mouse DNA2 and then analysed by dot blotting using a 32P-labelled probe corresponding to telomere sequence (telomere probe) or the Alu sequence (Alu probe). (B) The ChIP and input signals were quantified using the Image J program. The ChIP signal was normalized with the corresponding input signal and amount of ChIP DNA relative to the total amount of input was calculated. The values are the average±s.d of three independent assays. (C–E) ChIP assays on MEF cells with or without knockdown of mouse DNA2. (C) Representative image of dot blotting analysis of the input or ChIP DNA hybridized to a 32P-labelled probe corresponding to telomeres or Alu sequence. (D) Quantification of the ChIP DNA, which was normalized with the corresponding input DNA. The values are the average±s.d of three independent assays. (E) The input or ChIP DNA was analysed by real-time PCR. The single copy gene 36B4 was used to determine DNA2 binding to non-telomeric DNA region. The values are the average±s.d of three independent assays. (F) Representative IF-FISH images showing DNA2 is localized to telomeres in MEFs. The image of extended depth field of eight Z-stacking images (0.275 μM thickness per slice) was shown. Individual Z-stacking images were shown in Supplementary Figure S1. White arrows indicate DNA2 (red) co-localization with telomere DNA (green). Bottom panel shows enlarged images from the boxes areas.
Source data for this figure is available on the online supplementary information page.
Figure 2.
DNA2 interacts with telomeric proteins TRF1 and TRF2. Co-IP of endogenous DNA2 with TRF1 (A) and TRF2 (B) in MEF cells. GADPH was used as the internal control. (C) Co-IP was performed using TRF1 or TRF2 antibody in MEF cells. GADPH was used as the internal control. Flag-DNA2 and Myc-TRF1 (D) or Flag-DNA2 and Myc-TRF2 (E) were transiently co-expressed in 293T cells. Co-IP was performed using anti-FLAG antibody. The pulled-down Flag-DNA2 and Myc-TRF1 (D) or Flag-DNA2 and Myc-TRF2 (E) were analysed on Western blots using an antibody against Flag tag or Myc tag, respectively. GADPH was used as the internal control. (F) Co-localization of DNA2 with TRF1 or TRF2 in primary MEF cells with or without knockdown of TRF1 or TRF2. The bottom panel under the images of the control MEF cells is the enlarged view of the white boxed area of the image.
Source data for this figure is available on the online supplementary information page.
DNA2+/− MEF cells display elevated FTs, STAs, and other telomeric abnormalities
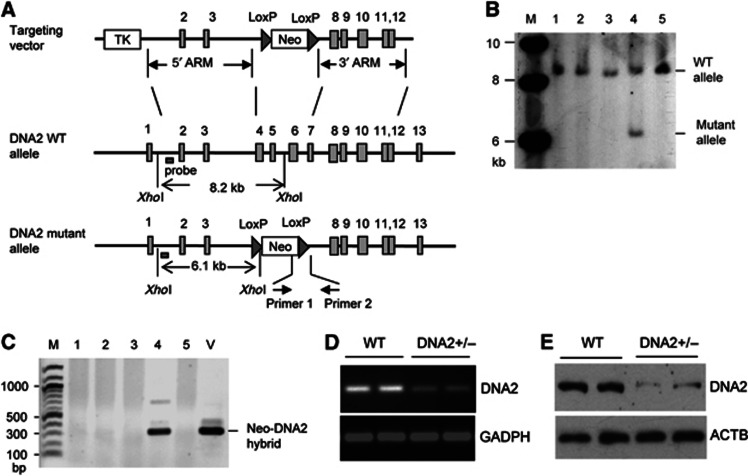
The fact that mammalian DNA2 localized to telomeres suggests that DNA2 may play a role in maintaining telomere integrity. To investigate the functions of DNA2 at telomeres, we knocked out DNA2 in the mouse genome using a gene targeting approach (Figure 3A) as previously described (Zheng et al, 2007b). The genotype of DNA2-knockout mice was confirmed by both Southern blotting and PCR (Figure 3B and C). No live homozygous DNA2-knockout mice were obtained, and all homozygous DNA2-knockout embryos died before the E7.5 stage, suggesting that DNA2 is essential for embryonic development. Heterozygous DNA2-knockout mice were viable. To determine if DNA2 haploinsufficiency caused telomere instabilities, we derived MEF cells from embryos of the heterozygous DNA2-knockout (DNA2+/−) mice (E13.5). The haploinsufficiency for DNA2 expression in the DNA2+/− MEF cells was confirmed by semi-quantitative RT-PCR and by western blot (Figure 3D and E).
Figure 3.
Establishment of DNA2-knockout mice. (A) Scheme for construction of the gene targeting vector for deletion the exons 4–7 of the mouse DNA2 genes. (B) Southern blot analysis to screen DNA2 heterozygous-knockout (DNA2+/−) ES clones. Genomic DNA was digested by XhoI. The DNA2 allele was analysed by Southern blot analysis. The WT allele (lanes 1–3, 5) gave a band of 8.2 kb, while the DNA2+/− allele (lane 4) gave a 8.2- and a 6.1-kb bands. (C) Genotyping DNA2 alleles of ES cells by PCR. PCR was conducted using the 5′ primer corresponding to the neomycin gene and the 3′ primer corresponding to the sequence within intron 7 of DNA2. Only PCR on genomic DNA with the DNA2 mutant allele could produce a DNA fragment of 300 bp (lane 4). The targeting vector was used as a positive control (lane 5). Confirmation of haploinsufficiency of DNA2+/− MEF cells (E13.5, P0) by RT-PCR (D) and western blot (E).
Source data for this figure is available on the online supplementary information page.
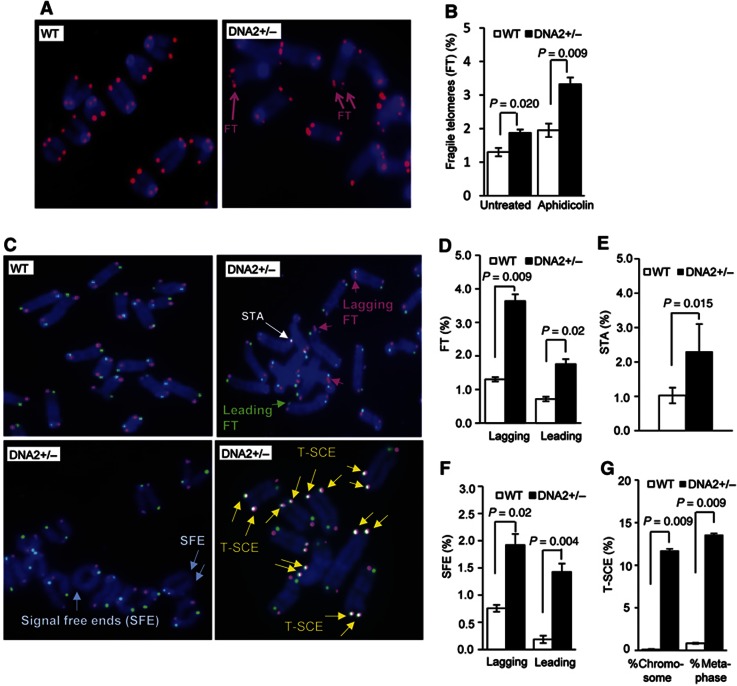
Compared with WT MEF cells, DNA2+/− MEF cells showed a significant increase in FTs (Figure 4A and B), which have been identified as hallmarks of telomere replication defects (Chan et al, 2009; Sfeir et al, 2009). The number of FTs observed in DNA2+/− MEFs was similar to that observed in WT cells that were treated with aphidicolin, which inhibits DNA polymerases and causes replication stress (Figure 4B), suggesting that DNA2 is required for efficient telomere replication. Moreover, FTs were further increased in the DNA2+/− MEFs when they were treated with aphidicolin (Figure 4B). In addition, we observed <1% of chromosome breaks in both WT and DNA2+/− MEF cells under normal culture conditions (Supplementary Figure S2). Although the aphidicolin treatment induced chromosome breaks in both WT and DNA2+/− MEF cells, greater increases were observed in the DNA2+/− MEF cells (Supplementary Figure S2). In addition, we found that the primary DNA2+/− MEF cells had a nucleotide incorporation rate similar to WT cells (Supplementary Figure S3). These results suggested that DNA2 might either be redundant with another function, as in yeast, or may not be a primary factor for DNA replication under normal physiology, but may play a critical role in counteracting DNA replication stress caused by telomeric DNA sequences or by genotoxic insults.
Figure 4.
DNA2 deficiency results in telomere defects. (A) Telomeres in WT and DNA2+/− MEF cells were analysed by FISH using the telomere-specific PNA probe, Cy3-(TTAGGG)3 (red). DNA was counterstained by DAPI (blue). FTs are indicated by pink arrows. (B) Percentage of FTs in WT and DNA2+/− cells untreated or treated with 0.2 μM aphidicolin for 24 h. Values are means±s.e.m. from five independent assays. In each assay, ∼3200 telomere ends were analysed. P-values were calculated using the two-tailed Wilcoxon–Mann–Whitney U test. (C) Representative CO-FISH images of WT and DNA2+/− MEF cells. Leading and lagging FTs are delineated by green and pink arrows, respectively. STA, telomere loss (SFE), and T-SCE are indicated by white, light blue, and yellow arrows, respectively. Quantification of telomeres that were fragile (D), STA (E), SFE (F), and T-SCE (G). Values are means±s.e.m. from independent assays. At least 4000 telomere ends were analysed for each experiment. P-values were calculated using the two-tailed Wilcoxon–Mann–Whitney U test.
We next employed chromosome-orientation FISH (CO-FISH) to determine if FTs caused by DNA2 deficiency occurred at telomeres replicated by leading- or lagging-strand synthesis. We found that FTs were increased on both leading and lagging strands in DNA2+/− MEF cells (Figure 4C and D). We also showed that sister telomere associations (STAs), indicated by overlapping green and red FISH signals from sister telomeres, were more frequent in DNA2+/− MEF cells than in the WT cells (Figure 4C and E). In addition, the number of signal free ends (SFEs), representing telomere loss, was increased on chromosome ends at both the leading and lagging strands in DNA2+/− MEF cells (Figure 4C and F). We also found that telomere sister-chromatid exchange (T-SCE) frequently occurred in DNA2+/− MEF cells, while T-SCE was nearly undetectable in the WT cells (Figure 4C and G), suggesting that homologous recombination was also elevated in DNA2-deficient cells.
DNA2 nuclease cleaves G4 within DNA bubble or flap structures
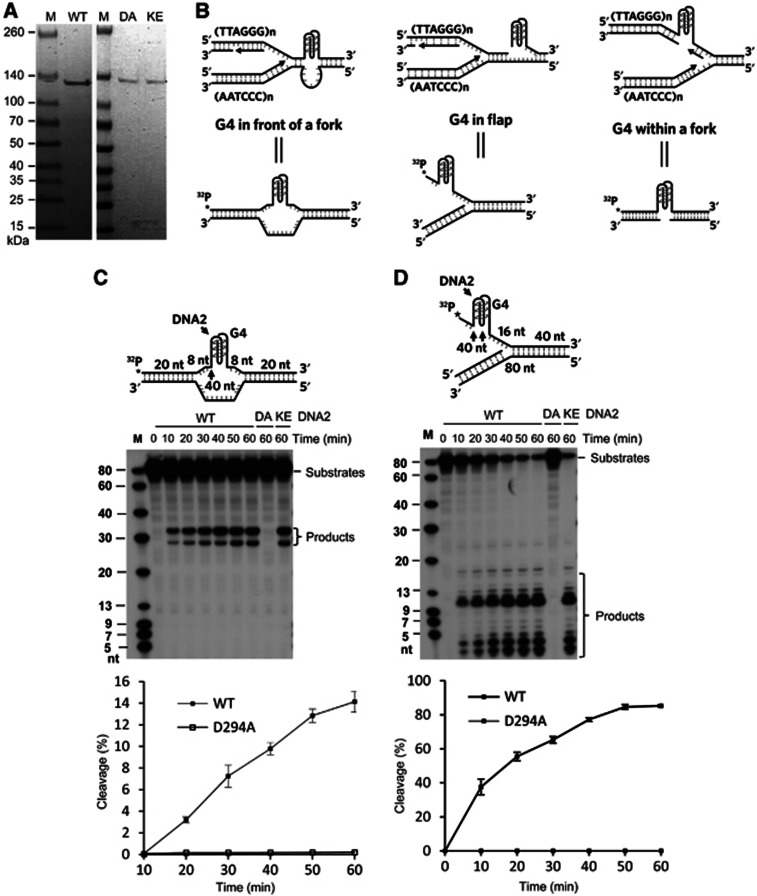
The observation that mammalian DNA2 deficiency increased the number of FTs under both normal growth and replication stress conditions suggests that mammalian DNA2 is required to reduce replication stresses and promote efficient telomere replication. One major obstacle when replicating telomere repeats is the potential formation of G4 structures at the TTAGGG template strand. When a G4 structure forms in the telomeric DNA template and is not resolved, it will impede the progression of the fork (Bochman et al, 2012). Helicases such as BLM and RTEL1 have been implicated in inhibiting the formation of telomeric G4 and suppressing FTs (Sfeir et al, 2009; Vannier et al, 2012). As DNA2 is a nuclease/helicase with binding, unwinding, and cleavage activity on both inter- or intra-molecular G4 (Masuda-Sasa et al, 2008), and G4 is readily formed by the TTAGGG repeats in vitro and antibodies specific for G4 react with human telomeres in vivo (Biffi et al, 2013), we proposed that mammalian DNA2 may play a role in promoting telomere replication by suppressing or reversing G4 formation in telomeres. To test our hypothesis, we purified full-length recombinant human DNA2 (Figure 5A) and assayed the ability of DNA2 to resolve telomeric G4 in model DNA substrates. Three model substrates designed to mimic G4 structures formed at different scenarios during replication were constructed. One substrate contained a G4 motif in a bubble (termed bubble substrate), mimicking a G4 in a template that would potentially block fork progression (Figure 5B). The second substrate contained a G4 motif in a single-stranded 5′ flap (termed flap substrate) (Figure 5B). The ssDNA flap structure could arise in different DNA repair pathways including long-patch base excision repair, the nucleotide excision repair, non-homologous end joining, or homologous recombination (Zheng et al, 2011). The un-repaired DNA flap, nicks, or gaps would collapse replication forks (Xu et al, 2011). The third substrate contained a gap on the strand opposite to the one containing a G4 structure (gap substrate) (Figure 5B), mimicking the G4 formed within the fork between two Okazaki fragments. Remarkably, DNA2 cleaved the G4 in the bubble substrate, producing DNA bands of about 30 nt (Figure 5C), indicating a role for the endonucleolytic activity of DNA2. Interestingly, the size of the products corresponded to cleavages at the junction between ssDNA and the runs of 3Gs in the G4 structure as well as inside the G4 structure, probably in the most 5′ loop (Figure 5C). However, we did not detect obvious cleavage of regular bubble DNA substrates by human DNA2 (Supplementary Figure S4A). This is consistent with previous report that yeast DNA2 does not cleave bubble substrates (Kao et al, 2004). In addition, DNA2 actively cleaved the G4 containing flap, also with major cleavage sites located both immediately to 5′ and inside the G4 structure (Figure 5D). This cleavage pattern was distinct from the cleavage of regular 5′ flap DNA substrates by DNA2 (Supplementary Figure S4B). These findings suggested that DNA2 could specifically recognize and cleave G4 DNA structures within DNA bubbles or flaps. However, DNA2 did not cleave the gapped G4 substrate that lacks ssDNA adjacent to the G4 (Supplementary Figure S5). This may be important, because cleavage of G4 within a replication fork would generate DSBs, which would lead to unwanted repair or recombination.
Figure 5.
DNA2 nuclease removes telomeric G4 in vitro. (A) Purification of recombinant human WT and mutant DNA2 proteins (D294A, K671E). Coomassie brilliant blue staining of purified Flag-tagged WT DNA2 (left) and nuclease-deficient DNA2 mutant D294A and helicase-deficient DNA2 mutant K671E (right). (B) Three different telomeric G4 structures potentially formed during replication and corresponding synthetic DNA substrates mimicking these structures. DNA2 cleaves G4 bubble DNA substrate (C) and G4 flap substrates (D). The numbers shown on the model substrates indicate the nucleotide lengths of DNA segments. G4 bubble or flap substrates 5′-end labelled with 32P (1 pmol) were incubated with purified human WT DNA2, nuclease-deficient mutant D294A (DA), or helicase-deficient mutant K671E (KE) at 37°C for the indicated time periods. The reactions were analysed by 15% denaturing PAGE. The DNA2 cleavage sites are indicated by arrows on the corresponding model substrates. The bottom panels in C and D show quantification of the cleavage. Values are means±s.e.m. of three independent assays. Oligo sequences for constructing the model substrates are included in Supplementary Table S1.
Source data for this figure is available on the online supplementary information page.
Next, we determined whether both the nuclease and helicase activities of DNA2 played an important role in G4 cleavage. The nuclease-deficient mutant, D294A (DA), and the helicase-deficient DNA2 mutant, K671E (KE) (Masuda-Sasa et al, 2006) were purified (Figure 5A) and used in the G4 cleavage assay. The DA but not the KE mutant completely abolished the ability of DNA2 to cleave the telomeric G4 bubble and flap substrates (Figure 5C and D), indicating that the unwinding activity of DNA2 is dispensable and that the endonuclease activity of DNA2 is sufficient for cleaving the G4 structures.
G4 stabilizing molecules significantly increase FTs in DNA2+/− MEF cells
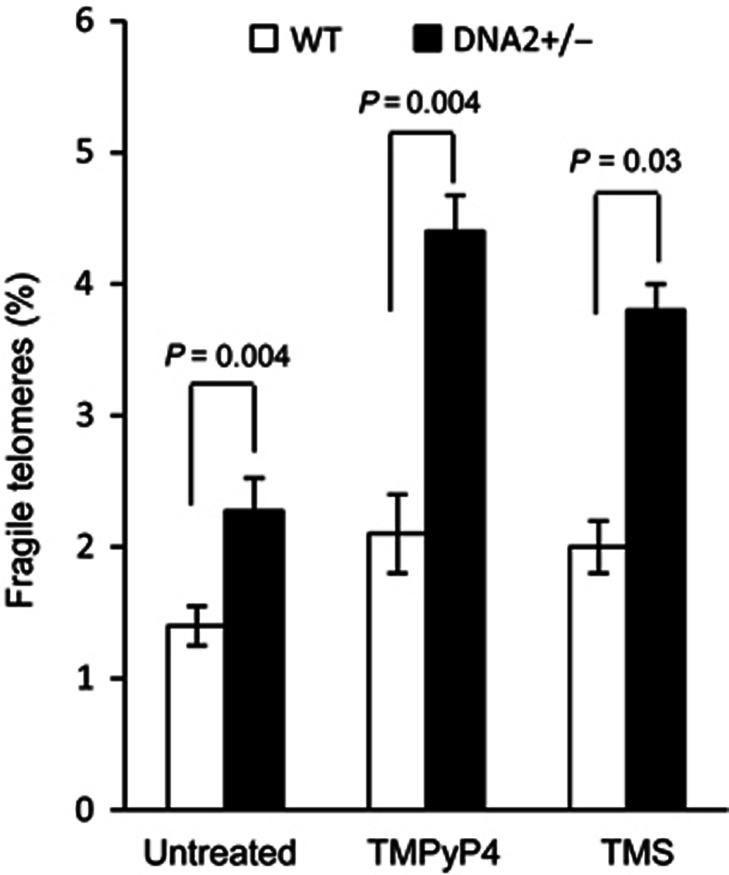
Our results suggested that the DNA2 nuclease resolves telomeric G4 structures in the bubble and in the 5′ flap by cleaving G4. This is distinct from G4 unwinding mediated by BLM and RTEL1. The repair of G4 in the telomere template may be important in preventing replication fork stalling or collapse. To support the role of DNA2 in removing G4 structures, we treated WT and DNA2+/− MEF cells with the small molecules TMPyP4 and telomestatin, which have been shown to stabilize G4 structures and cause dysfunctional telomeres (Rosu et al, 2003; Wu et al, 2008; Vannier et al, 2012). There was a moderate increase in the number of FTs in WT MEF cells treated with TMPyP4 or telomestatin (Figure 6). In striking contrast, TMPyP4 or telomestatin significantly increased the number of FTs in DNA2+/− MEF cells (Figure 6). We noted that the numbers of TMPyP4- or telomestatin-induced FTs in DNA2+/− MEF cells was similar to those of aphidicolin-induced FTs (Figure 4B). However, unlike aphidicolin, TMPyP4 or telomestatin did not lead to significant internal chromosome breaks (Supplementary Figure S2), which is consistent with the hypothesis that telomeric DNA sequences have a higher potential to form G4 structures than non-telomeric DNA sequence, given the large number of repeats (Gilson and Geli, 2007; Bochman et al, 2012). Altogether, our results suggest that G4 formation does occur at telomeres and that mammalian DNA2 plays an essential role in suppressing the replication stress caused by G4 formation.
Figure 6.
G4 stabilization molecules TMPyP4 and telomestatin induce more FT in DNA2+/− cells than in WT cells. WT or DNA2+/− MEF cells were left untreated or treated with TMPyP4 (50 μM, 24 h) or telomestatin (1 μM, 24 h). Levels of FTs in WT and DNA2+/− MEF cells were scored. Values are means±s.e.m. from five independent assays. P-values were calculated using the two-tailed Wilcoxon–Mann–Whitney U test.
DNA2 deficiency results in telomere DNA damage and chromosome segregation errors
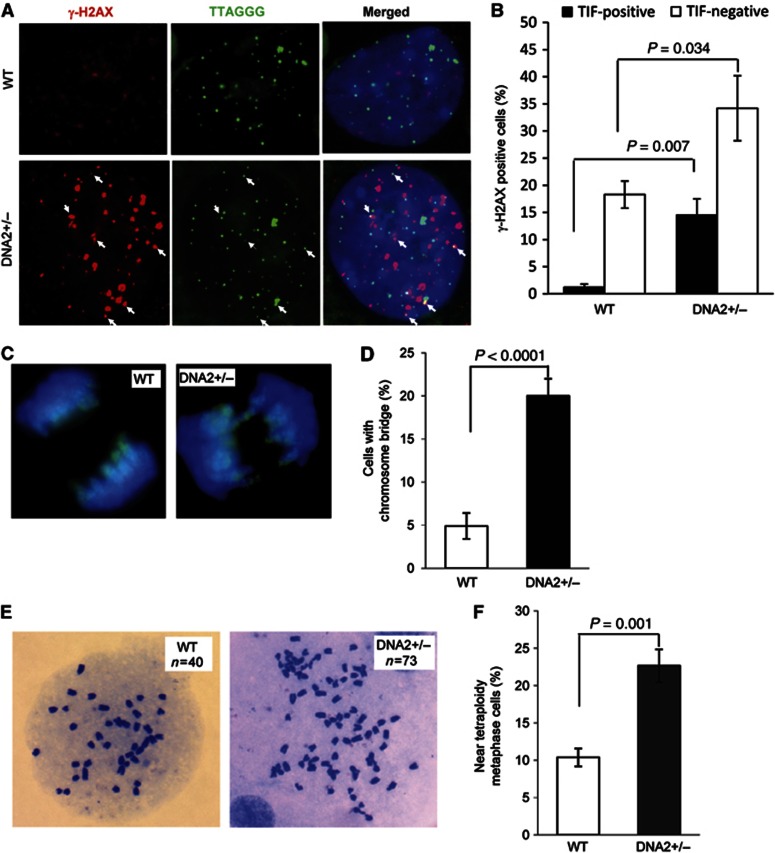
An increase in FTs in DNA2+/− MEF cells may result in the accumulation of telomere DNA damage in the cells. In agreement, γ-H2AX foci at telomeres (telomere dysfunction induced foci or TIF) were observed in 14.5% of DNA2+/− MEFs, a drastic increase compared with WT (1.2% TIF-positive cells) (Figure 7A and B). In addition, DNA2+/− MEF cells showed increased γH2AX foci at non-telomere regions by two-fold (Figure 7A and B), suggesting that DNA2 deficiency induced DNA damage in both telomeric and non-telomeric regions. We also found a marked increase in the number of anaphase chromosome bridges in DNA2+/− MEFs, compared with the WT control (20% versus 5%) (Figure 7C and D). Meanwhile, more than 20% of DNA2+/− metaphase cells were near tetraploid, compared with 10% in WT cells (Figure 7E and F). These results suggest that DNA2 deficiency-induced DNA damage, both at telomeric and non-telomeric regions, may perturb chromosome segregation and lead to aneuploidy and tetraploidy.
Figure 7.
DNA2 deficiency results in TIF and induces polyploidy. (A) Representative image showing co-localization of γ-H2AX foci (red) with telomeres (green) in DNA2+/− MEF cells. White arrows indicate the γ-H2AX foci at telomeres (yellow). (B) Percentage of γ-H2AX-positive cells in WT and DNA2+/− MEFs. Cells with no less than five γ-H2AX nuclear foci were scored as γ-H2AX-positive cells. Cells containing ≥5 γ-H2AX co-localization with telomeres are defined as TIF positive. Values are means±s.e.m. of five independent assays. Two-tailed t-test was used for calculation of the P-value. Approximately 150 cells were analysed in each assay. (C) DNA2+/− sMEF cells display anaphase chromosome bridges. Anaphase cells were detected by immunostaining using a monoclonal antibody against phosphorylated histone 3 (green). DNA was stained with DAPI (blue). (D) Percentage of anaphase cells with a chromosome bridge. (E) Representative metaphase spread of a diploid WT and a near-tetraploid DNA2+/− MEF cells. n Indicates chromosome numbers in each image. (F) Percentage of near-tetraploid cells in primary WT and DNA2+/− MEF cells. Values are means±s.e.m. of three independent assays. In each assay, more than 100 metaphase nuclei were analysed.
DNA2 deficiency causes dysfunctional telomeres contributing to the development of aneuploidy-associated cancer
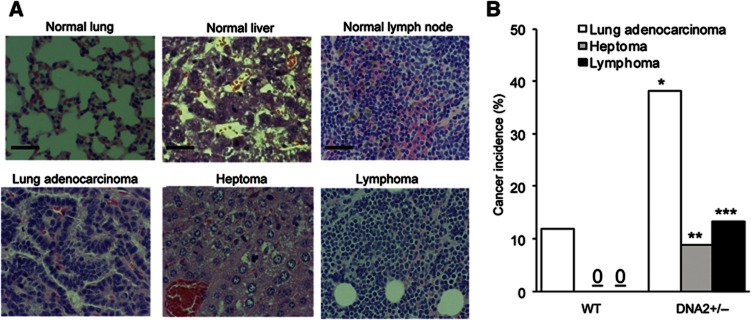
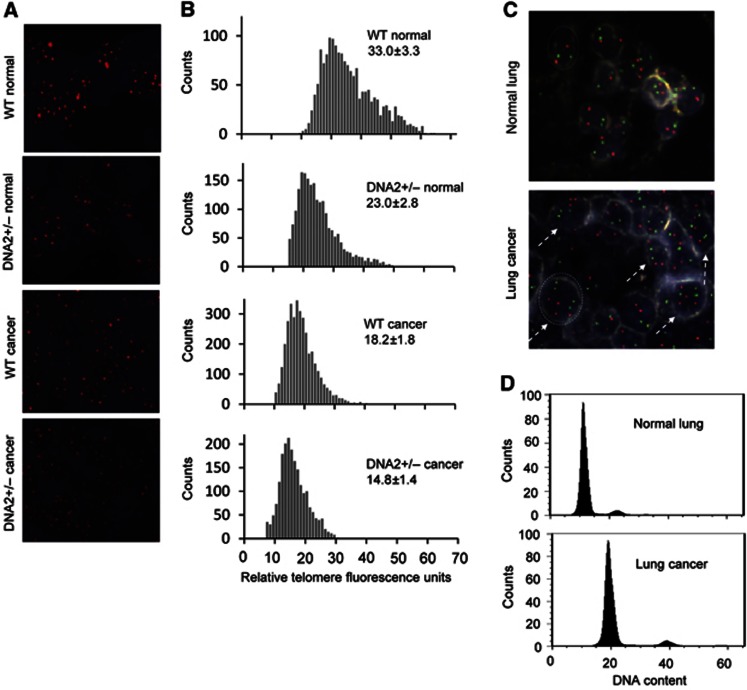
Next, we investigated if dysfunctional telomeres induced by DNA2 deficiency contributed to genomic instability and cancer development in vivo. We found that 62% of DNA2+/− mice (n=62) developed cancers including lung adenocarcinoma (39%, P=0.004), heptoma (10%, P=0.064), and lymphoma (12%, P=0.036), whereas only 12% of WT littermates (n=39) developed lung adenocarcinoma and no cancers were observed in other organs (Figure 8A and B). Furthermore, we employed quantitative telomere FISH to evaluate telomere lengths in vivo. We found that telomere lengths in normal DNA2+/− lung cells were considerably shorter than those in WT lung cells (Figure 9A and B), suggesting that DNA2 deficiency resulted in telomere dysfunction in vivo. Telomeres in lung cancer cells derived from DNA2+/− mice were significantly shorter than those in lung cancer cells derived from WT mice and those in the corresponding normal control cells (Figure 9A and B). To investigate if cancer developed in the DNA2+/− mice was aneuploid, we employed dual-colour FISH to analyse the numbers of chromosome 2 and chromosome 8 in tissue sections of WT normal lungs or DNA2+/− lung adenocarcinoma. The adenocarcinoma cells but not the normal lung cells carried three or more copies of chromosome 2 and/or chromosome 8 (Figure 9C), indicating that the lung adenocarcinoma was associated with aneuploidy. To validate the aneuploid feature of tumours, we then isolated cells from the WT normal lungs and a pool of lung adenocarcinomas and then used FACS to determine their ploidy status. Consistent with the FISH results, we found that the cells from adenocarcinoma were near-polyploid (Figure 9D). Taken together, these data suggest a positive correlation among DNA2 deficiency-induced telomere dysfunction, genome instabilities, and cancer development.
Figure 8.
DNA2+/− mice develop cancer at high frequency. Thirty-nine WT and 62 DNA2+/− littermates (16–20 m) were euthanized and anatomic and histopathological analyses were conducted on major organs. (A) Representative microimages of the normal lung, liver, and lymph nodes from WT mice and lung adenocarcinoma, heptoma, lymphoma developed in DNA2+/− mice. Scale bar: 50 μm. (B) Overall cancer incidence of WT and DNA2+/− mice. *P=0.004, **P=0.064, and ***P=0.036, Fisher exact test.
Figure 9.
DNA2+/− mice develop cancers displaying dysfunctional telomeres and aneuploid DNA contents. (A) Representative quantitative telomere FISH images on 5-μm sections of normal or lung cancer tissues of WT and DNA2+/− mice. (B) Relative telomere length from Q-FISH. Five randomly selected views were recorded on each sample (n=2). The relative fluorescence intensity for each FISH signal was analysed by ImagePro. The distribution of relative telomere length represented by the relative fluorescence intensity was displayed, and the mean±s.e.m. of the relative fluorescence intensity was calculated. (C) Dual-colour FISH on tissue section of the normal lung or lung adenocarcinoma. Nuclei were stained with DAPI. Each nucleus had clear edge with relatively light colour. White dashed lines indicate the edge of a typical nucleus. Aneuploid cells were indicated by dashed arrows. (D) FACS analysis of cells isolated from normal lung and lung adenomcarcinoma (a pool of three tumours of 3–5 mm diameters) of DNA2+/− mice. The data was processed using the FlowJo software. Comparison of the G1/G0 peak of lung adenocarcinoma with that of the normal lung cells (diploid) indicated most of the cells in lung adenocarcinoma were polyploid.
Discussion
Telomere DNA has been recognized as a difficult-to-replicate region, and inefficient duplication of the TTAGGG repeats results in rapid telomere shortening and telomere instabilities. Telomeric G4 has been suggested to play roles in impeding replication fork movement at telomeres (Gilson and Geli, 2007; Sfeir et al, 2009; Bochman et al, 2012; Vannier et al, 2012), though direct evidence for an inhibitory role for G-rich sequences has to date only been documented at internal sequences (Paeschke et al, 2011). Helicase RTEL1 and the RecQ helicases such as BLM, WRN, and RECQL4 have been proposed to reduce the formation of telomeric G4 and suppress the FT phenotype (Gilson and Geli, 2007; Sfeir et al, 2009; Ghosh et al, 2012; Vannier et al, 2012). However, the mechanisms underlying how these different helicases act to resolve telomeric G4 are unclear. In this study, we report a novel DNA2 nuclease-dependent pathway that resolves telomeric G4 structures to promote efficient telomere DNA replication. We have found that mammalian DNA2 localizes at telomeres and interacts with shelterin components TRF1 and TRF2. We have generated both DNA2-deficient cells and mice, and found that deficiency in DNA2 results in defective telomere replication, leading to elevated FTs, telomeres loss, and telomere DNA damage response. Using purified DNA2 protein, we demonstrate that DNA2 nuclease cleaves G4 structure formed within telomeric DNA sequences. The cleavage relies on DNA2 nuclease activity and the DNA2 helicase activity is not required. Consistently, DNA2-deficient MEFs showed a significant increase of FTs after treatments with G4 stabilization molecules, suggesting that DNA2 nuclease removes G4 structures to reduce replication stress at telomeres and thereby preserve telomere integrity. DNA2+/− mice develop aneuploidy-associated cancer with short telomeres. Together, these results suggest that mammalian DNA2 promotes efficient telomere replication and maintains telomere stability, which in turn contributes to tumour suppression.
The DNA2 nuclease-mediated cleavage of G4 structures revealed in this and in our previous studies is distinct from G4 unwinding. Mammalian DNA2 nuclease recognized and cleaved telomeric G4 DNA in a helicase-independent manner in vitro, as shown by using the helicase-dead DNA2 mutant protein, resulting in the nucleolytic removal of telomeric G4 formed in template DNA as well as the G4 formed in 5′ flap structures. The resulting DNA gap in the template could then presumably be repaired by the conventional ssDNA repair machinery. We propose that distinct pathways may exist to allow mammalian cells to efficiently remove various G4 structures arising from the TTAGGG repeats. In addition, it is plausible that under certain conditions, it may be difficult for helicases such as BLM, WRN, or RTEL1 to unwind the G4 structure, and DNA2 nuclease, working with ssDNA repair proteins, may cleave and repair the G4 structures to avoid replication fork stalling. To our knowledge, this is the first observation that telomeric G4 made up of telomeric repeats can be cleaved within a pseudo-replication bubble or on a 5′ flap, rather than simply within a ssDNA segment with both free 5′ and 3′ ends and rather than unwinding by a helicase/nuclease also required for stabilizing mammalian telomeres in vivo. Our data therefore provide novel insights into the molecular contribution of mammalian DNA2 to telomere maintenance, and add a new dimension to the mechanisms that cells can employ to deal with either telomeric or internal G4 structures.
DNA2 mutations, including a frame-shift mutation that disrupts the DNA2 gene, have been reported in cancer patients (Lee et al, 2010). Our current studies provide a positive correlation between these DNA2 mutations and cancer development. Furthermore, we suggest DNA2 acts as a tumour suppressor by maintaining the integrity of telomeres and counteracting DNA replication stresses induced by aphidicolin or other DNA damaging agents. FTs induced by DNA2 deficiency may cause a telomere DNA damage response, leading to telomere defects such as STAs or telomere loss. The telomere defects in turn may result in chromosome segregation errors and consequently tetraploidy and/or aneuploidy, which is a hallmark of human cancers (Weaver et al, 2007). Tetraploidy and aneuploidy have been linked to varying forms of chromosome instabilities (Weaver et al, 2007). Thus, telomere instabilities induced by DNA2 deficiency may directly or indirectly cause genome instabilities. This is consistent with previous studies showing that deficiency in telomeric protein Pot1 induces telomere dysfunction and results in tetraploidy and cancer (Wu et al, 2006; Davoli and de Lange, 2012). Meanwhile, DNA2 likely maintains the stability of non-telomeric sequences, and its deficiency impairs the cell’s capacity to counteract DNA replication stresses and results in accumulation of chromosome breaks, leading to chromosome deletions, translocations, and aneuploidy. Chromosome deletions and translocations have been frequently observed in human cancer and suggested to play a causal role in cancer development (Wang et al, 2005;Nussenzweig and Nussenzweig, 2010). In support of the hypothesis that DNA2 serves as a tumour suppressor, DNA2+/− mice has higher frequencies of cancer, which is viewed as a disease that arises from the accumulation of genome instabilities. Notably, the function of DNA2 as a tumour suppressor appears contradictory to a recent study showing that DNA2 was overexpressed in human cancers and played an important role in supporting cancer progression (Peng et al, 2012). We suggest that this phenomenon reflects the distinct role of DNA2 in normal cells and cancer cells. In normal cells, DNA2 reduces DNA replication stresses to maintain telomere integrity and to suppress cancer development. However, in cancer cells, oncogenesis induces high replication stresses that cause cellular senescence and apoptosis. In this case, DNA2 may work to reduce replication stresses to promote cell survival. A DNA repair gene with opposite roles as both a tumour suppressor and oncogene is not uncommon. Fen1 was previously shown to both suppress and support cancer development (Zheng et al, 2007b; Singh et al, 2008; Zheng et al, 2012). Recent studies suggest that RNF8, an E3 ubiquitin ligase that is involved in telomere DNA damage responses, can also both suppress and promote cancers (Li et al, 2010; Peuscher and Jacobs, 2011). Further studies may define the distinct roles of DNA2 as a tumour suppressor in normal cells but as a cancer promoter in the cancer-initiating cells.
Materials and methods
Generation of heterozygous mice for DNA2 knockout
To construct knockout vector for the mouse DNA2 allele, two DNA fragments corresponding to the mouse DNA2 gene were inserted into the poly-linker A and B, respectively, on the gene targeting vector, PKO scrambler NTK (Invitrogen, Carlsbad, CA). One DNA fragment (5′ arm) covered DNA sequences from intron 1 to intron 3 of the mouse DNA2 gene, and the other DNA fragment (3′ arm) corresponded to DNA sequences from intron 7 to intron 12 of the mouse DNA2 gene. The knockout vector encoding was electroporated into embryonic stem cells of the 129S1 genetic background. Recombination between the knockout vector and the WT DNA2 allele resulted in a mutant DNA2 allele, which deletes the exons 4–7 and disrupts the mouse DNA2 gene. DNA2+/− ES cells were selected by neomycin marker and confirmed with Southern blotting analysis using the probes 1 and 2. Male chimeric mice generated via FFAA ES cells (neo−) were crossed with female wt mice (129S1 genetic background) to transmit the mutation through the germline to produce heterozygous mice on 129S1 genetic background. The neomycin selection marker, which is flanked with LoxP sequences, was removed by cross the F1 DNA2+/− mice with the transgenic mice expressing Cre-recombinase (129S1 genetic background), generating DNA2+/− mice free of the neomycin gene.
Expression and purification of recombinant human and mouse DNA2 proteins
To express Flag-tagged human DNA2, the DNA fragment encoding full-length human or mouse DNA2 was amplified by PCR and inserted into the p3xflag CMV7.1 plasmid (Sigma). The expression vector, named as pCMV7-hDNA2 or mDNA2, was confirmed by sequencing. D294A and K671E for hDNA2 were produced by site-directed mutagenesis. The expression vectors were transfected into the 293T cells using Lipofectamine 2000 (Invitrogen). Recombinant WT and mutant DNA2 proteins were purified by a published affinity chromatograph protocol (Kim et al, 2006). In brief, whole cell lysates were incubated with the agarose beads conjugated with flag antibody (Sigma) for at least 6 h in cold room. After extensively washing with a buffer containing 50 mM Tris-Cl (pH 7.5) and 500 mM NaCl, the bound proteins were eluted with Flag peptide. The purity of DNA2 proteins was analysed by 4–15% gradient SDS–polyacrlamide electrophoresis (SDS–PAGE) and Coomassie brilliant blue staining.
siRNA knockdown
siRNA oligos against mouse DNA2, TRF1, TRF2, or control siRNA oligos were purchased from Bioland. siRNA oligos were transfected into mouse MEF cells using the RNAimax transfection reagent (Invitrogen) according to the instruction from Invitrogen.
CHIP assay
ChIP was performed as previously described (Loayza and De Lange, 2003). Briefly, primary MEF cells were fixed in 1% formaldehyde in PBS for 30 min at room temperature. The cells were lysed and sonicated. The sonicated lysates were pre-cleared with protein G agarose beads and incubated with the rabbit polyclonal antibody against the Flag tag (Sigma), actin (Santa Cruz) (non-specific control), or mouse DNA2 antibody (customized made by Genscript Corp, Piscataway, NJ). The specificity of the mouse DNA2 antibody was tested by purified recombinant mouse DNA2 and the mouse nuclear extracts from WT and DNA2 knockdown MEF cells (Supplementary Figure S6). The precipitates were dissolved in 10 μl water and dot-blotted onto Hybond N+ membranes. The membranes were hybridized with the 32P-labelled telomere-specific probe (TTAGGG)7 and the 32P-labelled Alu probe.
Co-immunoprecipitation assays
A non-specific mouse antibody or a mouse monoclonal antibody against the Flag tag (Sigma) was conjugated to protein A agarose beads. The beads were incubated with nuclear extracts at 4°C overnight and washed with PBS buffer containing 0.2% Tween-20. The precipitated proteins were analysed with Western blot. The polyclonal antibody against TRF1 or TRF2 is a generous gift from Dr de Lange. The monoclonal antibody against TRF1 or TRF2 is from Santa Cruz Biotechnologies. The monoclonal antibody against the Myc-tag or GADPH is from GeneTex.
IF microscopy
To stain the foci of DNA2/TRF1 or DNA2/TRF2 in MEF, cells were fixed with methanol at −20°C for 30 min and treated with ice cold acetone for 15 s. The cells were then incubated with a rabbit polyclonal antibody against DNA2 and a mouse monoclonal antibody again TRF1 or TRF2 (Santa Cruz) following the protocol as previously described (Zheng et al, 2007a). Nuclei were stained with DAPI. The slide was examined under a fluorescence microscope (AX70, Olympus).
FISH and CO-FISH
FISH and CO-FISH on telomeres were performed as described previously (Bailey et al, 2001). For FISH, cells were treated with colcemid, collected by trypsinization, swollen in 0.075 M KCl at 37°C for 15 min, and fixed in a freshly prepared 3:1 mix of methanol:glacial acetic acid. Slides containing metaphase spreads were then denatured and hybridized to peptide nucleic acid (PNA) probe Alexa488-OO-(TTAGGG)3 in order to visualize telomeres. For CO-FISH, cells were cultured in the presence of 10 μM BrdU and BrdC (3:1) for one population doubling before colcemid treatment. Slides containing metaphase spreads were incubated with Hoechst 33258 for 15 min, exposed to UV light for 30 min, treated with ExoIII (Promega) for 10 min at room temperature, and then sequentially hybridized to PNA Alexa488-OO-(TTAGGG)3 and Cy3-OO-(CCCTAA)3 probes at the room temperature for 2 h. Slides were then washed and dehydrated in ethanol series. DNA was counterstained with DAPI, and images were taken under Zeiss AxioImager M2 epifluorescence microscope. To determine the ploidy status of cells in normal and cancer tissues, dual-colour FISH for chromosomes 2 and 8 was conducted with FISH probes that corresponded to the mouse chromosomes 2qA1 and 8qA1. Slides were analysed with a BioView Imaging system (BioView, UK).
Immunofluorescence-FISH
IF-FISH was performed as described previously (Huang et al, 2012). Briefly, cells were grown in chamber slides and fixed either in 100% methanol at −20°C (for DNA2 staining) or 4% paraformaldehyde at room temperature, permeabilized with 0.15% Triton X-100 (for γ-H2AX), blocked with 1% BSA at 37°C for 1 h in humidified chamber, incubated with anti-DNA2 or anti-γ-H2AX for 1 h at 37 °C, washed with PBS three times, and then incubated with Dylight 549-conjugated secondary antibody (ThermoFisher) at 37°C for 1 h. Following DNA2 or γ-H2AX staining, the slides were refixed with 4% paraformaldehyde for 10 min, and hybridized to Alexa488-conjugated TTAGGG PNA telomere probe. The slides were denatured for 5 min at 90°C, incubated overnight at the room temperature, and washed with 70% formamide in 10 mM Tris (pH 7.5) for 15 min twice, followed by three times of wash in 0.1 M Tris (pH 7.5)/0.15 M NaCl/0.08% Tween-20 for 5 min each. Slides were then dehydrated in ethanol series, DNA was counterstained with DAPI, and Z-stack images were taken at a 0.275-μM thickness per slice under Zeiss AxioImager M2 epifluorescence microscope.
G4 model substrates and G4 cleavage assay
To make the G4 bubble substrate, the G4-B oligo was labelled with 32P at the 5′ end and annealed to the G4-B-T oligo at 1:1 ratio. To make the G4 flap substrate, the G4-F oligo was labelled with 32P at the 5′ end and annealed to the G4-F-T oligo and G4-F-T-B at 1:2:1 ratio. To make the G4 gap substrate, the G4-B oligo was labelled with 32P at the 5′ end and annealed to the G4-G-C1 and the G4-G-C2 oligos at 1:2:2 ratio. The oligo sequences were specified in Supplementary Table S1. The G4 bubble, flap, and gap substrates were annealed according to a previously described protocol that allow the formation of G4 structure (Vallur and Maizels, 2010). The formation of G4, which migrates faster than corresponding non-G4 bubble, flap, or gap, was confirmed by non-denaturing PAGE (Supplementary Figure S7). The substrates were then gel purified from non-denaturing PAGE and resuspended in a Tris–HCl buffer (pH 7.5) containing 10 mM EDTA and 10 mM KCl. To assay the nuclease activity, 100 fmol (Figure 5C) or 15 fmol (Figure 5D) WT or mutant DNA2 proteins was incubated with DNA substrates in the reaction buffer containing 50 mM Hepes-KOH (pH 7.5), 5 mM MgCl2, 2 mM DTT, and 0.1 mg/ml BSA at 37°C for a time period as specified. The DNA substrates and products were analysed with a 15% denaturing PAGE.
Histopathology and statistical analysis
Tissues were fixed in 10% formalin and tissue sections stained with hematoxylin and eosin (H&E). Slides were analysed in a double-blind fashion. The two-tails Fisher exact analysis was used to analyse if a disease phenotype was statistically significant. All protocols that involved animals were approved by the Research Animal Care Committee of City of Hope in compliance with the Public Health Service Policy of the United States.
Supplementary Material
Acknowledgments
We thank T de Lange for providing polyclonal antibodies against TRF1 or TRF2 and vectors expressing Myc-tagged mouse TRF1 and TRF2. We thank V Bedell D in the City of Hope Cytogenetics Core Facility for technical assistance with quantitative FISH analysis of tissue sections. We thank M Lee and B Armstrong in the City of Hope Microscopy Core Facility for assistance with IF. SS was supported by Poncin Trust Fellowship. This work was supported by NIH grants R01CA085344 to BS, and R15GM099008 and R21AG041375 to WC.
Author contributions: LZ, WC, BS conceived, designed, and supervised the study. WL, HD, JH, and LZ performed biochemical assays on recombinant DNA2 and characterized WT and DNA2 mutant mice. CL, MZ and JC performed recombinant DNA2 expression and purification. KS performed purification of telomestatin and contributed to the assay of FTs in the presence of the G4 stabilization molecules. SS, LZ, and WC performed experiments in vitro and in vivo to analyse telomeres in WT and DNA2 mutant cells. QH performed histological analyses. WL, SS, JC, LZ, WC, and BS analysed and interpreted data. LZ, WC, JC, and BS wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Artandi SE, Chang S, Lee SL, Alson S, Gottlieb GJ, Chin L, DePinho RA (2000) Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature 406: 641–645 [DOI] [PubMed] [Google Scholar]
- Badie S, Escandell JM, Bouwman P, Carlos AR, Thanasoula M, Gallardo MM, Suram A, Jaco I, Benitez J, Herbig U, Blasco MA, Jonkers J, Tarsounas M (2010) BRCA2 acts as a RAD51 loader to facilitate telomere replication and capping. Nat Struct Mol Biol 17: 1461–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae SH, Bae KH, Kim JA, Seo YS (2001) RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature 412: 456–461 [DOI] [PubMed] [Google Scholar]
- Bae SH, Kim DW, Kim J, Kim JH, Kim DH, Kim HD, Kang HY, Seo YS (2002) Coupling of DNA helicase and endonuclease activities of yeast Dna2 facilitates Okazaki fragment processing. J Biol Chem 277: 26632–26641 [DOI] [PubMed] [Google Scholar]
- Bailey SM, Cornforth MN, Kurimasa A, Chen DJ, Goodwin EH (2001) Strand-specific postreplicative processing of mammalian telomeres. Science 293: 2462–2465 [DOI] [PubMed] [Google Scholar]
- Barefield C, Karlseder J (2012) The BLM helicase contributes to telomere maintenance through processing of late-replicating intermediate structures. Nucleic Acids Res 40: 7358–7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi G, Tannahill D, Mcfferty J, Balasubramanian S (2013) Quantitative visualization of DNA G-quadruplex structures in human cells. Nature Chemistry 5: 182–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH, Greider CW, Szostak JW (2006) Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med 12: 1133–1138 [DOI] [PubMed] [Google Scholar]
- Bochman ML, Paeschke K, Zakian VA (2012) DNA secondary structures: stability and function of G-quadruplex structures. Nat Rev Genet 13: 770–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti D, Martina M, Clerici M, Lucchini G, Longhese MP (2009) Multiple pathways regulate 3′ overhang generation at S. cerevisiae telomeres. Mol Cell 35: 70–81 [DOI] [PubMed] [Google Scholar]
- Budd ME, Campbell JL (1995) A yeast gene required for DNA replication encodes a protein with homology to DNA helicases. Proc Natl Acad Sci U S A 92: 7642–7646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd ME, Campbell JL (1997) A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol Cell Biol 17: 2136–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd ME, Reis CC, Smith S, Myung K, Campbell JL (2006) Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase delta. Mol Cell Biol 26: 2490–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC (2010) DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature 467: 112–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying S, Hickson ID (2009) Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol 11: 753–760 [DOI] [PubMed] [Google Scholar]
- Choe W, Budd M, Imamura O, Hoopes L, Campbell JL (2002) Dynamic localization of an Okazaki fragment processing protein suggests a novel role in telomere replication. Mol Cell Biol 22: 4202–4217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe L, Verdun RE, Haggblom CI, Karlseder J (2004) Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science 306: 1951–1953 [DOI] [PubMed] [Google Scholar]
- Davoli T, De Lange T (2012) Telomere-driven tetraploidization occurs in human cells undergoing crisis and promotes transformation of mouse cells. Cancer Cell 21: 765–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lange T (2005) Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19: 2100–2110 [DOI] [PubMed] [Google Scholar]
- Duxin JP, Dao B, Martinsson P, Rajala N, Guittat L, Campbell JL, Spelbrink JN, Stewart SA (2009) Human Dna2 is a nuclear and mitochondrial DNA maintenance protein. Mol Cell Biol 29: 4274–4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxin JP, Moore HR, Sidorova J, Karanja K, Honaker Y, Dao B, Piwnica-Worms H, Campbell JL, Monnat RJ Jr, Stewart SA (2012) Okazaki fragment processing-independent role for human Dna2 enzyme during DNA replication. J Biol Chem 287: 21980–21991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal G, Muniyappa K (2005) Saccharomyces cerevisiae Mre11 is a high-affinity G4 DNA-binding protein and a G-rich DNA-specific endonuclease: implications for replication of telomeric DNA. Nucleic Acids Res 33: 4692–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh AK, Rossi ML, Singh DK, Dunn C, Ramamoorthy M, Croteau DL, Liu Y, Bohr VA (2012) RECQL4, the protein mutated in Rothmund-Thomson syndrome, functions in telomere maintenance. J Biol Chem 287: 196–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E, Geli V (2007) How telomeres are replicated. Nat Rev Mol Cell Biol 8: 825–838 [DOI] [PubMed] [Google Scholar]
- Gu P, Min JN, Wang Y, Huang C, Peng T, Chai W, Chang S (2012) CTC1 deletion results in defective telomere replication, leading to catastrophic telomere loss and stem cell exhaustion. EMBO J [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunes C, Rudolph KL (2013) The role of telomeres in stem cells and cancer. Cell 152: 390–393 [DOI] [PubMed] [Google Scholar]
- Han H, Hurley LH, Salazar M (1999) A DNA polymerase stop assay for G-quadruplex-interactive compounds. Nucleic Acids Res 27: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Dai X, Chai W (2012) Human Stn1 protects telomere integrity by promoting efficient lagging-strand synthesis at telomeres and mediating C-strand fill-in. Cell Res 22: 1681–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juranek SA, Paeschke K (2012) Cell cycle regulation of G-quadruplex DNA structures at telomeres. Curr Pharm Des 18: 1867–1872 [DOI] [PubMed] [Google Scholar]
- Kao HI, Campbell JL, Bambara RA (2004) Dna2p helicase/nuclease is a tracking protein, like FEN1, for flap cleavage during Okazaki fragment maturation. J Biol Chem 279: 50840–50849 [DOI] [PubMed] [Google Scholar]
- Karanja KK, Cox SW, Duxin JP, Stewart SA, Campbell JL (2012) DNA2 and EXO1 in replication-coupled, homology-directed repair and in the interplay between HDR and the FA/BRCA network. Cell Cycle 11: 3983–3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kim HD, Ryu GH, Kim DH, Hurwitz J, Seo YS (2006) Isolation of human Dna2 endonuclease and characterization of its enzymatic properties. Nucleic Acids Res 34: 1854–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Burgers PM (2013) Lagging strand maturation factor Dna2 is a component of the replication checkpoint initiation machinery. Genes Dev 27: 313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Lee M, Kang HJ, Kim DH, Kang YH, Bae SH, Seo YS (2013) The N-terminal 45-kDa domain of Dna2 targets the enzyme to secondary-structured DNA. J Biol Chem 288: 9468–9481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Kim YR, Yoo NJ, Lee SH (2010) Mutation and expression of DNA2 gene in gastric and colorectal carcinomas. Korean J Pathol 44: 5 [Google Scholar]
- Li L, Halaby MJ, Hakem A, Cardoso R, El Ghamrasni S, Harding S, Chan N, Bristow R, Sanchez O, Durocher D, Hakem R (2010) Rnf8 deficiency impairs class switch recombination, spermatogenesis, and genomic integrity and predisposes for cancer. J Exp Med 207: 983–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Gilbert W (1994) The yeast KEM1 gene encodes a nuclease specific for G4 tetraplex DNA: implication of in vivo functions for this novel DNA structure. Cell 77: 1083–1092 [DOI] [PubMed] [Google Scholar]
- Loayza D, De Lange T (2003) POT1 as a terminal transducer of TRF1 telomere length control. Nature 424: 1013–1018 [DOI] [PubMed] [Google Scholar]
- Martinez P, Thanasoula M, Munoz P, Liao C, Tejera A, McNees C, Flores JM, Fernandez-Capetillo O, Tarsounas M, Blasco MA (2009) Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev 23: 2060–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda-Sasa T, Imamura O, Campbell JL (2006) Biochemical analysis of human Dna2. Nucleic Acids Res 34: 1865–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda-Sasa T, Polaczek P, Peng XP, Chen L, Campbell JL (2008) Processing of G4 DNA by Dna2 helicase/nuclease and replication protein A (RPA) provides insights into the mechanism of Dna2/RPA substrate recognition. J Biol Chem 283: 24359–24373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaoka H, Nishiyama A, Saito M, Ishikawa F (2012) Xenopus laevis Ctc1-Stn1-Ten1 (xCST) complex is involved in priming DNA synthesis on single-stranded DNA template in Xenopus egg extract. J Biol Chem 287: 619–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC (2011) BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev 25: 350–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, Ira G, Sung P (2010) Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature 467: 108–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenzweig A, Nussenzweig MC (2010) Origin of chromosomal translocations in lymphoid cancer. Cell 141: 27–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan RJ, Karlseder J (2010) Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol 11: 171–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeschke K, Capra JA, Zakian VA (2011) DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 145: 678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeschke K, Juranek S, Simonsson T, Hempel A, Rhodes D, Lipps HJ (2008) Telomerase recruitment by the telomere end binding protein-beta facilitates G-quadruplex DNA unfolding in ciliates. Nat Struct Mol Biol 15: 598–604 [DOI] [PubMed] [Google Scholar]
- Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ (2005) Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat Struct Mol Biol 12: 847–854 [DOI] [PubMed] [Google Scholar]
- Palm W, De Lange T (2008) How shelterin protects mammalian telomeres. Annu Rev Genet 42: 301–334 [DOI] [PubMed] [Google Scholar]
- Peng G, Dai H, Zhang W, Hsieh HJ, Pan MR, Park YY, Tsai RY, Bedrosian I, Lee JS, Ira G, Lin SY (2012) Human nuclease/helicase DNA2 Alleviates replication stress by promoting DNA end resection. Cancer Res 72: 2802–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peuscher MH, Jacobs JJ (2011) DNA-damage response and repair activities at uncapped telomeres depend on RNF8. Nat Cell Biol 13: 1139–1145 [DOI] [PubMed] [Google Scholar]
- Rizzo A, Salvati E, Porru M, D'Angelo C, Stevens MF, D'Incalci M, Leonetti C, Gilson E, Zupi G, Biroccio A (2009) Stabilization of quadruplex DNA perturbs telomere replication leading to the activation of an ATR-dependent ATM signaling pathway. Nucleic Acids Res 37: 5353–5364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rog O, Miller KM, Ferreira MG, Cooper JP (2009) Sumoylation of RecQ helicase controls the fate of dysfunctional telomeres. Mol Cell 33: 559–569 [DOI] [PubMed] [Google Scholar]
- Ronchi D, Di Fonzo A, Lin W, Bordoni A, Liu C, Fassone E, Pagliarani S, Rizzuti M, Zheng L, Filosto M, Ferro MT, Ranieri M, Magri F, Peverelli L, Li H, Yuan YC, Corti S, Sciacco M, Moggio M, Bresolin N et al. (2013) Mutations in DNA2 Link progressive myopathy to mitochondrial DNA instability. Am J Hum Genet 92: 293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosu F, Gabelica V, Shin-ya K, De Pauw E (2003) Telomestatin-induced stabilization of the human telomeric DNA quadruplex monitored by electrospray mass spectrometry. Chem Commun (Camb) 21: 2702–2703 [DOI] [PubMed] [Google Scholar]
- Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A (2001) In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc Natl Acad Sci USA 98: 8572–8577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, De Lange T (2009) Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Yang M, Dai H, Yu D, Huang Q, Tan W, Kernstine KH, Lin D, Shen B (2008) Overexpression and hypomethylation of flap endonuclease 1 gene in breast and other cancers. Mol Cancer Res 6: 1710–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Chen Q, Yatsunyk LA, Nicoludis JM, Garcia MS, Kranaster R, Balasubramanian S, Monchaud D, Teulade-Fichou MP, Abramowitz L, Schultz DC, Johnson FB (2011) Rudimentary G-quadruplex-based telomere capping in Saccharomyces cerevisiae. Nat Struct Mol Biol 18: 478–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JA, Wang F, Chaiken MF, Kasbek C, Chastain PD 2nd, Wright WE, Price CM (2012) Human CST promotes telomere duplex replication and general replication restart after fork stalling. Embo J 31: 3537–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauchi T, Shin-ya K, Sashida G, Sumi M, Okabe S, Ohyashiki JH, Ohyashiki K (2006) Telomerase inhibition with a novel G-quadruplex-interactive agent, telomestatin: in vitro and in vivo studies in acute leukemia. Oncogene 25: 5719–5725 [DOI] [PubMed] [Google Scholar]
- Tomita K, Kibe T, Kang HY, Seo YS, Uritani M, Ushimaru T, Ueno M (2004) Fission yeast Dna2 is required for generation of the telomeric single-strand overhang. Mol Cell Biol 24: 9557–9567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallur AC, Maizels N (2010) Distinct activities of exonuclease 1 and flap endonuclease 1 at telomeric g4 DNA. PLoS One 5: e8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannier J-B, Pavicic-Kaltenbrunner V, Petalcorin MI, Hao D, Boulton SJ (2012) RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell 149: 795–806 [DOI] [PubMed] [Google Scholar]
- Wang Y, Putnam CD, Kane MF, Zhang W, Edelmann L, Russell R, Carrion DV, Chin L, Kucherlapati R, Kolodner RD, Edelmann W (2005) Mutation in Rpa1 results in defective DNA double-strand break repair, chromosomal instability and cancer in mice. Nat Genet 37: 750–755 [DOI] [PubMed] [Google Scholar]
- Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW (2007) Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell 11: 25–36 [DOI] [PubMed] [Google Scholar]
- Weitzmann MN, Woodford KJ, Usdin K (1996) The development and use of a DNA polymerase arrest assay for the evaluation of parameters affecting intrastrand tetraplex formation. J Biol Chem 271: 20958–20964 [DOI] [PubMed] [Google Scholar]
- Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, Deng Y, Behringer RR, Chang S (2006) Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell 126: 49–62 [DOI] [PubMed] [Google Scholar]
- Wu Y, Shin-ya K, Brosh RM Jr. (2008) FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol 28: 4116–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Zheng L, Dai H, Zhou M, Hua Y, Shen B (2011) Chemical-induced cancer incidence and underlying mechanisms in Fen1 mutant mice. Oncogene 30: 1072–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Lenain C, Bauwens S, Rizzo A, Saint-Leger A, Poulet A, Benarroch D, Magdinier F, Morere J, Amiard S, Verhoeyen E, Britton S, Calsou P, Salles B, Bizard A, Nadal M, Salvati E, Sabatier L, Wu Y, Biroccio A et al. (2010) TRF2 and apollo cooperate with topoisomerase 2alpha to protect human telomeres from replicative damage. Cell 142: 230–242 [DOI] [PubMed] [Google Scholar]
- Zheng L, Dai H, Qiu J, Huang Q, Shen B (2007a) Disruption of the FEN-1/PCNA interaction results in DNA replication defects, pulmonary hypoplasia, pancytopenia, and newborn lethality in mice. Mol Cell Biol 27: 3176–3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Dai H, Zhou M, Li M, Singh P, Qiu J, Tsark W, Huang Q, Kernstine K, Zhang X, Lin D, Shen B (2007b) Fen1 mutations result in autoimmunity, chronic inflammation and cancers. Nat Med 13: 812–819 [DOI] [PubMed] [Google Scholar]
- Zheng L, Dai H, Zhou M, Li X, Liu C, Guo Z, Wu X, Wu J, Wang C, Zhong J, Huang Q, Garcia-Aguilar J, Pfeifer GP, Shen B (2012) Polyploid cells rewire DNA damage response networks to overcome replication stress-induced barriers for tumour progression. Nat Commun 3: 815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Jia J, Finger LD, Guo Z, Zer C, Shen B (2011) Functional regulation of FEN1 nuclease and its link to cancer. Nucleic Acids Res 39: 781–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Shen B (2011) Okazaki fragment maturation: nucleases take centre stage. J Mol Cell Biol 3: 23–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Zhou M, Guo Z, Lu H, Qian L, Dai H, Qiu J, Yakubovskaya E, Bogenhagen DF, Demple B, Shen B (2008) Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol Cell 32: 325–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G (2008) Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.