Abstract
Damaged DNA, generated by the abstraction of one of five hydrogen atoms from the 2′-deoxyribose ring of the nucleic acid, can contain a variety of lesions, some of which compromise physiological processes. Recently, DNA damage, resulting from the formation of a C3′-thymidinyl radical in DNA oligomers, was found to be dependent on nucleic acid structure. Architectures relevant to DNA replication were observed to generate larger amounts of strand-break and 1-(2′-deoxy-β-D-threo-pentofuranosyl)thymidine formation than that observed for duplex DNA. To understand how this damage can affect the integrity of DNA, the impact of C3′-thymidinyl radical derived lesions on DNA stability and structure was characterized using biophysical methods. DNA architectures evaluated include duplex DNA (dsDNA), single 3′ or 5′-overhangs (OvHgs), and forks. Thermal melting analysis and differential scanning calorimetry measurements indicate that an individual 3′-OvHg is more destabilizing than a 5′-OvHg. The presence of a terminal 3′ or 5′ phosphate decreases the ΔG 25 to the same extent, while the effect of the phosphate at the ss-dsDNA junction of OvHgs is dependent on sequence. Additionally, the effect of 1-(2′-deoxy-β-D-threo-pentofuranosyl)thymidine is found to depend on DNA architecture and proximity to the 3′ end of the damaged strand.
1. Introduction
Maintaining DNA integrity is essential to facilitate proper physiological processes including DNA replication, repair, and transcription. The disruption of replication by reactive oxygen species (ROS) has been linked to a variety of diseases and disorders [1–3]. Causative in the development of disease via this mechanism is the stalling or collapse of replication forks. When this occurs, atypical secondary structures form which must be resolved and/or repaired before replication can resume [4]. Additionally, it has been shown that elevated levels of oxidative stress during replication result in a prolonged S-phase and if left unrepaired, can lead to apoptosis [5].
Double strand-breaks (DSBs), being one of the most lethal DNA damaging events resulting from ROS, are known to form through oxidative damage and have recently been reported to be replication induced [6]. Replication induced DSBs are proposed to result from a single strand-break (SSB) and/or other damage lesions generated in replicating DNA [6]. Under conditions of oxidative stress, SSBs can result from hydrogen atom abstraction at the 2′-deoxyribose ring in oligonucleotides. When a single hydrogen atom is removed by a ROS from any of the five carbons of the sugar moiety (Figure 1(a)), a highly reactive sugar radical is produced (Figure 1(b)). When trapping of the radical by a hydrogen atom donor does not occur, a strand-break is usually generated (Figure 1(c)). It is believed that the above-mentioned creation of stalled and collapsed replication forks is related to such sugar damaging events. The resulting substrates then signal spontaneous homologous recombination [7] and nonhomologous end-joining [8].
Figure 1.
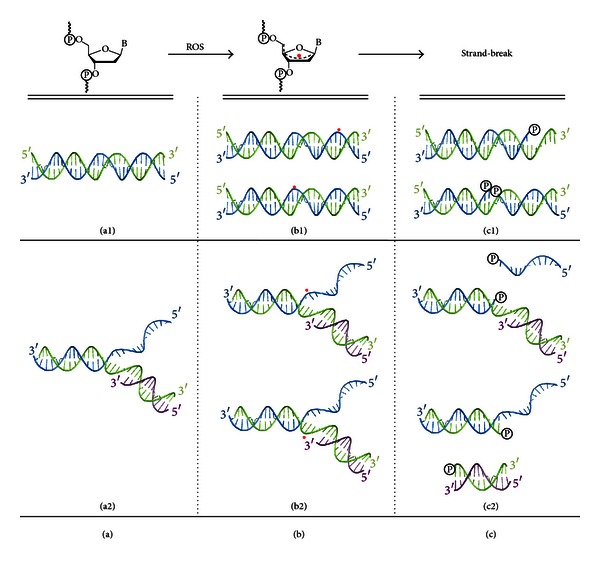
Hydrogen atom abstraction at a 2′-deoxyribose moiety in DNA replication relevant architectures. (a) Model replication substrates: duplex (a1) and flap (a2). (b) 2′-Deoxyribose radical intermediates in duplex (b1) and flap substrates (b2). (c) Strand-break products resulting from oxidative sugar damage in duplex (c1) and flap substrates (c2).
Our laboratory is interested in determining the role of DNA structure in preserving genomic integrity through protection of the nucleic acid from oxidative damage. We have observed that the selective generation of radical intermediates in DNA architectures that act as models of substrates which form during the process of replication results in elevated levels of strand-break products when compared to duplex DNA. The formation of 3′ and 5′-overhangs (OvHgs) containing a phosphate at the ss-dsDNA junction, as well as duplex substrates containing a terminal phosphate was observed [9]. Additionally, the formation of 1-(2′-deoxy-β-D-threo-pentofuranosyl)thymidine (2′-deoxyxylothymidine, 3, Figure 2) was observed as a result of trapping of the C3′-thymidinyl radical from the alpha face of the nucleotide by glutathione (GSH). This damage product was also formed readily in replication relevant architectures, while nearly absent in duplex DNA [9]. Together, these results highlight the protection against oxidative damage offered by the duplex structure of DNA when the C3′-thymidinyl radical is the reactive intermediate [9].
Figure 2.
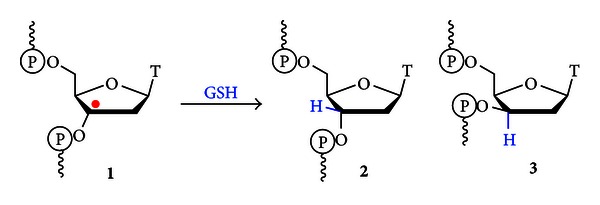
Reduction of the C3′-thymidinyl radical by GSH. Formation of substrates containing the repaired 2′-deoxyribose (2) and the 1-(2′-deoxy-β-D-threo-pentofuranosyl)thymidine (3) damage lesion were observed [9].
The effect of oxidatively generated damage lesions on the structure and stability of DNA substrates has received limited attention [10, 11]. Herein, we analyze the impact of damage lesions on the structure and stability of DNA architectures, which model those generated during replication. The impact of the presence of these lesions on replication derived model architectures reveals how DNA damage alters the stability of nucleotide substrates associated with replication.
2. Materials and Methods
2.1. Oligonucleotides
Oligonucleotides containing 3 (Z) were synthesized, using established methods, on an ABI 391 automated DNA synthesizer [12]. The synthesis of Sequence 1: 5′-TTTTTZTTTCAGGTTGCG-3′ and Sequence 2: 5′-GCGTTGGACTTTZTTTTT-3′ was performed on a 0.2 μm scale using polystyrene columns (Glen Research, Sterling, VA). Oligonucleotides were cleaved from the resin and the nucleobase and phosphate protecting groups removed by heating at 55°C overnight in aqueous ammonium hydroxide (28–30%). Oligonucleotide purification was accomplished using oligonucleotide purification cartridges (OPC, Glen Research, Sterling, VA) and the samples were quantified (ε = 158,100 L/mole∗cm) using UV absorbance at 260 nm on an Agilent 8453 UV-Visible Spectrophotometer. Sample purity was assessed by tandem ion-exchange/reverse-phase high-performance liquid chromatography and the identity confirmed using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). All unmodified oligonucleotides were either synthesized or purchased (Integrated DNA Technologies, Coralville, IA, USA).
2.2. UV Melting
Melting experiments were conducted on a Varian Cary 3E UV-Vis spectrophotometer equipped with a temperature controller. Absorbance changes for oligonucleotides (2 μM) in 25 mM chelex-100 treated phosphate buffer (pH 7.3) containing 60 mM potassium acetate and 6 mM magnesium acetate were recorded as a function of temperature from 10–80°C at a rate of 0.5°C/min at 260 nm. Each measurement was performed in triplicate. The average of three scans was taken and reported errors were calculated using standard deviation. The T m was obtained using the first derivative method. Calculated ΔT m was determined by subtracting the experimental values obtained for the 2′-deoxyribose containing construct from the corresponding constructs containing 3.
2.3. Circular Dichroism
The circular dichroism spectra for all architectures reported were collected using an AVIV 62DS CD Spectrometer at 25°C, using a quartz cuvette with a path length of 1 mm in a thermostatted holder. Spectra were collected from 320 to 200 nm at a rate of 20 nm/min using a bandwidth of 1 nm. Four scans were accumulated and averaged. Oligonucleotides were measured in phosphate buffer (pH 7.3) at (5 μM). Raw data (Θ) were converted to molar ellipticity ([Θ]) ([Θ] = Θ /(10∗C∗l)) and smoothed using a moving average of three. Cells were cleaned and dried between sample runs according to previously established methods [13].
2.4. Differential Scanning Calorimetry
Calorimetric excess heat capacity (ΔC p) versus temperature (T) profiles was plotted for each DNA architecture using data obtained on a VP Micro-Calorimeter following established methods [14]. Concentration of oligonucleotide samples was 0.2 mM in phosphate buffer (pH 7.3). All samples were stirred and degassed using a MicroCal ThermoVac for 16 minutes at 25°C. Samples were loaded and scanned at a rate of 1°C/min from 10°C to 100°C. Three scans per sample were collected and analyzed using Origin software. Values for ΔH DSC, ΔS, and ΔH νH were determined utilizing the equations listed below [15]:
| (1) |
where C p(ex) is the excess heat capacity, ΔH DSC is the enthalpy obtained directly from the raw data that has been baseline corrected and normalized, ΔS is the entropy calculated without assuming a two-state melting transition, ΔH νH is the calculated van't Hoff enthalpy, and C p(m) is the transition maximum value for C p(ex) [15]. ΔG at 25°C was obtained using the following Gibbs free energy equation:
| (2) |
Calculations for ΔΔH DSC, ΔΔS, and ΔΔG 25 were obtained by subtracting the values obtained for the core sequence from the corresponding products of interest. Reported errors were calculated using standard deviation.
3. Results and Discussion
3.1. Design of Architectures
During DNA replication, both the leading and lagging strand templates undergo a variety of architectural changes including the formation and unwinding of duplexes, as well as the transient formation of overhangs, flaps, and forks. Our previous work as well as that of others [9, 16, 17] has shown that the structure of DNA has a profound impact on the fate of 2′-deoxyribose radicals. After determining the structures of the lesions formed through independent generation of the C3′-thymidinyl radical, we investigated their impact on the structure and stability of DNA architectures that form via this type of damage. The interplay between DNA structure and damage outcomes is an important paradigm in our understanding of the ability of DNA to protect itself from damage events and the ability to repair damage when it occurs. The substrates utilized in these studies were designed to allow for the evaluation of the effects of both 3 and the strand-break products observed in our previous work.
Generally, the presence of short overhangs or terminal phosphates increases the stability of DNA constructs when compared to the corresponding blunt-ended duplexes [18–22]. In previous work, the effect of overhangs, otherwise known as “dangling ends,” on duplex stability was determined utilizing self-complimentary DNAs that produce a “dangling end” at both ends of the substrate (Figure 3). This design requires consideration of the thermodynamic contribution of two overhangs to DNA stability, which was compensated for by either halving the values of ΔΔG [20] or accounting for the changes induced by both [19, 21]. It is our hypothesis that the presence of “dangling ends” on both ends of the core DNA sequence has an influence on the T m and/or thermodynamics of melting. This is supported by the results reported in Benight et al. who used substrates containing both a 5′ and 3′-OvHg as well as dangling ends containing two 5′-OvHgs. These substrates indicated that the thermodynamic impact of the overhang was dependent upon the DNA substrates used [23]. The substrates utilized in these studies were designed to allow for the direct evaluation of the effects introduced by a single overhang, phosphate or 3 [9].
Figure 3.
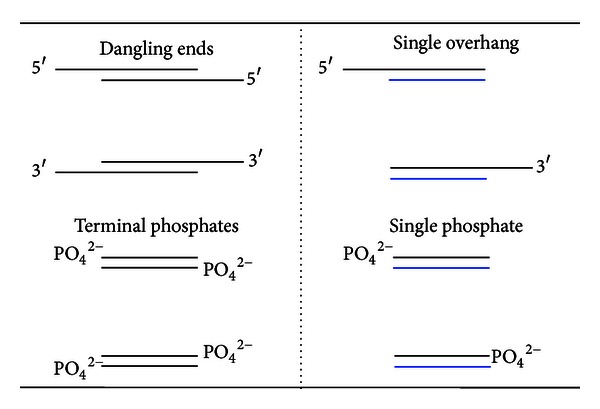
Structural differences in the design of some previously used substrates (left) and herein (right) in characterizing the thermodynamic contribution of a single-stranded DNA region or terminal phosphate (PO4 2−) on the stability of the core dsDNA. Oligonucleotides designed for dangling ends and terminal phosphates are self-complementary, while the evaluation of a single overhang or phosphate does not require self-complementary sequences.
3.2. Stability Studies of Architectures Containing 3
The melting temperatures for duplex, 5′ and 3′-OvHgs and fork architectures were determined for unmodified architectures as well as those containing 3. Incorporation of 3 into these architectures revealed that the replacement of a single 2′-deoxyribose with 3 decreases the stability of the duplex region (Table 1). Previously, it was shown that DNA duplexes containing 3 in a single location demonstrate decreased stability when compared to the unmodified duplex. This indicates that the inversion of configuration at the C3′-position of 2′-deoxyribose to 2′-deoxyxylose at a single nucleoside significantly destabilizes the DNA duplex [10, 24]. We made the same observation in the case of DNA duplexes 1D and 2D, obtaining decreases in T m of 5.1°C and 5.3°C, respectively. However, when 3 is found at an overhang or fork junction, the effect is significantly decreased. Melting temperatures of 5′-OvHgs 1A and 1B and fork 1E & 1F show no appreciable change when 3 is placed at the junction. Alternatively, the 3′-OvHg 2B and fork 2F containing 3 at the same position demonstrate a decrease in T m of 1.4°C and 0.7°C, respectively. As indicated by the respective melting temperatures of 2B and 2F, the overall effect of 3 appears to have less of an impact on stability in less stable substrates. These results suggest that the destabilizing effect of 3 is not only dependent on DNA architecture, but also on the location of the modification relative to the 5′ and 3′ ends of the oligonucleotide.
Table 1.
Comparison of T m values for unmodified and 3 containing DNA architectures relevant to replication. The position of 3 is indicated by “Z”.
| ID | Base Sequence | T m (°C) | ΔT m (°C) | |
|---|---|---|---|---|
| 1 | 5′-CGCAACCTGAAAX | |||
| 3′-GCGTTGGACTTTY | ||||
| 5′-OvHg | ||||
| 1A | X = OH | Y = TTTTTT | 51.6 ± 0.3 | — |
| 1B | X = OH | Y = ZTTTTT | 51.6 ± 0.1 | 0 |
| dsDNA | ||||
| 1C | X = AAAAAA | Y = TTTTTT | 57.9 ± 0.1 | — |
| 1D | X = AAAAAA | Y = ZTTTTT | 52.8 ± 0.1 | −5.1 |
| Fork | ||||
| 1E | X = GACTGT | Y = TTTTTT | 47.3 ± 0.3 | — |
| 1F | X = GACTGT | Y = ZTTTTT | 47.3 ± 0.2 | 0 |
|
| ||||
| 2 | 5′-GCGTTGGACTTTX | |||
| 3′-CGCAACCTGAAAY | ||||
| 3′-OvHg | ||||
| 2A | X = TTTTTT | Y = OH | 51.6 ± 0.3 | — |
| 2B | X = ZTTTTT | Y = OH | 50.2 ± 0.3 | −1.4 |
| dsDNA | ||||
| 2C | X = TTTTTT | Y = AAAAAA | 58.9 ± 0.1 | — |
| 2D | X = ZTTTTT | Y = AAAAAA | 53.6 ± 0.2 | −5.3 |
| Fork | ||||
| 2E | X = TTTTTT | Y = GACTGT | 48.6 ± 0.2 | — |
| 2F | X = ZTTTTT | Y = GACTGT | 47.9 ± 0.2 | −0.7 |
3.3. CD Analysis of Architectures Containing 3
To determine the effect of 3 on the overall conformation of these substrates, CD analysis of duplex, 5′ and 3′-OvHgs and fork DNA architectures containing 3 was performed (Figure 4). Previously, it was observed that 3 has a minimal effect on the secondary structure of duplex DNA [10]. All architectures, with and without 3, were observed to be predominately B-form as indicated by the presence of a positive band around 280 nm and a negative band around 245 nm [25]. Thus, the presence of 3 does not have a significant effect on the global conformation of these substrates. In comparing the architectures for Sequence 1 to those of Sequence 2, more pronounced differences between 2′-deoxyribose versus 3 containing architectures of Sequence 2 were observed as indicated by decreases in the minima at 245 nm and maxima at 280 nm. In duplex substrates 2C and 2D, the presence of 3 causes a decrease in the intensity of the minima at 245 nm and the maxima at 280 nm. In the 3′-OvHg 2B, the presence of 3 significantly decreases the intensity of the minimum at 245 nm, but has minimal to no affect on the maximum at 280 nm. Additionally, fork 2F shows no change at the minimum while a decrease in the maximum is observed. Taken together, the presence of 3 at a single location has more of an effect on base pairing interactions in duplex DNA, OvHg, and fork architectures of Sequence 2, than Sequence 1.
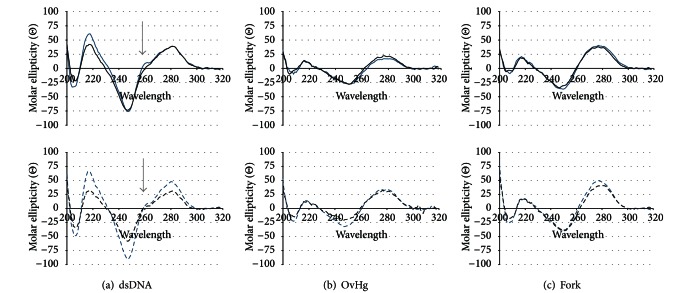
Figure 4.
CD spectra comparing the effect of 3 on the secondary structure of replication relevant architectures. The top row shows the CD spectra for constructs of Sequence 1 (solid lines), while the bottom row shows the CD spectra for Sequence 2 (dashed lines). Constructs containing unmodified oligomers are represented in blue, while the constructs containing 3 are in black. (a) duplexes, (b) 5′ and 3′ OvHgs, (c) fork.
In considering the architectures for Sequence 1, the comparisons between those containing unmodified oligomers and those containing 3, 5′-OvHg 1A and 1B and fork 1E and 1F, indicate no significant difference in overall conformation. Interestingly, the region between 240 and 300 nm in the spectra was obtained for duplexes 1C and 1D; there are also minimal differences with the exception of the shoulder at 260 nm. In the presence of 3, this shoulder is lost. This is an important observation as the presence of this shoulder is characteristic of poly A•T duplex regions [26]. However, this shoulder is not lost in the analysis of the duplex containing 3 of Sequence 2 (2D). This indicates that poly A•T base pairing in duplex DNA is affected by the presence of 3 in Sequence 1, but not in Sequence 2, suggesting a sequence dependent effect on duplex structure by the presence of a single 3.
3.4. Summary: Affects of 3 on Structure and Stability of Replication Relevant Architectures
Thermal melting and CD analysis are powerful methods for the evaluation of changes in DNA stability and structure. Here we demonstrate differences in substrate stability and structure introduced by the presence of 3 in duplex, OvHg and fork architectures. This is the first time that 5′-OvHg, 3′-OvHg, and fork constructs containing 3 have been reported. These results show that 3 destabilizes all DNA architectures investigated. It appears that 3 has a greater effect when located closer to the 3′ end of the strand. In Sequence 1, where 3 is located near the 5′ end of the strand, the T m and CD analysis of 5′-OvHgs 1A and 1B and forks 1E and 1F show no change in stability and insignificant differences in conformation. This indicates that 3 does not disrupt base pairing in the duplex region 3′ of 3. In the case of 3′-OvHg 2B and fork 2F, where 3 is located near the 3′ end of the oligonucleotide, both decreases in stability and conformational changes are evident, indicating that 3 effects base pairing 5′ of the lesion. In duplex DNA, the presence of 3 causes the characteristic poly A•T shoulder to be lost in the CD spectra of 1D, but maintained in 2D. If 3 affects base-pairing interactions in both the 5′ and 3′ directions, it is expected that both duplex constructs of Sequences 1 and 2 containing 3 would lose the poly A•T shoulder in the CD spectra. Provided that there are only five base pairs 5′ of 3 in 1D, and twelve base pairs 5′ of 3 in 2D, the differences in the duplex CD spectra can be attributed to an effect of 3 on conformation towards the 5′ end, resulting in a greater disruption in the structure of Sequence 1. Interestingly, despite the conformation differences indicated through CD analysis between unmodified and 3 containing sequences, both duplex constructs containing 3 are destabilized to the same extent. Given that base context, the position of 3 and design of the architecture are conserved between Sequence 1 and 2, and the observed changes can only be attributed to 3.
3.5. Melting Temperature Analysis of Architectures Containing Overhangs and Phosphates
Strand-breaks resulting from 2′-deoxyribose oxidation are often associated with phosphorylated 3′ and 5′ ends. The effects of such lesions on the stability of model replication forks for both Sequence 1 and 2 were determined (Table 2). The presence of a 5′ or 3′ terminal phosphate on duplexes 1H and 2H decreases the T m values by 0.4°C and 0.9°C, respectively. Previously, it was shown that changes in T m caused by a 3′ or 5′ terminal phosphate on duplex DNA are dependent on counter-ion concentration [18]. At comparable counter ion concentrations, Bower et al. observed a decrease in T m in the presence of a 5′ or 3′ phosphate by 0.8°C and 0.3°C, respectively [18]. Besides the difference in experimental design, our experiments have the terminal phosphate residing beside an A•T bp, while in the work by Bower et al. it resides beside a G•C bp. The small difference in destabilization observed between the 5′ and 3′ phosphates herein and those reported by Bower et al. suggests that the terminal base-pair adjacent to the phosphate may contribute to the effects of the terminal phosphates.
Table 2.
Comparison of T m values for replication relevant DNA architectures containing individual 3′ and 5′ overhangs, a terminal phosphate, and presence of both an overhang and phosphate. The presence of a 3′ or 5′ phosphate (PO4), hydroxyl (OH) and a 6 mer overhang is indicated as an “X” or “Y” at the end of the core dsDNA sequences. ΔT m values were obtained by subtracting the T m of each construct from the T m value of the core duplex.
| ID | Base Sequence | T m (°C) | ΔT m (°C) | |
|---|---|---|---|---|
| 1 | 5′-CGCAACCTGAAAX | |||
| 3′-GCGTTGGACTTTY | ||||
| 1G | X = OH | Y = OH | 51.7 ± 0.2 | — |
| 1H | X = OH | Y = PO4 | 51.3 ± 0.0 | −0.4a |
| 1I | X = AAAAAA | Y = OH | 48.9 ± 0.3 | −2.8b |
| 1J | X = AAAAAA | Y = PO4 | 47.9 ± 0.1 | −3.4b |
| −1.0a | ||||
| 1K | X = GACTGT | Y = OH | 49.8 ± 0.3 | −1.9b |
| 1L | X = GACTGT | Y = PO4 | 48.7 ± 0.2 | −2.6b |
| −1.1a | ||||
|
| ||||
| 2 | 5′-GCGTTGGACTTTX | |||
| 3′-CGCAACCTGAAAY | ||||
| 2G | X = OH | Y = OH | 52.9 ± 0.2 | — |
| 2H | X = PO4 | Y = OH | 52.0 ± 0.2 | −0.9a |
| 2I | X = OH | Y = AAAAAA | 51.8 ± 0.1 | −1.1b |
| 2J | X = PO4 | Y = AAAAAA | 50.5 ± 0.3 | −1.5b |
| −1.3a | ||||
| 2K | X = OH | Y = GACTGT | 51.6 ± 0.2 | −1.3b |
| 2L | X = PO4 | Y = GACTGT | 50.4 ± 0.2 | −1.6b |
| −1.2a | ||||
aIndicates the ΔT m introduced by an individual phosphate. ΔT m was determined by subtracting substrate G from H, I from J and K from L. bIndicates the ΔT m introduced by an individual overhang. ΔT m was determined by subtracting substrate G from I, G from K, H from J, and H from L.
The presence of a 6 mer 3′-OvHg (1I & 1 K) decreases the T m of the core duplex by 1.9–2.8°C, while a 5′-OvHg (2I & 2 K) causes a less significant decrease of 1.1–1.3°C. It has been widely demonstrated that overhangs consisting of one nucleotide, regardless of polarity, are stabilizing [20]. This stabilization is attributed to base stacking between the unpaired nucleoside and the neighboring base pair [22]. Overhangs consisting of two nucleotides have also been found to increase stability [19]. Our results indicate that the presence of either a 6 mer 3′ or 5′-OvHg decreases core duplex stability. Interestingly, the range in ΔT m for the 5′-OvHgs is significantly smaller than that of the 3′-OvHgs. This suggests that the base context of the single-stranded region influences the T m of the 3′-OvHg more than the 5′-OvHg. Additional studies using 3 mer, 4 mer, and 5 mer OvHgs will be pursued to further evaluate the effect of overhang length on T m.
In using the constructs with a phosphate at the ss-dsDNA junction, both the effect of the overhang and phosphate were evaluated to determine how their combined presence influences core duplex stability. Compared to the ΔT m introduced by the OvHgs alone, we observed that a 3′-OvHg decreases the T m by an additional 0.6°C when a phosphate is present at the ss-dsDNA junction (1J & 1L), while the 5′-OvHg decreases duplex stability by an additional 0.3°C in the presence of a phosphate (2J & 2L). As seen previously in the case of OvHgs without a phosphate, the range of destabilization introduced by the 5′-OvHgs with a phosphate at the ss-dsDNA junction is lower than that of the comparable 3′-OvHgs. This further suggests that the decrease in stability is influenced by the base context of the 3′-OvHg region, but not by the 5′-OvHg region. Additionally, the decrease in T m introduced by the 5′ phosphate at the ss-dsDNA junction (1J & 1L) is more than twice that observed for the terminal 5′ phosphate (1H), while the 3′ phosphate at the ss-dsDNA junction (2J & 2L) has only a slightly greater effect on T m than the terminal 3′ phosphate (2H). Together, these results indicate that the 3′-OvHg and 3′ phosphate have a larger effect on the stability of the duplex region than the corresponding 5′-OvHg and 5′ phosphate. Furthermore, decreases in duplex stability are observed when both an overhang and phosphate are present.
3.6. DSC Analysis of Architectures Containing Overhangs and Phosphates
Differential scanning calorimetry of C3′-thymidinyl radical derived single strand-break products of the model replication fork was performed to thermodynamically characterize the effects of OvHgs and phosphates on the core duplex DNA as indicated in Table 3. Previously, using dangling ends, the presence of a 5′ phosphate was shown to decrease the ΔG of formation for duplex DNA, while the 3′ phosphate introduces minimal to no change [18]. Additionally, favorable changes in ΔG of formation and melting have been observed for both 3′ and 5′ dangling ends up to two dangling bases, indicating stabilization [19–22]. Interestingly, dangling ends ranging from 3 to 10 bases have been reported to be stabilizing in the case of the 5′ end and destabilizing in the case of the 3′ end [23]. Our results indicate that both the 5′ and 3′-OvHgs are destabilizing. The presence of a 3′-OvHg destabilizes by an average of 0.93 kcal/mol, while the 5′-OvHg destabilizes by 0.16 kcal/mol. The presence of a single 3′ or 5′ terminal phosphate also causes destabilization of the duplex. These results show no measurable difference in ΔΔG 25 between 3′ versus 5′ terminal phosphates, indicating that the presence of a terminal phosphate, increases the ΔG 25 by an average of +0.38 kcal/mol. Additionally, the destabilization introduced by a 3′-OvHg in the presence of the phosphate at the ss-dsDNA junction compared to the 5′-OvHg in the presence of the phosphate at the same position resulted in no significant difference in ΔΔG 25 for constructs 1L and 2L. Alternatively, in constructs 1J and 2J, the 3′-OvHg with the phosphate at the ss-dsDNA junction has no effect on DNA stability, while the comparable 5′-OvHg destabilizes by 0.80 kcal/mol. Lastly, the effect introduced by the phosphate at the ss-dsDNA junction of overhangs is found to be destabilizing in the case of the 3′ phosphate in 2J and 2L (+0.82 kcal/mol), stabilizing in the case of the 5′ phosphate in 1J (−0.76 kcal/mol) and not significant in 1L. Together, these results show that all strand-break products, consisting of an individual overhang, phosphate, or both, contain less stable duplexes for both Sequence 1 and 2, as indicated by unfavorable ΔΔG 25 values in all cases except for 1J.
Table 3.
Thermodynamic parameters obtained from differential scanning calorimetry for replication relevant DNA architectures containing individual 3′ and 5′ overhangs, a terminal phosphate, and presence of both an overhang and phosphate. The presence of a 3′ or 5′ phosphate (PO4), hydroxyl (OH) and a 6 mer overhang is indicated as an “X” or “Y” in the core dsDNA sequences.
| ID | Base Sequence | T m (°C) | ΔH
DSC
(kcal/mol) |
ΔH
vH
(kcal/mol) |
ΔS
(cal/mol∗K) |
ΔG 25 (kcal/mol) | ΔT m (°C) | ΔΔH
DSC
(kcal/mol) |
ΔΔS
(cal/mol∗K) |
ΔΔG
25
(kcal/mol) |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 5′-CGCAACCTGAAAX | ||||||||||
| 3′-GCGTTGGACTTTY | |||||||||||
| 1G | X = OH | Y = OH | 60.91 ± 0.10 | −77.28 ± 0.82 | −62.39 ± 0.29 | −231.4 ± 2.5 | −8.34 ± 0.07 | — | — | — | — |
| 1H | X = OH | Y = PO4 | 60.62 ± 0.05 | −74.67 ± 0.21 | −62.34 ± 0.11 | −223.7 ± 0.7 | −8.01 ± 0.02 | −0.29a | +2.61a | +7.7a | +0.33a |
| 1I | X = AAAAAA | Y = OH | 59.01 ± 0.05 | −70.93 ± 0.12 | −63.90 ± 0.08 | −213.6 ± 0.4 | −7.29 ± 0.01 | −1.90b | +6.35b | +17.8b | +1.05b |
| 1J | X = AAAAAA | Y = PO4 | 57.79 ± 0.02 | −80.85 ± 0.39 | −60.66 ± 0.12 | −244.3 ± 1.2 | −8.05 ± 0.04 | −2.83b | −6.18b | −20.6b | −0.04b |
| −1.22a | −9.92a | −30.7a | −0.76a | ||||||||
| 1K | X = GACTGT | Y = OH | 58.72 ± 0.07 | −73.83 ± 0.40 | −61.58 ± 0.07 | −222.5 ± 1.2 | −7.53 ± 0.05 | −2.19b | +3.45b | +8.9b | +0.81b |
| 1L | X = GACTGT | Y = PO4 | 58.16 ± 0.03 | −74.32 ± 0.73 | −62.22 ± 0.36 | −224.3 ± 2.2 | −7.47 ± 0.07 | −2.46b | +0.35b | −0.6b | +0.54b |
| −0.56a | −0.49a | −1.8a | +0.06a | ||||||||
| 2 | 5′-GCGTTGGACTTTX | ||||||||||
| 3′-CGCAACCTGAAAY | |||||||||||
| 2G | X = OH | Y = OH | 61.88 ± 0.08 | −88.31 ± 0.44 | −61.38 ± 0.34 | −263.6 ± 1.4 | −9.76 ± 0.03 | — | — | — | — |
| 2H | X = PO4 | Y = OH | 61.37 ± 0.05 | −86.31 ± 0.65 | −59.81 ± 0.36 | −258.0 ± 1.9 | −9.42 ± 0.08 | −0.51a | +2.00a | +5.6a | +0.34a |
| 2I | X = OH | Y = AAAAAA | 61.04 ± 0.02 | −88.48 ± 0.36 | −68.32 ± 0.11 | −264.8 ± 1.1 | −9.58 ± 0.04 | −0.84b | −0.17b | −1.2b | +0.18b |
| 2J | X = PO4 | Y = AAAAAA | 60.15 ± 0.13 | −81.38 ± 0.15 | −68.64 ± 0.16 | −244.2 ± 0.4 | −8.62 ± 0.04 | −1.22b | +4.93b | +13.8b | +0.80b |
| −0.89a | +7.10a | +20.6a | +0.96a | ||||||||
| 2K | X = OH | Y = GACTGT | 60.37 ± 0.01 | −90.36 ± 0.50 | −64.46 ± 0.33 | −270.9 ± 1.5 | −9.62 ± 0.06 | −1.51b | −2.05b | −7.3b | +0.14b |
| 2L | X = PO4 | Y = GACTGT | 59.88 ± 0.07 | −84.91 ± 0.30 | −65.66 ± 0.20 | −255.0 ± 0.9 | −8.93 ± 0.04 | −1.49b | +1.40b | +3.0b | +0.49b |
| −0.49a | +5.45a | +15.9a | +0.69a | ||||||||
aIndicates the effects introduced by an individual phosphate. ΔΔH DSC, ΔΔS, and ΔΔG 25 were determined by subtracting substrate G from H, I from J, and K from L. bIndicates the effects introduced by an individual overhang. ΔΔH DSC, ΔΔS, and ΔΔG 25 were determined by subtracting substrate G from I, G from K, H from J, and H from L.
3.7. Summary: Effect of Overhangs and Phosphates on the Stability of Replication Relevant Architectures
As previously mentioned, all architectures investigated, except for 1J, are less stable as indicated by the ΔT m and ΔΔG 25 values for UV and DSC analyses. Here one can determine the effects of an individual 3′ or 5′ phosphate and an individual 3′ or 5′-OvHg. Both UV melting and DSC results indicate that 3′-OvHgs are more destabilizing than 5′-OvHgs. 3′-OvHgs are more destabilizing and lower the T m by ~1°C more than the 5′-OvHgs. Additionally, 3′-OvHgs (1I & 1 K) demonstrate an increase in both entropy and enthalpy, while 5′ OvHg 2 K demonstrates a decrease in entropy and enthalpy. In the case of the 3′-OvHg, the observed destabilization is enthalpic in nature, while in the case of the 5′-OvHg, destabilization is entropic in nature. Thus, based on the small sampling of DNA OvHg constructs investigated here, these results indicate that both a single 3′- and 5′-OvHg of six nucleotides are destabilizing, but the 3′-OvHg is more destabilizing and enthalpy driven, while the 5′-OvHg is entropy driven.
With respect to the terminal phosphates, both UV melting and DSC analyses indicate that the 3′ phosphate (2H) decreases the T m to a greater extent than the 5′ phosphate (1H). By averaging the T m values of the UV melting and DSC experiments, it is observed that the 3′ phosphate decreases the T m by 0.70°C, about twice the change induced by the 5′ phosphate, 0.37°C. Interestingly, no measurable difference in ΔG 25 is observed for an individual 3′ or 5′ terminal phosphate, indicating that polarity does not affect the destabilization introduced by a terminal phosphate. The destabilization observed in the presence of a single 3′ or 5′ terminal phosphate is enthalpy driven, thermodynamically following the trend found for the 3′-OvHgs.
In evaluating the constructs containing both an overhang and phosphate, the destabilization introduced by the overhang region in the presence of a phosphate at the ss-dsDNA junction appears to be heavily dependent on base context and polarity. This is demonstrated by the lack of differences observed in ΔΔG 25 values for constructs 1L and 2L, indicating a 6 mer overhang of mixed base context to be destabilizing to the same extent, regardless of polarity. While in the case of constructs 1J and 2J, the 3′-OvHg introduces no measurable change in ΔΔG 25, and the comparable 5′-OvHg destabilizes by +0.80 kcal/mol. Intriguingly, the effect of the phosphate at the ss-dsDNA junction appears to be sequence dependent as indicated by the destabilization introduced by the 3′ phosphate in 2J and 2L, stabilization by the 5′ phosphate in 1J, and the lack of effect on stability in the case of the 5′ phosphate in 1L.
3.8. Possible Impact of C3′-Thymidinyl Radical Damage Products on DNA Replication
The formation of 3 in DNA oligomers has not yet been verified in vivo. Given the high levels of formation of 3 in vitro, there is a strong possibility that this lesion also forms in biological systems [9]. The effects of this lesion on DNA stability and structure, as well as previously reported effects of 1-(2′-deoxy-β-D-threo-pentofuranosyl)nucleotides on the enzymatic cleavage of the sugar-phosphate backbone [27], indicate that 3 will have a significant impact on genomic integrity if left unrepaired. Thus, the fidelity of replication processes should be investigated to determine the physiological consequences of this lesion. Currently, detailed structural studies investigating the effect of 3 on DNA structure are being pursued to understand fully the structural impact of the presence of this lesion.
Alternatively, evaluating the implications of 3′ and 5′-OvHgs and phosphates on DNA stability related to replication may provide insight into the thermodynamic contributions of DNA to facilitate protein-DNA binding and selectivity. The counter-ion condensation (CC) theory has been applied to DNA and DNA-protein complexes and found to be reliable in determining the driving forces for DNA-protein interactions [28]. Specifically, it has been determined that the binding energy required for DNA-protein interactions can be divided into electrostatic and nonelectrostatic components [28]. The electrostatic portion is completely entropic in nature, while the nonelectrostatic portion, which is responsible for specificity, corresponds to the binding enthalpy [28]. Given the CC theory and the results reported herein for strand-break product stability, DNA stability has the potential to be utilized to establish the driving force of binding for replication proteins that recognize and bind to these architectures. Two DNA replication relate proteins that may heavily rely on differences in duplex stability in replication relevant architectures are the single-stranded DNA binding proteins (SSBs) and DNA polymerases.
Single-stranded DNA binding proteins, which are generally classified as nonspecific in binding, are essential in both replication and repair processes. These proteins have demonstrated some intriguing preferences for DNA polarity depending on species. The human single-stranded DNA binding protein, known as Replication Protein A (RPA), has been reported to preferentially bind to 3′-OvHgs [29] and demonstrates a directionality of binding with respect to the 3′ and 5′ ends of the bound DNA [30–32]. Additionally, the single-stranded DNA binding protein of T4 demonstrates a preference for 5′-OvHgs, while that of E. coli demonstrates a binding preference for 3′-OvHgs [29]. Given that the destabilization of the 3′-OvHg is found to be enthalpy driven, this may suggest that the hRPA and E. coli single-stranded DNA binding proteins preferential bind 3′-OvHgs to form more favorable nonelectrostatic interactions. If this is the case, then the energy barrier to forming these non-electrostatic DNA-protein interactions may be lower in the case of 3′-OvHg, than with 5′-OvHg, as the non-electrostatic component is the driving force of 3′OvHg destabilization. This hypothesis is in good agreement with the literature, of which calorimetric studies of the E. coli single-stranded DNA binding protein binding to DNA have been reported to be enthalpically driven [33]. DNA-protein binding is expected to be favorable with respect to both DNA and protein, while the inherent decrease in enthalpy introduced by the 3′-OvHg may increase binding for the E. coli single-stranded DNA binding protein by suppressing the energy barrier of binding.
A wide variety of DNA polymerases exist for replication and repair of DNA. Recently, it was reported that Pol I in E. coli (Klenow) preferentially binds to DNA replication architectures as compared to those generated from strand-breaks, suggesting that the Klenow polymerase can selectively associate with DNA replication substrates [34]. Specifically, the presence of a 3′ phosphate at the ss-dsDNA junction was observed to decrease the binding affinity by 0.9–1.5 kcal/mol, depending on the presence of magnesium [34]. Our results indicate that the presence of a 3′ phosphate at the ss-dsDNA junction destabilizes to the extent of 0.82 kcal/mol, suggesting that the presence of the 3′ phosphate and its impact on DNA stability accounts for a significant amount of destabilization in the DNA-protein binding complex. Furthermore, the destabilization introduced by this 3′ phosphate is enthalpy driven, suggesting that the binding of Klenow is likely enthalpy dependent. Previous results reported the binding of Klenow at physiological temperatures to be enthalpy driven [35], being in good agreement with our hypothesis. This further supports the significance of DNA damage lesions and their impact on DNA stability in understanding the formation and stability of DNA-protein interactions.
4. Conclusions
The impact of strand-break products on DNA stability in model DNA architectures mimicking those generated through 2′-deoxyribose oxidation at the C3′-position during replication has been analyzed. The results obtained demonstrate that both an individual 3′ or 5′-OvHg of six nucleotides is destabilizing, with the 3′-OvHg destabilizing to a greater extent than the 5′-OvHg. Also, the presence of an individual 3′ or 5′ terminal phosphate was observed to be destabilizing, but polarity was not found to affect the magnitude of destabilization. Intriguingly, enthalpy is the driving force of destabilization for the 3′-OvHg and both 3′ and 5′ terminal phosphates, while the 5′-OvHg is entropy driven. Alternatively, the effects on substrate stability of OvHgs containing a phosphate at the ss-dsDNA junction are highly dependent on base context and polarity. In addition to these strand-break substrates, we determined the effects of 3 on the structure and stability of dsDNA, fork, 3′ and 5′-OvHg. The presence of 3 was observed to have a greater impact on both stability and structure when located closer to the 3′ end of the oligonucleotide strand. Taken together, evaluating the impact of lesions previously observed as a result of C3′-radical formation in replication relevant architectures provides insight into how these DNA damage lesions alter the stability and structure of replication associated nucleotide substrates, directly expanding the current scope of how oxidatively generated sugar damage impacts DNA integrity. With these results reported herein and our previous results in determining the impact of DNA structure on the fate of the C3′-thymidinyl radical, the role of proteins can now be evaluated to determine the interplay between oxidative sugar damage and protein binding.
Conflict of Interests
The authors declare that they do not have any conflict of interests with the content of the paper.
Acknowledgments
The authors would like to thank Dr. Max O. Funk at The University of Toledo for providing access to instrumentation for the UV melting analysis and differential scanning calorimetry. Additionally, They like to thank Dr. Michael Y. Ogawa at Bowling Green State University for proving access to the AVIV CD spectrometer. The authors would also like to thank the National Science Foundation for providing funding for instrumentation used for MALDI-TOF MS analysis (DBI-923184).
References
- 1.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408(6809):239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 2.Loft S, Poulsen HE. Cancer risk and oxidative DNA damage in man. Journal of Molecular Medicine. 1996;74(6):297–312. doi: 10.1007/BF00207507. [DOI] [PubMed] [Google Scholar]
- 3.Krishnan KJ, Greaves LC, Reeve AK, Turnbull DM. Mitochondrial DNA mutations and aging. Annals of the New York Academy of Sciences. 2007;1100:227–240. doi: 10.1196/annals.1395.024. [DOI] [PubMed] [Google Scholar]
- 4.Masai H, Tanaka T, Kohda D. Stalled replication forks: making ends meet for recognition and stabilization. BioEssays. 2010;32(8):687–697. doi: 10.1002/bies.200900196. [DOI] [PubMed] [Google Scholar]
- 5.Burhans WC, Weinberger M. DNA replication stress, genome instability and aging. Nucleic Acids Research. 2007;35(22):7545–7556. doi: 10.1093/nar/gkm1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harper JV, Anderson JA, O’Neill P. Radiation induced DNA DSBs: contribution from stalled replication forks? DNA Repair. 2010;9(8):907–913. doi: 10.1016/j.dnarep.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Saleh-Gohari N, Bryant HE, Schultz N, Parker KM, Cassel TN, Helleday T. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Molecular and Cellular Biology. 2005;25(16):7158–7169. doi: 10.1128/MCB.25.16.7158-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pitcher RS, Brissett NC, Picher AJ, et al. Structure and function of a mycobacterial NHEJ DNA repair polymerase. Journal of Molecular Biology. 2007;366(2):391–405. doi: 10.1016/j.jmb.2006.10.046. [DOI] [PubMed] [Google Scholar]
- 9.Amato NJ, Bryant-Friedrich AC. The impact of structure on oxidatively generated DNA damage products resulting from the C3′-thymidinyl radical. ChemBioChem. 2013;14(2):187–190. doi: 10.1002/cbic.201200723. [DOI] [PubMed] [Google Scholar]
- 10.Lahoud G, Fancher J, Grosu S, Cavanaugh B, Bryant-Friedrich A. Automated synthesis, characterization, and structural analysis of oligonucleotide C-3′-radical precursors. Bioorganic and Medicinal Chemistry. 2006;14(8):2581–2588. doi: 10.1016/j.bmc.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 11.Zaliznyak T, Lukin M, de los Santos C. Structure and stability of duplex DNA containing (5′S)-5′, 8-cyclo-2′-deoxyadenosine: an oxidatively generated lesion repaired by NER. Chemical Research in Toxicology. 2012;25(10):2103–2111. doi: 10.1021/tx300193k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosemeyer H, Seela F. 1-(2′-deoxy-β-D-xylofuranosyl)thymine building-blocks for solid-phase synthesis and properties of oligo(2′-deoxyxylonucleotides) Heleticava Chimica Acta. 1991;74(4):748–760. [Google Scholar]
- 13.Bishop GR, Chaires JB. UNIT 7.11 characterization of DNA structures by circular dichroism. Current Protocols in Nucleic Acid Chemistry. 2003 doi: 10.1002/0471142700.nc0711s11. [DOI] [PubMed] [Google Scholar]
- 14.Pilch DS. UNIT 7.4 calorimetry of nucleic acids. Current Protocols in Nucleic Acid Chemistry. 2001 doi: 10.1002/0471142700.nc0704s00. [DOI] [PubMed] [Google Scholar]
- 15.Spink CH. Differential scanning calorimetry. Methods in Cell Biology. 2008;84:115–141. doi: 10.1016/S0091-679X(07)84005-2. [DOI] [PubMed] [Google Scholar]
- 16.Giese B, Dussy A, Meggers E, Petretta M, Schwiter U. Conformation, lifetime, and repair of 4′-DNA radicals. Journal of the American Chemical Society. 1997;119(45):11130–11131. [Google Scholar]
- 17.Hwang JT, Greenberg MM. Kinetics and stereoselectivity of thiol trapping of deoxyuridin-1′-yl in biopolymers and their relationship to the formation of premutagenic α- deoxynucleotides. Journal of the American Chemical Society. 1999;121(18):4311–4315. [Google Scholar]
- 18.Bower M, Summers MF, Kell B, Hoskins J, Wilson WD. Synthesis and characterization of oligodeoxyribonucleotides containing terminal phosphates. NMR, UV spectroscopic and thermodynamic analysis of duplex formation of [d(PGGAATTCC)]2, [d(GGAATTCCP)]2 and [d(PGGAATTCCP)]2. Nucleic Acids Research. 1987;15(8):3531–3547. doi: 10.1093/nar/15.8.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Senior M, Jones RA, Breslauer KJ. Influence of dangling thymidine residues on the stability and structure of two DNA duplexes. Biochemistry. 1988;27(10):3879–3885. doi: 10.1021/bi00410a053. [DOI] [PubMed] [Google Scholar]
- 20.Bommarito S, Peyret N, SantaLucia J., Jr. Thermodynamic parameters for DNA sequences with dangling ends. Nucleic Acids Research. 2000;28(9):1929–1934. doi: 10.1093/nar/28.9.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guckian KM, Schweitzer BA, Ren RXF, et al. Experimental measurement of aromatic stacking affinities in the context of duplex DNA. Journal of the American Chemical Society. 1996;118(34):8182–8183. doi: 10.1021/ja961733f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isaksson J, Chattopadhyaya J. A uniform mechanism correlating dangling-end stabilization and stacking geometry. Biochemistry. 2005;44(14):5390–5401. doi: 10.1021/bi047414f. [DOI] [PubMed] [Google Scholar]
- 23.Dickman R, Manyanga F, Brewood GP, et al. Thermodynamic contributions of 5′- and 3′-single strand dangling-ends to the stability of short duplex DNAs. Journal of Biophysical Chemistry. 2012;3(1):1–15. [Google Scholar]
- 24.Seela F, Heckel M, Rosemeyer H. 122. Xylose-DNA containing the four natural bases. Helvetica Chimica Acta. 1996;79(5):1451–1461. [Google Scholar]
- 25.Kypr J, Kejnovská I, Renčiuk D, Vorlíčková M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Research. 2009;37(6):1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greve J, Maestre MF, Levin A. Circular dichroism of adenine and thymine containing synthetic polynucleotides. Biopolymers. 1977;16(7):1489–1504. doi: 10.1002/bip.1977.360160709. [DOI] [PubMed] [Google Scholar]
- 27.Petrauskene OV, Yakovleva JN, Alekseev YI, Subach FV, Babkina OV, Gromova ES. DNA duplexes containing altered sugar residues as probes of EcoRII and MvaI endonuclease interactions with sugar-phosphate backbone. Journal of Biomolecular Structure and Dynamics. 2000;17(5):857–870. doi: 10.1080/07391102.2000.10506574. [DOI] [PubMed] [Google Scholar]
- 28.Privalov PL, Dragan AI, Crane-Robinson C. Interpreting protein/DNA interactions: distinguishing specific from non-specific and electrostatic from non-electrostatic components. Nucleic Acids Research. 2011;39(7):2483–2491. doi: 10.1093/nar/gkq984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delagoutte E, Heneman-Masurel A, Baldacci G. Single-stranded DNA binding proteins unwind the newly synthesized double-stranded DNA of model miniforks. Biochemistry. 2011;50(6):932–944. doi: 10.1021/bi101583e. [DOI] [PubMed] [Google Scholar]
- 30.Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385(6612):176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 31.Iftode C, Borowiec JA. 5′→3′ molecular polarity of human replication protein A (hRPA) binding to pseudo-origin DNA substrates. Biochemistry. 2000;39(39):11970–11981. doi: 10.1021/bi0005761. [DOI] [PubMed] [Google Scholar]
- 32.Kolpashchikov DM, Khodyreva SN, Khlimankov DY, Wold MS, Favre A, Lavrik OI. Polarity of human replication protein A binding to DNA. Nucleic Acids Research. 2001;29(2):373–379. doi: 10.1093/nar/29.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozlov AG, Lohman TM. Calorimetric studies of E. coli SSB protein-single-stranded DNA interactions. Effects of monovalent salts on binding enthalpy. Journal of Molecular Biology. 1998;278(5):999–1014. doi: 10.1006/jmbi.1998.1738. [DOI] [PubMed] [Google Scholar]
- 34.Yang Y, LiCata VJ. Interactions of replication versus repair DNA substrates with the Pol I DNA polymerases from Escherichia coli and Thermus aquaticus . Biophysical Chemistry. 2011;159(1):188–193. doi: 10.1016/j.bpc.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 35.Datta K, Wowor AJ, Richard AJ, LiCata VJ. Temperature dependence and thermodynamics of klenow polymerase binding to primed-template DNA. Biophysical Journal. 2006;90(5):1739–1751. doi: 10.1529/biophysj.105.071837. [DOI] [PMC free article] [PubMed] [Google Scholar]