Abstract
Spinal muscular atrophy (SMA) is an autosomal recessive disease affecting ∼1 in 10,000 live births. The most striking component is the loss of α-motor neurons in the ventral horn of the spinal cord, resulting in progressive paralysis and eventually premature death. There is no current treatment paradigm other than supportive care, though the past 15 years has seen a striking advancement in understanding of both SMA genetics and molecular mechanisms. A variety of disease-modifying interventions are rapidly bridging the translational gap from the laboratory to clinical trials, including the application of antisense oligonucleotide (ASO) therapy for the correction of aberrant RNA splicing characteristic of SMA. Survival motor neuron (SMN) is a ubiquitously expressed 38-kD protein. Humans have two genes that produce SMN, SMN1 and SMN2, the former of which is deleted or nonfunctional in the majority of patients with SMA. These two genes are nearly identical with one exception, a C to T transition (C6T) within exon 7 of SMN2. C6T disrupts a modulator of splicing, leading to the exclusion of exon 7 from ∼90% of the mRNA transcript. The resultant truncated Δ7SMN protein does not oligomerize efficiently and is rapidly degraded. SMA can therefore be considered a disease of too little SMN protein. A number of cis-acting splice modifiers have been identified in the region of exon 7, the steric block of which enhances the retention of the exon and a resultant full-length mRNA sequence. ASOs targeted to these splice motifs have shown impressive phenotype rescue in multiple SMA mouse models.
Introduction
Spinal muscular atrophy (SMA) is a neuromuscular disease that is characterized by dysfunction and eventually death of α-motor neurons in the spinal cord ventral horn (Crawford and Pardo, 1996). SMA is considered common among National Institutes of Health–designated “orphan” diseases (http://rarediseases.info.nih.gov/) with a prevalence of 1 case per 10,000 live births (Pearn, 1978; Prior et al., 2010). Rapid disease progression and the severity of paralysis-related morbidity leaves SMA as one of the leading causes of genetic infant mortality (Roberts et al., 1970). There is no current treatment except for supportive care, although some promising therapies are preparing for phase 1/2a clinical trials.
SMA is classified into a spectrum of disease severity (Type I through Type IV; Zerres and Rudnik-Schoneborn, 1995). Type I patients are never able to sit upright and typically show rapid loss of motor tone before 6 months of age. Death from respiratory insufficiency or other associated comorbidities is typical before 1 year without aggressive intervention including positive-pressure ventilation and enteric tube feeds. Alternatively, type II patients can sit upright but not ambulate without assistance, and those with type III can ambulate but lose this function in passing years. Type IV patients may only have evidence of neuromuscular decline in later decades (Piepers et al., 2008).
Advances in SMA genetics support a molecular classification scheme that directly correlates with the range of clinical phenotypes. These disease insights have generated multiple proposed treatment paradigms. All species require the critical survival motor neuron (SMN) protein, and all cells require at least a basal production of SMN. The deletion of Smn results in embryonic lethality in mice and death of that cell if removed (Cifuentes-Diaz et al., 2001; Frugier et al., 2000; Schrank et al., 1997; Vitte et al., 2004). Humans have two similar genes that produce SMN, SMN1 and SMN2, the latter of which is unique to our species (Rochette et al., 2001). SMN2 is therefore postulated to be a relatively recent mutation since no other species has two SMN-producing genes with the exception of chimpanzees (with two copies of SMN1).
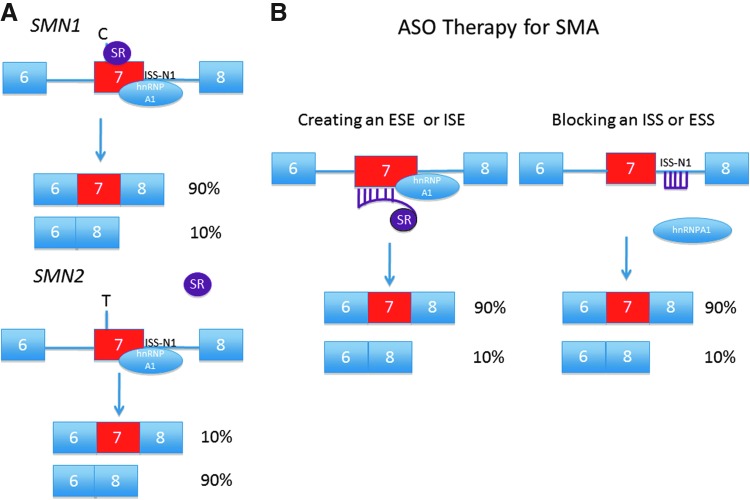
SMN2 essentially differs from SMN1 by a single nucleotide (C6T) within exon 7. While the coded amino-acid sequence is unchanged, the mutation does disrupt a modulator of RNA splicing (Lorson et al., 1999; Monani et al., 1999). Efficiency of exon 7 retention is radically altered with ∼90% of the SMN2 RNA transcript undergoing alternative splicing-out of the exon (Fig. 1A; Gennarelli et al., 1995). The Δ7SMN protein does not oligomerize efficiently and is rapidly degraded by cellular enzymatic activity (Burnett et al., 2009; Lorson et al., 1998). SMN1 is deleted or nonfunctional in the majority of patients with SMA. Affected individuals survive by virtue of the low levels of FLSMN produced by SMN2, but it is clear that these levels are insufficient for normal cell maintenance (Coovert et al., 1997; Lefebvre et al., 1997; Parsons et al., 1996). Different cell and tissue populations likely require unique basal and peak levels of SMN (and at different time points) in order to meet metabolic demands. This could be one reason why the motor neuron unit is so exquisitely susceptible to dysfunction (Burghes and Beattie, 2009). Regardless of the vagaries of timing and location of SMN depletion, SMA is at its core a disease of insufficient SMN.
FIG. 1.
(A) SMN1 and SMN2 splice dynamics. The C to T change within SMN2 exon 7 position 6 either disrupts a splice enhancer or creates a splice silencer. The SMN1 transcript binds both the splice activator ASF/SF2 (SR protein) and the splice repressor hnRNPA1 (at intron 7 ISS-N1). The resulting mRNA transcript includes exon 7. SMN2 does not bind the splice activator, and the majority of mRNA transcript lacks exon 7. SMN1 is missing or nonfunctional in SMA, and the patient survives on the small amount of full-length SMN transcript produced by SMN2. (B) SMN2 gene splicing can be modified by either bifunctional ASOs that promote binding of SR proteins, or through steric block of hnRNPA1 binding at splice suppressor sites (e.g., ISS-N1). Figure adapted from Burghes and McGovern (2010). SMA, spinal muscular atrophy; SMN, survival motor neuron; ASO, antisense oligonucleotide; ISS, intron splice suppressor; ESS, exon splice suppressor; ESE, exon splice enhancer; ISE, intron splice enhancer.
A direct modifier of disease severity is therefore the relative quantity of FLSMN produced inefficiently by SMN2. An inverse correlation is seen between SMN2 copy number and clinical disease classification from type I (single SMN2 copy) through type IV (more than three copies) (McAndrew et al., 1997; Parsons et al., 1998). This copy-number scheme is supported by the multiple murine models of disease (Monani et al., 2000a; Schmid and DiDonato, 2007). Mice have only one gene that produces SMN; animals without native Smn and with two inserted copies of SMN2 (i.e., “severe mouse” Smn−/−; SMN2+/+) display a striking motor decline and die by postnatal day 5 (P5) if on a complete null background, and survive to P10 if the deletion allele is capable of making the mouse Smn transcript lacking exon 7 (Hsieh-Li et al., 2000; Monani et al., 2000b). To effect a longer survival and thus facilitate therapeutic pharmacological trials, the SMNΔ7 mouse incorporates an additional transgene expressing SMN2 cDNA (Smn−/−; SMN2+/+; SMNΔ7+/+) which prolongs survival to P15 (Le et al., 2005). Δ7SMN is capable of interacting with FLSMN and providing some minimal function. The amount of FLSMN produced by an SMN2 gene is critical in determining the severity of SMA phenotype (Prior et al., 2009; Vezain et al., 2010). More intermediate mouse models have increasing copies of SMN2; these animals develop ear and tail necrosis but not necessarily weakness (Hsieh-Li et al., 2000). Necrosis does not occur in patients with mild SMA, and clear electrophysiological abnormalities indicating reduced motor neuron number and sprouting are not available.
Five percent of SMN1 mutations are small mutations. Some of these are missense mutations that can produce both severe and mild phenotypes in a low SMN2 copy background, depending on whether the mutation can complement the FLSMN produced by SMN2 (Burghes and Beattie 2009; Workman et al., 2009). The major function of SMN in complex with other proteins is the assembly of Sm proteins onto snRNA, which is then transported into the nucleus and plays a critical function in gene splicing (Pellizzoni et al., 1998). SMN reduction drastically alters snRNP assembly and results in gene splicing changes in spinal cord tissue of Δ7SMA mice (Baumer et al., 2009; Gabanella et al., 2007). It is unclear if alternative splicing is a primary driver of disease or rather a secondary effect of generalized organism decline. Further characterization of the splicing changes unique to snRNP dysfunction in SMA, specifically within the motor neuron, should shed light on the molecular and cellular pathways affected. Future investigation will define the critical splicing changes that contribute to all or part of the SMA phenotype.
The facilitation of RNA axonal transport by SMN is debated and under investigation. Shorter axons are observed with knockdown of SMN in zebrafish as well as in motor neuron cultures from SMA mice (Jablonka et al., 2006; McWhorter et al., 2003; Rossoll et al., 2003). However, SMA mice do not show axonal growth defects (McGovern et al., 2008). There is disrupted synaptic morphology and function in these animals, which is corrected by central nervous system (CNS) SMN expression (Gavrilina et al., 2008; Gogliotti et al., 2012; Martinez et al., 2012; Park et al., 2010). It is therefore indeterminate what direct function SMN has with respect to axonal and synaptic SMA pathology (Burghes and Beattie, 2009).
The above insights into the genesis of SMA due to the depletion of FLSMN have yielded two main concepts of therapy: first is the wholesale replacement of SMN above that afforded by SMN2 through the introduction of a construct expressing FLSMN, facilitated by viral vectors carrying the full-length construct. This method has yielded impressive results in severe mouse models after neonatal delivery (Dominguez et al., 2011; Foust et al., 2010; Passini et al., 2010; Valori et al., 2010). Investigation has continued in larger and more mature animals to determine rates of motor neuron transfection as well as to analyze biodistribution (Bevan et al., 2011; Marco Passini, personal communication, Families of SMA International SMA Research Group Meeting, June 23, 2012). While viral delivery of an intact SMN construct has progressed well and is approaching phase I/IIa clinical trials, impediments remain, including the ability to produce sufficient virus for a large clinical study.
An alternative method to increase FLSMN levels is to manipulate cis-acting RNA sequences to increase exon 7 incorporation during SMN2 RNA processing. As with other pre-mRNA transcripts, SMN2 contains multiple intronic and exonic conserved coding regions that recruit components of the splicing machinery (Bebee et al., 2010). Many of these sequences, both those that inhibit splicing (exon splice suppressor [ESS], intron splice suppressor [ISS]) and those that activate splicing (exon splice enhancer [ESE], intron splice enhancer [ISE]), have been identified adjacent to or within exon 7, and antisense oligonucleotides (ASOs) targeting these regions can alter RNA processing dynamics and increase FLSMN (Fig. 1B). Optimization of the target sequence and chemistry of ASO are ongoing; nevertheless, in vitro and recent in vivo investigations have shown this line of therapeutic attack can vastly increase the complement of FLSMN to levels seen with SMN1. Fortuitously, ASOs have shown impressive disease amelioration in murine models. As this model of treatment edges closer from the laboratory towards the clinic, important questions remain regarding dosing, toxicity, timing, and location of ASO delivery.
Splice Regulation of SMN2 Exon-7
Post-transcriptional mRNA processing is a complex process involving both local splicing elements as well as a trans acting modulators of splicing. A series of defined features are necessary but not altogether sufficient for splicing, including weakly conserved 5′ and 3′ splice-sites (ss), a polypyrimidine tract, and the branch point (Cartegni et al., 2002). Additional splice effectors (ESE, ESS, ISE, ISS) flanking the intron–exon junctions are especially important in the setting of alternative splicing or intrinsically weak splice-sites. These specialized regions can conscript proteins that can either positively or negatively influence spliceosome recruitment, aid in exon definition, and promote favorable or unfavorable tertiary structures.
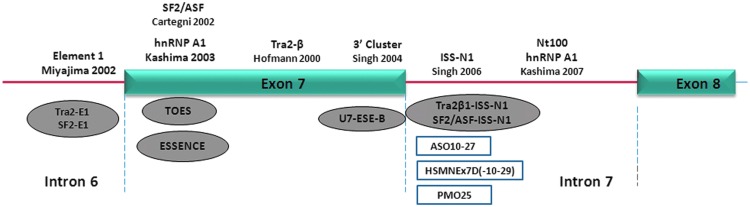
The translationally silent mutation at exon 7 C6T drastically alters exon inclusion from >90% in SMN1 to 10% in SMN2, leading to protein instability and rapid cellular degradation (Burnett et al., 2009; Lorson and Androphy, 2000; Lorson et al., 1998). The 3′ ss of both SMN1 and SMN2 are inherently weak due to a suboptimal polypyrimidine tract and intron 6 branch point (Lim and Hertel, 2001; Scholl et al., 2007), and therefore, the alternative splice inclusion of exon 7 is heavily dependent on the combinatorial effects of local enhancers and repressors. A systemic examination of the exon–intron region adjacent to and within exon 7 during the past decade has defined these alternative splicing mechanisms (Fig. 2).
FIG. 2.
Schematic representation of SMN2 exon 7 splicing regulatory elements. The functional elements regulating exon 7 splicing in SMN2 are depicted along the top. ASOs targeting these regulatory elements are listed at the bottom, including bifunctional ASOs (gray ovals) and conventional ASOs (open rectangles). Figure and legend reprinted by permission from Zhou et al. (2013).
A handful of ESEs, including both binding sites for the positive regulators SF2/ASF and TRA2β1, heavily influence exon inclusion. One of the most powerful, Conserved, rests within the middle of the 54-nucleotide exon 7. Conserved is an AG-rich TRA2β1 binding site that is both necessary and sufficient for basal levels of exon retention (Hofmann et al., 2000; Lorson and Androphy, 2000; Singh et al., 2004a, 2004b; Singh 2007). Blocking protein recognition of Conserved inhibits U2 snRNP recruitment, while elevated TRA2β1 expression increases exon retention (Hofmann et al., 2000; Martins de Araujo et al., 2009).
The splice modulatory effect of C6T suggests either ESE or ESS characteristics within this sequence adjacent to the 3′ ss. SF2/ASF protein binds to a hexamer motif overlapping position 6 in SMN1, defining the exon and recruiting spliceosomal machinery. The C6T substitution of SMN2 obliterates SF2/ASF binding, suggesting the abrogation of a strong ESE (Cartegni et al., 2002; Lorson and Androphy, 2000). Alternatively, C6T may conversely create an ESS by defining an efficient binding site for hnRNP A1, a protein that blocks spliceosome binding (Kashima and Manley, 2003). Refinement of the 5′ region splice mechanisms suggests that C6T may both decrease enhancer and increase suppressor protein binding, though impedance of the former mechanism likely plays a greater role in alternative splicing (Cartegni et al., 2006). An additional consequence of C6T is the creation of an extended inhibitory context (Exinct) that further represses exon inclusion (Singh et al., 2004a, 2004b).
Regional mRNA processing dynamics strongly affect exon 7 3′ ss efficiency. The exon 7 5′ ss is intrinsically weak and is flanked by dual suppressors including the exon 7 3′ cluster and intron 7 ISS-N1. Additional splice-suppressing elements have been identified within intron 6 and intron 7 that negatively influence SMN2 exon 7 inclusion (Kashima et al., 2007; Miyajima et al., 2002; Miyaso et al., 2003). ISS-N1, first identified by Singh et al. (2006) and further characterized by Hua et al. (2008), has been at the forefront of therapeutic ASO applications for exon 7 inclusion. The splice suppressor is located immediately beyond the conserved junction binding region for U1snRNA (intron-7 positions 10–24, CCAGCAUUAUGAAAG), and its adjacent hnRNP A1 and A2 motifs are instrumental in decreasing accessibility and thus binding of this positive splicing factor. ISS-N1 deletion in the SMN2 minigene increases FLSMN to SMN1 levels, an effect that is continued even after abolishment of multiple local splice enhancer elements. Patient-derived SMA cells (SMN−/−; SMN2+/+) show striking increases in FLSMN and SMN protein after application of a 2′-O-methyl phosphorothioate (2OMePS) ASO targeting ISS-N1. The splice modulation was effective in multiple types of cells in culture and did not show off-target splicing effects (Singh et al., 2006).
ASO Therapy
Deletion or dysfunction of SMN1 is a common denominator in all SMA cases, but it is the alternatively spliced SMN2 gene that both rescues from embryonic lethality and permits the SMA phenotype. The presence of the deficient SMN2 rescue gene, together with the duality of both weak 3′ and 5′ exon 7 splice sites and a multitude of local and regional splice silencers and enhancers, makes SMA uniquely suited for therapeutic ASO intervention. The goal of splice modulation for SMN2 is to increase exon 7 incorporation and thus FLSMN. Targeting ASOs should increase exon retention with high efficiency, have durable resistance to cellular degradation and robust target specificity, and have low toxicity and high penetrance to target cells including neurons within the CNS. A variety of sequences have been targeted by three different chemistries of ASO: 2OMePS, 2′-O-methoxyethyl (MOE), and morpholino.
Bifunctional ASOs
Sequence targeting within or adjacent to exon 7 can take advantage of the multitude of splice suppressors and activators by combining a sequence-specific ASO with a tethered oligomer that functions as a binding platform for positive splice activators. Two groups using different ASO chemistries (2OMePS and MOE) showed this proof-of-concept by targeting the putative ESE abolished by C6T transition, combined with a tailed splice-factor recruiting ASO (Cartegni and Krainer, 2003; Skordis et al., 2003). Splicing was restored to SMN1 levels, though the groups did not extend their investigations towards in vivo animal analysis. A similar 3′ ss exon 7 targeting scheme utilizing lentiviral germline transgenesis also drastically increased exon 7 retention and SMN protein levels (Marquis et al., 2007). The vector expression product, a bifunctional U7 snRNA with a positive splice-factor recruiting tail, increased median survival to 123 days in a severe SMA model (Smn−/−; SMN2+/+; Meyer et al., 2009).
Two studies have combined bifunctional ASOs targeting splice suppressor sequences with tethered splice activator recruiters of SR-like proteins (SF2/ASF, hTra2β1). Osman et al. (2012) selected steric block of the ISS-N1 sequence with a 2OMePS chemistry ASO. Delivery was by intracerebroventricular (ICV) injection at three time points before P5. SMN protein was increased up to 3.5-fold within the CNS, yet doses up to 60 μg ASO per injection extended median survival to only ∼20 days in the SMNΔ7 mouse. Similarly, Baughan et al. (2009), blocked the E1 splice suppressor upstream of exon 7 with a 2OMePS chemistry. Despite early 2-fold increases in SMN protein throughout the neuraxis after ICV injection, delayed analysis at 5 days showed little or no effect. Severe SMA mice (Smn−/−; SMN2+/+) had very modest increased survival and weight gain. Low ASO doses, multiple cerebral cannulations, and delayed initial treatment at P2 may have reduced the potential for disease amelioration (Baughan et al., 2009).
Other strategies have directed ASOs to the intron 7/exon-8 junction with the goal of reducing recognition of the exon 8 3′ ss, thereby shifting favor to the 3′ ss of exon 7 and thus increasing exon 7 retention (Lim and Hertel, 2001; Madocsai et al., 2005). A bifunctional ASO with a tail designed to recruit negative splice effectors (hnRNP A1/A2) can further facilitate splice suppression (Gendron et al., 2006). One such 2OMePS ASO design targeted SMN2 exon-8; ICV injection at low dose into the SMNΔ7 mouse increased exon 7 retention, though survival in SMA animals was not assessed (Dickson et al., 2008).
Suppressor Sequence Steric Block
Chemically modified ASOs that resist cellular endonuclease activity and operate independent of RNase-H degradation can be used as powerful steric blocks of splice suppressors, thereby promoting robust exon retention. Perhaps the most important splice modulator yet targeted by ASOs for SMA has been the ISS-N1 sequence adjacent to the exon-7 5′ ss. ASO microwalks have revealed strong splice modification when targeting intron positions 10–27, 10–29, 10–34, and 07–14 (Hua et al., 2008; Mitrpant et al., 2013; Porensky et al., 2012; Singh et al., 2009; Zhou et al., 2013).
Recent studies of ASO ISS-N1 therapy have continued the refinement of delivery methods, ASO chemistries, and dosing strategies, resulting in concurrent incremental disease amelioration in SMA mouse models. A 20-mer 2OMePS ICV injection into SMNΔ7 mice increased FLSMN throughout the CNS and improved righting and weight gain; survival was not assessed. The study was hindered by a total of 10 ICV cannulations delivering a relatively low dose (10 μg total) of ASO (Williams et al., 2009).
Dose elevation analysis of an 18-mer MOE into a mild mouse model (Smn−/−, four copies SMN2; ear and tail necrosis at 6 weeks of age) showed that continuous CNS infusion into adult animals of up to 100 μg/d were well tolerated and produced effective and long-lasting splice modulation (Hua et al., 2010). Embryonic and neonatal ICV bolus therapy (20 μg maximum dose) delayed necrosis onset up to 12 weeks. In a direct comparison of 2OMePS and MOE chemistries, the former showed increased spinal cord inflammatory markers as well as reduced capacity for exon 7 inclusion after CNS infusion. Further investigation of the MOE chemistry after ICV bolus delivery in the neonatal SMNΔ7 mouse showed strong exon 7 retention throughout the entire neuraxis (Passini et al., 2011). Despite improved weight gain and motor function, as well as increased α-motor neuron counts and concurrent neuromuscular junction architecture, median survival was extended only modestly (26 days). Interestingly, a >8-μg bolus dose led to rapid animal decline, suggesting acute CNS toxicity of the MOE chemistry in neonates.
Recent studies report long-lasting survival of >100 days after ASO treatment. Hua et al. (2011) found marginal increased survival (severe mouse, Smn−/−; SMN2+/+) after a 20-μg ICV bolus MOE treatment. Strikingly, multiple subcutaneous or intraperitoneal injections of large dose MOE (320 μg ASO/g) resulted in 248-day median survival, with some animals living >500 days. Peripheral tissue, particularly liver, showed large increases in SMN protein; brain and spinal cord also showed a 4-fold increase in exon 7 retention, suggesting ASO translocation across the immature blood–brain barrier (BBB) of the neonatal mouse. Recent research has found that even a modest elevation of SMN within the CNS can affect survival (Bowerman et al., 2012; Gavrilina et al., 2008; Marco Passini, personal communication, Families of SMA International SMA Research Group Meeting, June 23, 2012). One factor to consider with different routes of ASO administration is whether the same neurons are transduced and at what times. Our ASO investigation, using a 20-mer morpholino chemistry delivered by single-bolus ICV injection, also showed dose-dependent animal survival (SMNΔ7 mouse) to a median of 112 days without evidence of toxicity. CNS splice modulation was elevated through 60 days after P0 injection, suggesting strong endonuclease resistance (Porensky et al., 2012). Subsequent investigations (morpholino chemistry, 25-mer, ICV injection) have confirmed the efficacy for ISS-N1 targeting (Mitrpant et al., 2013; Zhou et al., 2013). These results are confirmation that ASO therapy can drastically increase survival in animal SMA models and should proceed forward as potential strong disease modifiers. Indeed, ISIS Pharmaceuticals is embarking on early clinical trials to assess safety and pharmacokinetics of single dose MOE ASO delivered by lumbar intrathecal injection for children with Type II and III SMA (Chirboga et al., 2013). It remains to be seen whether these patients, rather than newly diagnosed infants with SMA, will show molecular or clinical benefit with such delayed treatment.
When Is Treatment Effective?
SMA disease severity is largely dependent on the amount of full-length SMN protein, which, in the absence of SMN1, varies directly with the number of gene copies of SMN2. As previously described, disease classification (groups I–IV, with decreasing severity) correlates with SMN2 copy number (McAndrew et al., 1997; Monani et al., 2000b; Parsons et al., 1998; Schmid and DiDonato, 2007). One of the key differences among patient disease severity scoring is the age at which symptoms manifest. Electrodiagnostic studies in presymptomatic patients show early preservation of the motor unit, followed by a precipitous drop, then a much more gradual decline (Finkel, 2013; Swoboda et al., 2005). Surviving motor neurons likely sprout and compensate for the acute loss. These results have been mirrored in the SMNΔ7 mouse (W. Arnold, personal communication, November 2012). Motor neuron and synapse dysfunction have been identified as key early events in SMA mice (Baumer et al., 2009). Two groups have recently developed mouse models with an inducible SMN transgene; embryonic and early neonatal induction yielded much improved survival when compared to a delayed start (Le et al., 2011; Lutz et al., 2011). These studies suggest a critical temporal window for motor neuron loss.
Therapeutic trials in murine SMA models advocate for early intervention yielding the greatest survival benefit. Foust et al. (2010) used a self-complementary adeno-associated virus (scAAV9) to replace SMN in SMA pups, vastly improving motor function and survival. When the P1 treatment was delayed to P5 a much more modest effect was seen, and P10 dosed animals were indistinguishable from controls. ASO trials have shown similar results with delayed treatment. Embryonic ICV injection delayed onset of peripheral necrosis in mild SMA mice (four copies of SMN2) to a much greater extent than P1 dosing, despite similar rates of splice modulation (Hua et al., 2010). Both ICV and peripheral delayed injection of ISS-N1 blocking ASO showed decreased survival compared to P0 administration (Hua et al., 2011; Porensky et al., 2012). These and others results have shown that complete restoration of FLSMN yields only moderate phenotypic advantage when achieved at a delayed time point.
It is becoming increasing clear that therapeutic interventions, including ASO therapy, are most advantageous when delivered before motor neuron collapse. What is uncertain is the effect of increasing SMN in human patients who manifest disease over a time course that is an order of magnitude greater than murine models. Will increasing SMN stabilize and perhaps promote recovery in dysfunctional motor units, or will SMN therapy only protect the remaining healthy neurons? SMA is not currently part of any standard prenatal or neonatal screening, though the capacity for such exists. Until recently the argument against screening was predicated on the lack of effective therapy, a condition that should change as ASOs and gene therapy proceed toward human clinical trials.
Where Is Treatment Necessary?
SMA has been shown to be a disease first and foremost of the spinal cord motor neuron unit. The most glaring symptoms in human patients relate to a progressive voluntary motor paresis, leading to paralysis-related secondary morbidity. While all cell populations require a basal production of SMN and complete elimination of the protein from any organ leads to cellular and organism death, it is likely that the decreased levels within spinal cord neurons lead to the unique neuromuscular pathology of SMA (Burghes and Beattie, 2009). Furthermore, analysis of laser microdissected motor neurons from SMA mice reveals a lower production of FLSMN from SMN2 when compared to production levels in other neural populations, suggesting that motor neurons have a further reduction in SMN levels than other cell types (Ruggiu et al., 2012). A logical corollary is whether elevation of SMN within the motor neuron alone will resolve the spectrum of morbidity and mortality seen with SMA.
Recent studies have investigated both increased and decreased expression of SMN in mouse SMA models using tissue-specific promoters targeting skeletal muscle or CNS cells. Increasing SMN from basal rates produced by SMN2 within spinal cord ventral horn cells, including neurons and supporting glia, yields striking improvement in motor neuron retention, synaptic integrity, muscle physiology, and animal survival (Gavrilina et al., 2008; Martinez et al., 2012; Vicki McGovern, personal communication, Families of SMA International SMA Research Group Meeting, June 22, 2012). Focal SMN reduction in motor neurons and some glial populations results in an early SMA neuromuscular phenotype, though without the spectrum of morbidity and infant mortality seen in SMA mice with ubiquitously low levels of SMN (Park et al., 2010). Inversely, elevation of SMN solely in motor neurons will correct motor neuron counts and restore sensory-motor spinal circuitry, but does not have a major impact on SMA survival (Gogliotti et al., 2012; Mentis et al., 2011). These results suggest that dysfunction of the motor neuron is the major contributor to the neuromuscular symptoms of SMA, but that other neuronal cells likely have a strong effect on disease-related mortality. Increasing SMN in motor neurons is necessary but probably not sufficient to treat the full disease spectrum.
Skeletal muscle is also postulated to contribute to SMA pathology independent of motor neuron dysfunction. Inhibition of myostatin, itself a potent inhibitor of cell growth and regeneration, does not alter SMA survival in murine models (Rindt et al., 2012; Rose et al., 2009; Sumner et al., 2009). Skeletal muscle-specific SMN restoration did not affect mortality in one study and increased survival and myofiber size in another, suggesting that SMN may regulate muscle growth autonomously from CNS innervation (Gavrilina et al., 2008; Martinez et al., 2012). Future studies should clarify muscle–SMA dynamics by developing conditions that correct or create SMA, rather than showing only a lack of correction. Therefore, a future goal should be to demonstrate a physiologic impairment in force generation after decreasing muscle SMN to levels seen in SMA (with two copies of SMN2).
Similarly, multiple groups have documented dysfunction of the autonomic nervous system (ANS) in SMA models, largely manifest by cardiac dysfunction (Bevan et al., 2010; Gogliotti et al., 2012; Heier et al., 2010; Shababi et al., 2010). The autonomic instability and resulting bradyarrhythmias are at least partially corrected after systemic restoration of SMN (Bevan et al., 2010; Shababi et al., 2012). There is growing clinical evidence of autonomic dysfunction in SMA, again largely manifest by cardiac arrhythmia, though these reports are limited to retrospective or small cohort reports (Arai et al., 2005; Araujo Ade et al., 2009; Bach 2007; Finsterer and Stollberger, 1999; Hachiya et al., 2005). Intrinsic basal heart-rate differences between mice and humans may be a reason for the purported paucity of clinical bradyarrhythmia. The extent and frequency of autonomic dysfunction in human SMA are unknown, and a more comprehensive prospective clinical study would be of great benefit.
Therapeutic trials with both viral-SMN vectors and ASOs have offered conflicting evidence of the importance of extra-CNS SMN elevation. Vector delivery of SMN to the CNS but not peripheral tissue results in extended SMA survival, but peripheral injection with diffuse CNS, skeletal and cardiac muscle, and ANS transduction showed even more impressive survival (Foust et al., 2010; Passini et al., 2010). Likewise, ASO therapy delivered directly into the CNS, without evidence of subsequent peripheral splice correction, strongly modulated SMA phenotype in various mouse models (Hua et al., 2010; Mitrpant et al., 2013; Porensky et al., 2012; Zhou et al., 2013). In contrast, peripheral ASO injection, albeit at a very high dose and at multiple neonatal time points, also produced extended SMA mouse survival (Hua et al., 2011). The authors found concurrent elevated levels of liver-specific IGF-1 in treated mice, and postulate that strong splice modulation in the liver may be an important condition for prolonged survival.
Future therapeutic development of ASOs for SMA must consider whether mice can be more completely rescued (i.e., longer survival) with repeat CNS dosing and whether peripheral delivery is required. CNS delivery is essential, whether by bare ASOs injected directly by intrathecal or intraventricular cannulation or by tagged oligomers that can cross the BBB and subsequently escape into cerebrospinal fluid pathways for full neuraxis distribution. The importance of extra-CNS targeting, such as autonomic nervous system ganglia, requires further investigation. Optimal treatment strategies may require a hybridized delivery mechanism into multiple compartments.
Future Considerations/Outlook
ASO therapy for SMA has made great strides during the past decade, including optimization of target mRNA sequence and impressive effects in severe models of disease. ASO splice modulation, together with SMN gene therapy and small molecules, has altered the conversation of SMA intervention from symptom palliation toward disease prevention and stabilization. Many questions remain before efficacy in human patients can be assessed. As already detailed, questions yet to be answered include selection of the optimal oligomer chemistry, as well as extensive questions regarding frequency and location of dosing. ASOs have illuminated disease insights that were not previously appreciated; the most interesting and exciting is that additional neuron groups other than the motor neuron could be involved. The role of peripheral tissues is harder to ascertain.
A logical next step towards human clinical trials would be translation into a large animal with anatomy and physiology similar to humans. Timing, volume, and location of dosing could be optimized, and potential toxicity better observed. Moreover, recapitulation of the unique human morphology, including a fully intact BBB and large-volume cerebrospinal fluid space, would better simulate human applications. No large SMA models currently exist, though both transgenic and vector-induced porcine models are in development (Lorson et al., 2008; personal communication, Monique Lorson, Families of SMA International SMA Research Group Meeting, June 24, 2011; personal communication, Sandra Duque, Families of SMA International SMA Research Group Meeting, June 21, 2012). Whatever the path taken, it is clear that ASO therapy for SMA is rapidly approaching human applications.
Acknowledgments
We thank all members of The Ohio State Motor Neuron Group and the SMA research community for many interesting ideas and discussion points. In particular, we thank Dr. Vicki McGovern for her insight and expertise. The work in the Burghes laboratory was supported by the National Institute of Health R01 HD060586 (A.H.M.B.) and RC2 NS069476 (A.H.M.B.) and The Ohio State University Department of Neurological Surgery (P.N.P).
Author Disclosure Statement
The authors declare that they have no competing interests.
References
- Arai H. Tanabe Y. Hachiya Y., et al. Finger cold-induced vasodilatation, sympathetic skin response, and R-R interval variation in patients with progressive spinal muscular atrophy. J. Child Neurol. 2005;20:871–875. doi: 10.1177/08830738050200110301. [DOI] [PubMed] [Google Scholar]
- Araujo Ade Q. Araujo M. Swoboda K.J. Vascular perfusion abnormalities in infants with spinal muscular atrophy. J. Pediatr. 2009;155:292–294. doi: 10.1016/j.jpeds.2009.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach J.R. Medical considerations of long-term survival of Werdnig-Hoffmann disease. Am. J. Phys. Med. Rehabil. 2007;86:349–355. doi: 10.1097/PHM.0b013e31804b1d66. [DOI] [PubMed] [Google Scholar]
- Baughan T.D. Dickson A. Osman E.Y. Lorson C.L. Delivery of bifunctional RNAs that target an intronic repressor and increase SMN levels in an animal model of spinal muscular atrophy. Hum. Mol. Genet. 2009;18:1600–1611. doi: 10.1093/hmg/ddp076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumer D. Lee S. Nicholson G., et al. Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet. 2009;5:e1000773. doi: 10.1371/journal.pgen.1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebee T.W. Gladman J.T. Chandler D.S. Splicing regulation of the survival motor neuron genes and implications for treatment of spinal muscular atrophy. Front. Biosci. 2010;15:1191–1204. doi: 10.2741/3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan A.K. Hutchinson K.R. Foust K.D., et al. Early heart failure in the SMNDelta7 model of spinal muscular atrophy and correction by postnatal scAAV9-SMN delivery. Hum. Mol. Genet. 2010;19:3895–3905. doi: 10.1093/hmg/ddq300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan A.K. Duque S. Foust K.D., et al. Systemic gene delivery in large species for targeting spinal cord, brain, and peripheral tissues for pediatric disorders. Mol. Ther. 2011;19:1971–1980. doi: 10.1038/mt.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman M. Murray L.M. Beauvais A., et al. A critical smn threshold in mice dictates onset of an intermediate spinal muscular atrophy phenotype associated with a distinct neuromuscular junction pathology. Neuromuscul. Disord. 2012;22:263–276. doi: 10.1016/j.nmd.2011.09.007. [DOI] [PubMed] [Google Scholar]
- Burghes A.H. Beattie C.E. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat. Rev. Neurosci. 2009;10:597–609. doi: 10.1038/nrn2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett B.G. Munoz E. Tandon A., et al. Regulation of SMN protein stability. Mol. Cell Biol. 2009;29:1107–1115. doi: 10.1128/MCB.01262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L. Krainer A.R. Correction of disease-associated exon skipping by synthetic exon specific activators. Nat. Struct. Biol. 2003;10:120–125. doi: 10.1038/nsb887. [DOI] [PubMed] [Google Scholar]
- Cartegni L. Chew S.L. Krainer A.R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Cartegni L. Hastings M.L. Calarco J.A., et al. Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2. Am. J. Hum. Genet. 2006;78:63–77. doi: 10.1086/498853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirboga C. Swoboda K. Darras B., et al. Results of an open-label, escalating dose study to assess the safety, tolerability, and dose range finding of a single intrathecal dose of ISIS-SMNRx in patients with spinal muscular atrophy. Annual Academy of Neurology meeting abstract; 2013. p. S36.002. [Google Scholar]
- Cifuentes-Diaz C. Frugier T. Tiziano F.D., et al. Deletion of murine SMN exon 7 directed to skeletal muscle leads to severe muscular dystrophy. J. Cell. Biol. 2001;152:1107–1114. doi: 10.1083/jcb.152.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coovert D.D. Le T.T. McAndrew P.E., et al. The survival motor neuron protein in spinal muscular atrophy. Hum. Mol. Genet. 1997;6:1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- Crawford T.O. Pardo C.A. The neurobiology of childhood spinal muscular atrophy. Neurobiol. Dis. 1996;3:97–110. doi: 10.1006/nbdi.1996.0010. [DOI] [PubMed] [Google Scholar]
- Dickson A. Osman E. Lorson C.L. A negatively acting bifunctional RNA increases survival motor neuron both in vitro and in vivo. Hum. Gene Ther. 2008;19:1307–1315. doi: 10.1089/hum.2008.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez E. Marais T. Chatauret N., et al. Intravenous scAAV9 delivery of a codon-optimized SMN1 sequence rescues SMA mice. Hum. Mol. Genet. 2011;20:681–693. doi: 10.1093/hmg/ddq514. [DOI] [PubMed] [Google Scholar]
- Finkel R.S. Electrophysiological and motor function scale association in a pre-symptomatic infant with spinal muscular atrophy type I. Neuromuscul. Disord. 2013;23:112–115. doi: 10.1016/j.nmd.2012.09.006. [DOI] [PubMed] [Google Scholar]
- Finsterer J. Stollberger C. Cardiac involvement in Werdnig-Hoffmann's spinal muscular atrophy. Cardiology. 1999;92:178–182. doi: 10.1159/000006968. [DOI] [PubMed] [Google Scholar]
- Foust K.D. Wang X. McGovern V.L., et al. Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat. Biotechnol. 2010;28:271–274. doi: 10.1038/nbt.1610. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Frugier T. Tiziano F.D. Cifuentes-Diaz C., et al. Nuclear targeting defect of SMN lacking the C-terminus in a mouse model of spinal muscular atrophy. Hum. Mol. Genet. 2000;9:849–858. doi: 10.1093/hmg/9.5.849. [DOI] [PubMed] [Google Scholar]
- Gabanella F. Butchbach M.E. Saieva L., et al. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS One. 2007;2:e921. doi: 10.1371/journal.pone.0000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilina T.O. McGovern V.L. Workman E., et al. Neuronal SMN expression corrects spinal muscular atrophy in severe SMA mice while muscle-specific SMN expression has no phenotypic effect. Hum. Mol. Genet. 2008;17:1063–1075. doi: 10.1093/hmg/ddm379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron D. Carriero S. Garneau D., et al. Modulation of 5′ splice site selection using tailed oligonucleotides carrying splicing signals. BMC Biotechnol. 2006;6:5. doi: 10.1186/1472-6750-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennarelli M. Lucarelli M. Capon F., et al. Survival motor neuron gene transcript analysis in muscles from spinal muscular atrophy patients. Biochem. Biophys. Res. Commun. 1995;213:342–348. doi: 10.1006/bbrc.1995.2135. [DOI] [PubMed] [Google Scholar]
- Gogliotti R.G. Quinlan K.A. Barlow C.B., et al. Motor neuron rescue in spinal muscular atrophy mice demonstrates that sensory-motor defects are a consequence, not a cause, of motor neuron dysfunction. J. Neurosci. 2012;32:3818–3829. doi: 10.1523/JNEUROSCI.5775-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachiya Y. Arai H. Hayashi M., et al. Autonomic dysfunction in cases of spinal muscular atrophy type 1 with long survival. Brain Dev. 2005;27:574–578. doi: 10.1016/j.braindev.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Heier C.R. Satta R. Lutz C. Didonato C.J. Arrhythmia and cardiac defects are a feature of spinal muscular atrophy model mice. Hum. Mol. Genet. 2010;19:3906–3918. doi: 10.1093/hmg/ddq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann Y. Lorson C.L. Stamm S., et al. Htra2-beta 1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2) Proc. Natl. Acad. Sci. U. S. A. 2000;97:9618–9623. doi: 10.1073/pnas.160181697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh-Li H.M. Chang J.G. Jong Y.J., et al. A mouse model for spinal muscular atrophy. Nat. Genet. 2000;24:66–70. doi: 10.1038/71709. [DOI] [PubMed] [Google Scholar]
- Hua Y. Vickers T.A. Okunola H.L., et al. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet. 2008;82:834–848. doi: 10.1016/j.ajhg.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y. Sahashi K. Hung G., et al. Antisense correction of SMN2 splicing in the Cns rescues necrosis in a type III SMA mouse model. Genes Dev. 2010;24:1634–1644. doi: 10.1101/gad.1941310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y. Sahashi K. Rigo F., et al. Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 2011;478:123–126. doi: 10.1038/nature10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonka S. Karle K. Sandner B., et al. Distinct and overlapping alterations in motor and sensory neurons in a mouse model of spinal muscular atrophy. Hum. Mol. Genet. 2006;15:511–518. doi: 10.1093/hmg/ddi467. [DOI] [PubMed] [Google Scholar]
- Kashima T. Manley J.L. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat. Genet. 2003;34:460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- Kashima T. Rao N. Manley J.L. An intronic element contributes to splicing repression in spinal muscular atrophy. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3426–3431. doi: 10.1073/pnas.0700343104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le T.T. Pham L.T. Butchbach M.E., et al. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum. Mol. Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- Le T.T. McGovern V.L. Alwine I.E., et al. Temporal requirement for high SMN expression in SMA mice. Hum. Mol. Genet. 2011;20:3578–3591. doi: 10.1093/hmg/ddr275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre S. Burlet P. Liu Q., et al. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- Lim S.R. Hertel K.J. Modulation of survival motor neuron pre-mRNA splicing by inhibition of alternative 3′ splice site pairing. J. Biol. Chem. 2001;276:45476–45483. doi: 10.1074/jbc.M107632200. [DOI] [PubMed] [Google Scholar]
- Lorson C.L. Androphy E.J. An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum. Mol. Genet. 2000;9:259–265. doi: 10.1093/hmg/9.2.259. [DOI] [PubMed] [Google Scholar]
- Lorson C.L. Strasswimmer J. Yao J.M., et al. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat. Genet. 1998;19:63–66. doi: 10.1038/ng0598-63. [DOI] [PubMed] [Google Scholar]
- Lorson C.L. Hahnen E. Androphy E.J. Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. U. S. A. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorson M.A. Spate L.D. Prather R.S. Lorson C.L. Identification and characterization of the porcine (Sus scrofa) survival motor neuron (SMN1) gene: an animal model for therapeutic studies. Dev. Dyn. 2008;237:2268–2278. doi: 10.1002/dvdy.21642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz C.M. Kariya S. Patruni S., et al. Postsymptomatic restoration of SMN rescues the disease phenotype in a mouse model of severe spinal muscular atrophy. J. Clin. Invest. 2011;121:3029–3041. doi: 10.1172/JCI57291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madocsai C. Lim S.R. Geib T., et al. Correction of SMN2 pre-mRNA splicing by antisense U7 small nuclear RNAs. Mol. Ther. 2005;12:1013–1022. doi: 10.1016/j.ymthe.2005.08.022. [DOI] [PubMed] [Google Scholar]
- Marquis J. Meyer K. Angehrn L., et al. Spinal muscular atrophy: SMN2 pre-mRNA splicing corrected by a U7 snRNA derivative carrying a splicing enhancer sequence. Mol. Ther. 2007;15:1479–1486. doi: 10.1038/sj.mt.6300200. [DOI] [PubMed] [Google Scholar]
- Martinez T.L. Kong L. Wang X., et al. Survival motor neuron protein in motor neurons determines synaptic integrity in spinal muscular atrophy. J. Neurosci. 2012;32:8703–8715. doi: 10.1523/JNEUROSCI.0204-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins De Araujo M. Bonnal S. Hastings M.L., et al. Differential 3′ splice site recognition of SMN1 and SMN2 transcripts by U2af and U2 snRNP. RNA. 2009;15:515–523. doi: 10.1261/rna.1273209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAndrew P.E. Parsons D.W. Simard L.R., et al. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am. J. Hum. Genet. 1997;60:1411–1422. doi: 10.1086/515465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern V.L. Gavrilina T.O. Beattie C.E. Burghes A.H. Embryonic motor axon development in the severe SMA mouse. Hum. Mol. Genet. 2008;17:2900–2909. doi: 10.1093/hmg/ddn189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhorter M.L. Monani U.R. Burghes A.H. Beattie C.E. Knockdown of the survival motor neuron (SMN) protein in zebrafish causes defects in motor axon outgrowth and pathfinding. J. Cell Biol. 2003;162:919–931. doi: 10.1083/jcb.200303168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentis G.Z. Blivis D. Liu W., et al. Early functional impairment of sensory-motor connectivity in a mouse model of spinal muscular atrophy. Neuron. 2011;69:453–467. doi: 10.1016/j.neuron.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K. Marquis J. Trub J., et al. Rescue of a severe mouse model for spinal muscular atrophy by U7 snRNA-mediated splicing modulation. Hum. Mol. Genet. 2009;18:546–555. doi: 10.1093/hmg/ddn382. [DOI] [PubMed] [Google Scholar]
- Mitrpant C. Porensky P. Zhou H., et al. Improved antisense oligonucleotide design to suppress aberrant SMN2 gene transcript processing: towards a treatment for spinal muscular atrophy. PloS One. 2013;8:e62114. doi: 10.1371/journal.pone.0062114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyajima H. Miyaso H. Okumura M., et al. Identification of a cis-acting element for the regulation of SMN exon 7 splicing. J. Biol. Chem. 2002;277:23271–23277. doi: 10.1074/jbc.M200851200. [DOI] [PubMed] [Google Scholar]
- Miyaso H. Okumura M. Kondo S., et al. An intronic splicing enhancer element in survival motor neuron (SMN) pre-mRNA. J. Biol. Chem. 2003;278:15825–15831. doi: 10.1074/jbc.M209271200. [DOI] [PubMed] [Google Scholar]
- Monani U.R. Lorson C.L. Parsons D.W., et al. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- Monani U.R. Coovert D.D. Burghes A.H. Animal models of spinal muscular atrophy. Hum. Mol. Genet. 2000a;9:2451–2457. doi: 10.1093/hmg/9.16.2451. [DOI] [PubMed] [Google Scholar]
- Monani U.R. Sendtner M. Coovert D.D., et al. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in SMN(−/−) mice and results in a mouse with spinal muscular atrophy. Hum. Mol. Genet. 2000b;9:333–339. doi: 10.1093/hmg/9.3.333. [DOI] [PubMed] [Google Scholar]
- Osman E.Y. Yen P.F. Lorson C.L. Bifunctional RNAs targeting the intronic splicing silencer N1 increase SMN levels and reduce disease severity in an animal model of spinal muscular atrophy. Mol. Ther. 2012;20:119–126. doi: 10.1038/mt.2011.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G.H. Maeno-Hikichi Y. Awano T., et al. Reduced survival of motor neuron (SMN) protein in motor neuronal progenitors functions cell autonomously to cause spinal muscular atrophy in model mice expressing the human centromeric (SMN2) gene. J. Neurosci. 2010;30:12005–12019. doi: 10.1523/JNEUROSCI.2208-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons D.W. McAndrew P.E. Monani U.R., et al. An 11 base pair duplication in exon 6 of the SMN gene produces a type I spinal muscular atrophy (SMA) phenotype: further evidence for SMN as the primary SMA-determining gene. Hum. Mol. Genet. 1996;5:1727–1732. doi: 10.1093/hmg/5.11.1727. [DOI] [PubMed] [Google Scholar]
- Parsons D.W. McAndrew P.E. Iannaccone S.T., et al. Intragenic telsmn mutations: frequency, distribution, evidence of a founder effect, and modification of the spinal muscular atrophy phenotype by censmn copy number. Am. J. Hum. Genet. 1998;63:1712–1723. doi: 10.1086/302160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passini M.A. Bu J. Roskelley E.M., et al. CNS-targeted gene therapy improves survival and motor function in a mouse model of spinal muscular atrophy. J. Clin. Invest. 2010;120:1253–1264. doi: 10.1172/JCI41615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passini M.A. Bu J. Richards A.M., et al. Antisense oligonucleotides delivered to the mouse Cns ameliorate symptoms of severe spinal muscular atrophy. Sci. Transl. Med. 2011;3:72ra18. doi: 10.1126/scitranslmed.3001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearn J. Incidence, prevalence, and gene frequency studies of chronic childhood spinal muscular atrophy. J. Med. Genet. 1978;15:409–413. doi: 10.1136/jmg.15.6.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L. Kataoka N. Charroux B. Dreyfuss G. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mrna splicing. Cell. 1998;95:615–624. doi: 10.1016/s0092-8674(00)81632-3. [DOI] [PubMed] [Google Scholar]
- Piepers S. Van Den Berg L.H. Brugman F., et al. A natural history study of late onset spinal muscular atrophy types 3b and 4. J. Neurol. 2008;255:1400–1404. doi: 10.1007/s00415-008-0929-0. [DOI] [PubMed] [Google Scholar]
- Porensky P.N. Mitrpant C. McGovern V.L., et al. A single administration of morpholino antisense oligomer rescues spinal muscular atrophy in mouse. Hum. Mol. Genet. 2012;21:1625–1638. doi: 10.1093/hmg/ddr600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior T.W. Krainer A.R. Hua Y., et al. A positive modifier of spinal muscular atrophy in the SMN2 gene. Am. J. Hum. Genet. 2009;85:408–413. doi: 10.1016/j.ajhg.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior T.W. Snyder P.J. Rink B.D., et al. Newborn and carrier screening for spinal muscular atrophy. Am. J. Med. Genet. A. 2010;152A:1608–1616. doi: 10.1002/ajmg.a.33474. [DOI] [PubMed] [Google Scholar]
- Rindt H. Buckley D.M. Vale S.M., et al. Transgenic inactivation of murine myostatin does not decrease the severity of disease in a model of spinal muscular atrophy. Neuromuscul. Disord. 2012;22:277–285. doi: 10.1016/j.nmd.2011.10.012. [DOI] [PubMed] [Google Scholar]
- Roberts D.F. Chavez J. Court S.D. The genetic component in child mortality. Arch. Dis. Child. 1970;45:33–38. doi: 10.1136/adc.45.239.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochette C.F. Gilbert N. Simard L.R. SMN gene duplication and the emergence of the SMN2 gene occurred in distinct hominids: SMN2 is unique to Homo sapiens. Hum. Genet. 2001;108:255–266. doi: 10.1007/s004390100473. [DOI] [PubMed] [Google Scholar]
- Rose F.F., Jr. Mattis V.B. Rindt H. Lorson C.L. Delivery of recombinant follistatin lessens disease severity in a mouse model of spinal muscular atrophy. Hum. Mol. Genet. 2009;18:997–1005. doi: 10.1093/hmg/ddn426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossoll W. Jablonka S. Andreassi C., et al. SMN, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mrna in growth cones of motoneurons. J. Cell. Biol. 2003;163:801–812. doi: 10.1083/jcb.200304128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiu M. McGovern V.L. Lotti F., et al. A role for SMN exon 7 splicing in the selective vulnerability of motor neurons in spinal muscular atrophy. Mol. Cell Biol. 2012;32:126–138. doi: 10.1128/MCB.06077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid A. DiDonato C.J. Animal models of spinal muscular atrophy. J. Child Neurol. 2007;22:1004–1012. doi: 10.1177/0883073807305667. [DOI] [PubMed] [Google Scholar]
- Scholl R. Marquis J. Meyer K. Schumperli D. Spinal muscular atrophy: position and functional importance of the branch site preceding SMN exon 7. RNA Biol. 2007;4:34–37. doi: 10.4161/rna.4.1.4534. [DOI] [PubMed] [Google Scholar]
- Schrank B. Gotz R. Gunnersen J.M., et al. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc. Natl. Acad. Sci. U. S. A. 1997;94:9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shababi M. Habibi J. Yang H.T., et al. Cardiac defects contribute to the pathology of spinal muscular atrophy models. Hum. Mol. Genet. 2010;19:4059–4071. doi: 10.1093/hmg/ddq329. [DOI] [PubMed] [Google Scholar]
- Shababi M. Habibi J. Ma L., et al. Partial restoration of cardio-vascular defects in a rescued severe model of spinal muscular atrophy. J. Mol. Cell. Cardiol. 2012;52:1074–1082. doi: 10.1016/j.yjmcc.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N.K. Singh N.N. Androphy E.J. Singh R.N. Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron. Mol. Cell Biol. 2006;26:1333–1346. doi: 10.1128/MCB.26.4.1333-1346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N.N. Androphy E.J. Singh R.N. In vivo selection reveals combinatorial controls that define a critical exon in the spinal muscular atrophy genes. RNA. 2004a;10:1291–1305. doi: 10.1261/rna.7580704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N.N. Androphy E.J. Singh R.N. The regulation and regulatory activities of alternative splicing of the SMN gene. Crit. Rev. Eukaryot. Gene Expr. 2004b;14:271–285. doi: 10.1615/critreveukaryotgeneexpr.v14.i4.30. [DOI] [PubMed] [Google Scholar]
- Singh N.N. Shishimorova M. Cao L.C., et al. A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy. RNA Biol. 2009;6:341–350. doi: 10.4161/rna.6.3.8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R.N. Evolving concepts on human SMN pre-mRNA splicing. RNA Biol. 2007;4:7–10. doi: 10.4161/rna.4.1.4535. [DOI] [PubMed] [Google Scholar]
- Skordis L.A. Dunckley M.G. Yue B., et al. Bifunctional antisense oligonucleotides provide a trans-acting splicing enhancer that stimulates SMN2 gene expression in patient fibroblasts. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4114–4119. doi: 10.1073/pnas.0633863100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner C.J. Wee C.D. Warsing L.C., et al. Inhibition of myostatin does not ameliorate disease features of severe spinal muscular atrophy mice. Hum. Mol. Genet. 2009;18:3145–3152. doi: 10.1093/hmg/ddp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swoboda K.J. Prior T.W. Scott C.B., et al. Natural history of denervation in SMA: relation to age, SMN2 copy number, and function. Ann. Neurol. 2005;57:704–712. doi: 10.1002/ana.20473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valori C.F. Ning K. Wyles M., et al. Systemic delivery of scAAV9 expressing SMN prolongs survival in a model of spinal muscular atrophy. Sci. Transl. Med. 2010;2:35ra42. doi: 10.1126/scitranslmed.3000830. [DOI] [PubMed] [Google Scholar]
- Vezain M. Saugier-Veber P. Goina E., et al. A rare SMN2 variant in a previously unrecognized composite splicing regulatory element induces exon 7 inclusion and reduces the clinical severity of spinal muscular atrophy. Hum. Mutat. 2010;31:E1110–E1125. doi: 10.1002/humu.21173. [DOI] [PubMed] [Google Scholar]
- Vitte J.M. Davoult B. Roblot N., et al. Deletion of murine SMN exon 7 directed to liver leads to severe defect of liver development associated with iron overload. Am. J. Pathol. 2004;165:1731–1741. doi: 10.1016/S0002-9440(10)63428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J.H. Schray R.C. Patterson C.A., et al. Oligonucleotide-mediated survival of motor neuron protein expression in CNS improves phenotype in a mouse model of spinal muscular atrophy. J. Neurosci. 2009;29:7633–7638. doi: 10.1523/JNEUROSCI.0950-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman E. Saieva L. Carrel T.L., et al. A SMN missense mutation complements SMN2 restoring snRNPs and rescuing SMA mice. Hum. Mol. Genet. 2009;18:2215–2229. doi: 10.1093/hmg/ddp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerres K. Rudnik-Schoneborn S. Natural history in proximal spinal muscular atrophy. Clinical analysis of 445 patients and suggestions for a modification of existing classifications. Arch. Neurol. 1995;52:518–523. doi: 10.1001/archneur.1995.00540290108025. [DOI] [PubMed] [Google Scholar]
- Zhou H. Janghra N. Mitrpant C., et al. A novel morpholino oligomer targeting ISS-N1 improves rescue of severe spinal muscular atrophy transgenic mice. Hum. Gene Ther. 2013;24:331–342. doi: 10.1089/hum.2012.211. [DOI] [PMC free article] [PubMed] [Google Scholar]