The engulfment and clearance of apoptotic cells by neighboring cells or professional phagocytes is crucial to tissue homeostasis and the regulation of immune responses. The Arf-like GTPase Arl8, which localizes primarily to lysosomes, mediates phagolysosome formation to promote the efficient degradation of apoptotic germ cells in Caenorhabditis elegans.
Abstract
Efficient clearance of apoptotic cells by phagocytes is important for development, tissue homeostasis, and the prevention of autoimmune responses. Phagosomes containing apoptotic cells undergo acidification and mature from Rab5-positive early to Rab7-positive late stages. Phagosomes finally fuse with lysosomes to form phagolysosomes, which degrade apoptotic cells; however, the molecular mechanism underlying phagosome–lysosome fusion is not fully understood. Here we show that the Caenorhabditis elegans Arf-like small GTPase Arl8 (ARL-8) is involved in phagolysosome formation and is required for the efficient removal of apoptotic cells. Loss of function of arl-8 results in the accumulation of apoptotic germ cells. Both the engulfment of the apoptotic cells by surrounding somatic sheath cells and the phagosomal maturation from RAB-5- to RAB-7-positive stages occur in arl-8 mutants. However, the phagosomes fail to fuse with lysosomes in the arl-8 mutants, leading to the accumulation of RAB-7-positive phagosomes and the delayed degradation of apoptotic cells. ARL-8 localizes primarily to lysosomes and physically interacts with the homotypic fusion and protein sorting complex component VPS-41. Collectively our findings reveal that ARL-8 facilitates apoptotic cell removal in vivo by mediating phagosome–lysosome fusion during phagocytosis.
INTRODUCTION
Apoptosis, or programmed cell death, is a genetically regulated process of cell suicide. Under normal conditions, apoptotic cells are recognized and degraded by phagocytes (e.g., macrophages) before they release potentially toxic materials. Defective clearance of apoptotic cells can thus lead to various diseases, including autoimmune disorders (Elliott and Ravichandran, 2010). Phagocytic degradation of apoptotic cells is controlled by mechanisms that are well conserved from the nematode Caenorhabditis elegans to humans (Fadeel, 2003). C. elegans is a useful genetic system in which to study the mechanism controlling apoptotic cell phagocytosis (Reddien and Horvitz, 2004; Lettre and Hengartner, 2006). Two conserved signaling pathways have been proposed to mediate the recognition and internalization of apoptotic cells. In one pathway, the engulfment receptor CED-1, together with CED-7 (an ATP-binding cassette transporter), functions upstream of CED-6 (a phosphotyrosine-binding domain–containing adaptor protein) to regulate the recognition of apoptotic cells and transduce engulfing signals (Liu and Hengartner, 1998; Wu and Horvitz, 1998; Zhou et al., 2001). In the other pathway, the engulfment receptor PSR-1 acts upstream of the signaling proteins CED-2/CrkII, CED-5/Dock180, and CED-12/ELMO1 to activate CED-10/Rac1; this cascade leads to actin remodeling, which drives the growth of membrane pseudopods to enclose apoptotic cells (Fadok et al., 2000; Wang et al., 2003; Kinchen et al., 2005).
The internalized apoptotic cells are enclosed within membrane-bound compartments called phagosomes. Phagosomes undergo a “maturation process” that involves recruitment of early/late endosomal compartments and V-ATPases for acidification (Desjardins et al., 1994; Henry et al., 2004). The Rab GTPases Rab5 and Rab7 are sequentially recruited to phagosomes to coordinate early-to-late phagosome maturation (Duclos et al., 2000; Roberts et al., 2006; Seto et al., 2011). Rab5 promotes phagosomal fusion with early endosomes and regulates phagosome conversion to a Rab7-positive stage (Vieira et al., 2003). Studies in C. elegans identified several Rab GTPases as critical regulators of phagosome maturation (Kinchen et al., 2008; Lu et al., 2008; Mangahas et al., 2008; Yu et al., 2008; Guo et al., 2010; Kinchen and Ravichandran, 2010). The transition of phagosomes from the RAB-5 (Rab5)- to the RAB-7 (Rab7)-positive stage is regulated by the SAND-1 (Mon1)/CCZ-1 (Ccz1) complex (Kinchen and Ravichandran, 2010): GTP-bound RAB-5 interacts with the SAND-1/CCZ-1 complex, and the resultant RAB-5/SAND-1/CCZ-1 complex then facilitates RAB-7 activation/recruitment by dislodging GDP-bound RAB-7 from its complex with the GDP-dissociation inhibitor (GDI). RAB-7 is required for phagosome–lysosome fusion, which forms the phagolysosomes in which apoptotic cells are degraded by lysosomal hydrolytic enzymes (Kinchen et al., 2008; Yu et al., 2008). Studies suggested that the homotypic fusion and vacuolar protein sorting (HOPS) complex components may function downstream of RAB-7 and be involved in phagosome–lysosome fusion (Kinchen et al., 2008; Yu et al., 2008; Akbar et al., 2011). The HOPS complex, originally characterized in yeast as a factor in vacuolar fusion, consists of six subunits (a core of four subunits, VPS-11, -16, -18, and -33, with two accessory subunits, VPS-39 and -41). In yeast, the HOPS complex is an effector of Ypt7p (yeast Rab7) and binds the vacuolar soluble N-ethylmaleimide–sensitive factor attachment protein receptors (SNAREs) to drive vacuole fusion (Nickerson et al., 2009; Wickner, 2010; Epp et al., 2011).
Arl8 is a small GTPase that is highly conserved among multicellular organisms and localizes primarily to the lysosomes (Bagshaw et al., 2006; Hofmann and Munro, 2006; Nakae et al., 2010). Previous studies showed that Arl8 is involved in lysosomal movement along microtubules (Bagshaw et al., 2006; Hofmann and Munro, 2006) and endocytic traffic to lysosomes (Nakae et al., 2010). Arl8 was shown to interact with SifA and kinesin-interacting protein (SKIP), thereby recruiting kinesin-1 to lysosomes to facilitate lysosome migration toward the cell periphery (Rosa-Ferreira and Munro, 2011). In the context of microtubule-dependent membrane transport, C. elegans Arl8 (ARL-8) facilitates the axonal transport of presynaptic cargoes in neurons (Klassen et al., 2010). A recent article reported an unexpected role of Arl8 in Tobmorius RNA replication in Arabidopsis thaliana (Nishikiori et al., 2011).
In the present study, we identified ARL-8 as a critical component for efficient phagocytic degradation of apoptotic germ cells in C. elegans. We found that an arl-8 deletion mutant exhibited an accumulation of apoptotic germ cells. Apoptotic cells were engulfed by somatic sheath cells (large phagocytic cells) in the arl-8 mutants; however, degradation of the apoptotic cells was severely impaired. In arl-8 mutants, phagosomes containing apoptotic cells were arrested in a RAB-7–positive stage. Time-lapse imaging of live animals revealed that the fusion of lysosomes to phagosomes is defective in arl-8 mutants. ARL-8 localized primarily to lysosomes and physically interacted with the HOPS complex component VPS-41. We therefore propose that arl-8 acts to clear apoptotic cells by mediating phagolysosome formation.
RESULTS
arl-8 is required for cell corpse removal in the adult hermaphrodite gonad
In C. elegans, approximately half of the hermaphrodite germ cells undergo apoptosis and are phagocytosed by surrounding somatic sheath cells for degradation by lysosomal enzymes (Gumienny et al., 1999). We previously reported that C. elegans ARL-8 localizes primarily to lysosomes and mediates delivery of endocytosed macromolecules to the lysosomes in macrophage-like coelomocytes (Nakae et al., 2010). We found that arl-8(tm2504), a strong loss-of-function mutant (Nakae et al., 2010), exhibited an increased number of cell corpses in the adult gonad arms (Figure 1A, arrows). We scored cell corpses in one gonad arm of wild-type and arl-8 mutant animals every 12 h after the L4 stage; the number of cell corpses increased progressively in an age-dependent manner in arl-8 mutants (Figure 1B). There was no apparent difference between the wild-type and arl-8 mutant animals in the frequency of cell death in the U-turn region of the gonad (4.4 ± 0.84 and 4.5 ± 0.74 germ cell deaths in 180 min in wild-type and arl-8(tm2504) animals, respectively; n = 4), suggesting that the increase in the number of cell corpses in arl-8(tm2504) animals was not caused by elevated germ cell apoptosis. We next analyzed cell corpse duration to investigate whether arl-8(tm2504) mutants are defective in cell corpse removal. Most cell corpses were degraded within 50 min after their appearances in the wild type, whereas they persisted for more than 100 min in arl-8(tm2504) animals (Figure 1D), indicating that the clearance of cell corpses is impaired in arl-8(tm2504) animals. The arl-8(tm2504) persistent cell corpse phenotype was significantly rescued by expression of arl-8::venus under the ced-1 promoter, which drives gene expression specifically in the engulfing sheath cells (Figure 1C). These results suggest that arl-8 is required for the efficient removal of apoptotic cells by sheath cells. It remains to be determined whether arl-8(tm2504) animals are defective also for the removal of embryonic apoptotic cells, since the homozygous embryos from arl-8(tm2504) homozygous hermaphrodites arrest in early embryonic stages before programmed cell death is induced (unpublished data).
FIGURE 1:
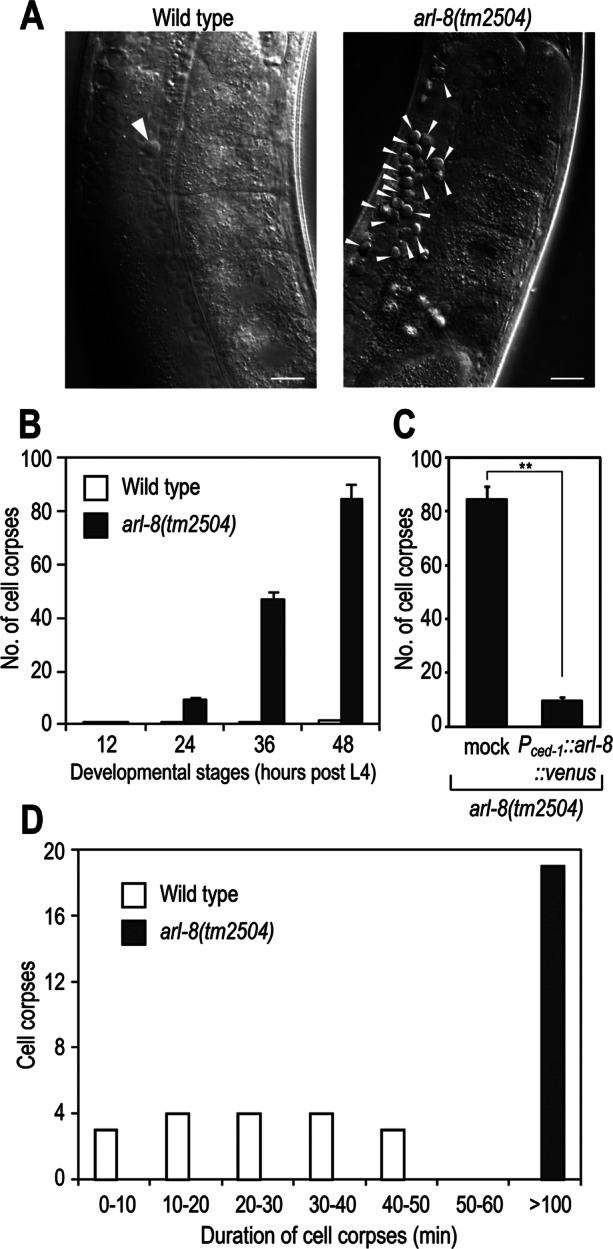
arl-8(tm2504) mutants are defective in the removal of germ cell corpses. (A) Nomarski images of gonad arms in wild-type or arl-8(tm2504) adult hermaphrodites 48 h after the L4 larval stage. Germ cell corpses are marked with arrowheads. Bars, 10 μm. (B) Germ cell corpses of one gonad arm from each animal were scored every 12 h after the L4 larval stage. Fifteen animals were scored for each strain, and data are expressed as mean ± SEM. (C) The increased germ cell corpse phenotype of arl-8(tm2504) animals was rescued by Pced-1::arl-8::venus. Germ cell corpses of one gonad arm from the indicated animals were scored after the L4 larval stage. More than 10 animals were scored for each strain, and the data are expressed as mean ± SEM. **p < 0.01 by Student's t test. (D) Histogram summarizing cell corpse duration the germline of the indicated animals. More than 15 cell corpses were monitored in each strain.
Phagosomes containing cell corpses are arrested at an RAB-7–positive stage in arl-8 mutants
Apoptotic germ cells are engulfed by sheath cells and subsequently degraded inside phagosomes. We first investigated whether the apoptotic cells were engulfed by the sheath cells in arl-8 mutants using acridine orange (AO), which preferentially stains acidic compartments and can be used to label the internalized cell corpses in living cells (Gumienny et al., 1999; Lu et al., 2008; Xiao et al., 2009). Most cell corpses in arl-8 mutants were stained by AO (Figure 2A), indicating that the cell corpses were engulfed by the sheath cells. Phagosomal acidification in arl-8 mutants was also confirmed by LysoTracker Red staining (Figure 2B). We next analyzed the phagosomal maturation of arl-8 mutants by examining the recruitment of Rab GTPases to the phagosomes. Studies showed that the phagosomes recruit RAB-5 and RAB-7 sequentially to their membranes: whereas RAB-5 is recruited transiently to early phagosomes, RAB-7 is present on phagosomes at late stages of phagosome maturation (Kinchen et al., 2008; Kinchen and Ravichandran, 2010; Yu et al., 2008). Time-lapse analysis showed that GFP::RAB-5 was recruited to nascent phagosomes in arl-8(tm2504) animals and detached from the phagosomes similarly to that in the control arl-8(tm2504)/nT1 animals (Figure 2C), although the average duration of RAB-5 at phagosomes of arl-8(tm2504) was slightly longer than that of arl-8(tm2504)/nT1 (39.2 ± 2.33 and 33.2 ± 1.91 min, respectively; n = 11). The number of RAB-5–positive phagosomes in arl-8 mutants was comparable to that observed in the wild type, whereas the number of RAB-7–positive phagosomes increased to a great degree in arl-8 mutants (Figure 2D). Although we cannot rule out the possibility that loss of arl-8 affects early stages of phagosome maturation, these results suggest that the phagosomes of arl-8 mutants undergo early maturation and arrest at a RAB-7–positive late stage.
FIGURE 2:
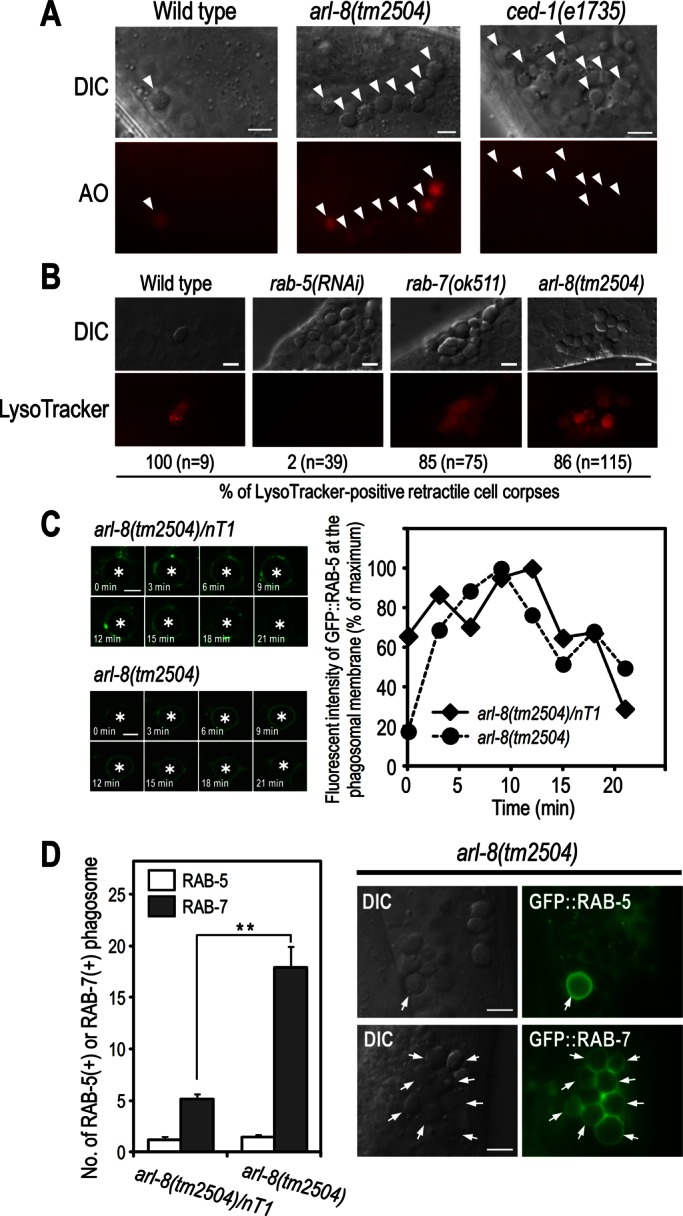
Phagosomes are arrested at an RAB-7–positive stage in arl-8(tm2504) mutants. (A) AO staining of germ cell corpses in the indicated animals. Germ cell corpses are marked with arrowheads. Most cell corpses were stained by AO in arl-8(tm2504) animals, whereas corpses were not stained in ced-1(e1735) animals in which cell corpses are not phagocytosed by sheath cells. Bars, 5 μm. (B) LysoTracker staining of germ cell corpses and quantitation of LysoTracker-positive cell corpses in the indicated animals. n, number of cell corpses scored. Bars, 5 μm. (C) Time-lapse images of GFP::RAB-5 expression at the phagosome (left) and quantitation of the fluorescence intensity (right) in arl-8(tm2504)/nT1 (as a control) and arl-8(tm2504) animals. Asterisks indicate germ cell corpses. Bars, 2.5 μm. (D) Quantitation of RAB-5–positive and RAB-7–positive phagosomes in the germline of the indicated animals (left). Fifteen animals were scored for each strain, and data are expressed as mean ± SEM. **p < 0.01 by Student's t test. Nomarski and fluorescence images of arl-8(tm2504) animals expressing GFP::RAB-5 or GFP::RAB-7 (right). Arrows indicate the germ cell corpses positive for the corresponding GFP-marker proteins. Bars, 5 μm.
ARL-8 is localized to lysosomes and recruited to phagosomes at late stages of phagosome maturation
To investigate the potential function of ARL-8 in phagosomal maturation, we analyzed the localization of ARL-8::green fluorescent protein (GFP) in sheath cells. ARL-8::GFP was detected in the punctate structures (arrows in Figure 3A) and also in the cytoplasm surrounding germ cells (arrowhead in Figure 3A). Consistent with reports showing that Arl8/ARL-8 localizes primarily to lysosomes (Bagshaw et al., 2006; Hofmann and Munro, 2006; Nakae et al., 2010), most ARL-8::GFP-labeled punctate structures colocalized with NUC-1::mCherry (NUC-1: a lysosomal nuclease involved in the degradation of apoptotic cell DNA; Wu et al., 2000; Guo et al., 2010). We observed some ARL-8::GFP-positive structures with little NUC-1::mCherry (open arrowhead in the inset of Figure 3A), suggesting that ARL-8 may also localize to other membrane compartments besides lysosomes (e.g., endosomes).
FIGURE 3:
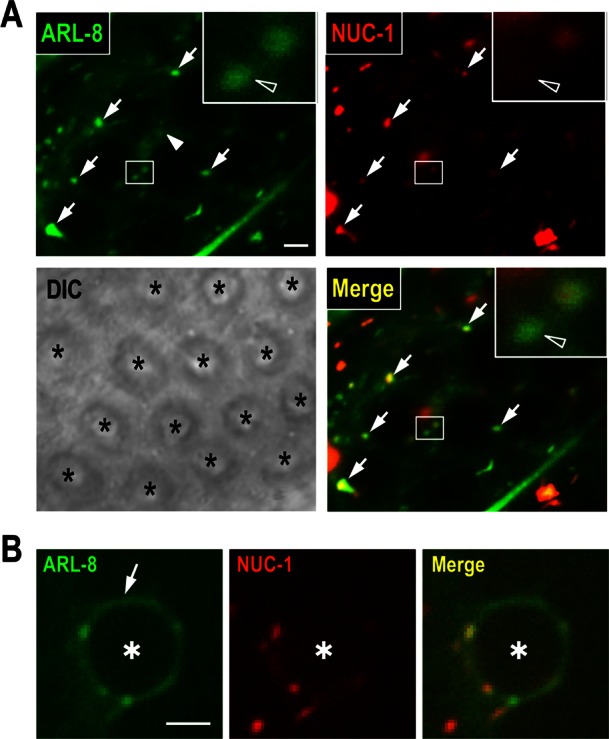
Analysis of ARL-8::GFP localization in sheath cells. (A) Fluorescence and differential interference contrast images of the gonad arm of a wild-type animal coexpressing ARL-8::GFP and NUC-1::mCherry. Asterisks indicate germ cells. ARL-8::GFP was observed in punctate structures (arrows) and cytoplasm surrounding germ cells (arrowhead). Insets, enlarged views of the areas indicated by white rectangles. The open arrowheads in the insets indicate puncta of ARL-8::GFP with little NUC-1::mCherry. Bar, 2 μm. (B) Fluorescence images of a phagosome containing a germ cell corpse (asterisk) in a wild-type animal coexpressing ARL-8::GFP and NUC-1::mCherry. ARL-8::GFP was observed at the phagosomal membrane (arrow). Bar, 2.5 μm.
ARL-8::GFP was also detected at phagosomal membranes (arrow in Figure 3B). To investigate the dynamics of ARL-8 localization to phagosomes, we performed time-lapse imaging of transgenic animals expressing both ARL-8::GFP and mCherry::RAB-5 (an early-stage marker of phagosome maturation). ARL-8::GFP signal was only faint at the RAB-5–labeled phagosomal membranes (0 min in Figure 4A). The ARL-8 signal then proceeded to form a smooth, continuous circle, whereas the RAB-5 signal on phagosomes decreased; phagosomes converted from RAB-5 positive/ARL-8 negative to RAB-5 negative/ARL-8 positive. Next we did time-lapse analysis using transgenic animals expressing both ARL-8::GFP and mCherry::RAB-7 (a late-stage maker of phagosomal maturation), showing that the timing of ARL-8::GFP localization to phagosomes was similar to that of mCherry::RAB-7 (Figure 4B). Both ARL-8 and RAB-7 persisted on the phagosomes until degradation of phagosomal contents (unpublished data). Collectively these results indicate that ARL-8 was localized to phagosomes at late stages of phagosome maturation.
FIGURE 4:
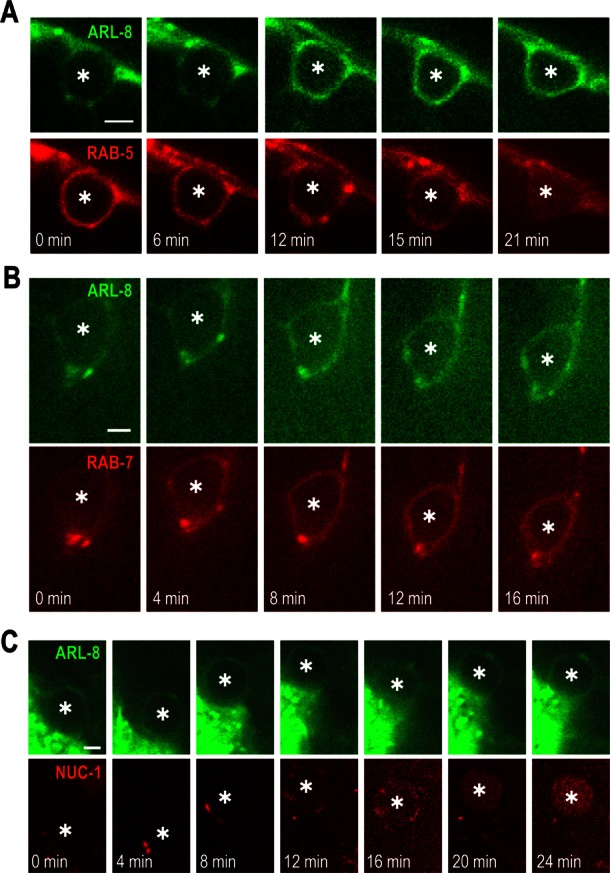
ARL-8 is recruited to phagosomes at late stages of phagosome maturation. Time-lapse images of fluorescence signal at a phagosome containing a germ cell corpse (asterisk) in a wild-type animal coexpressing ARL-8::GFP and mCherry::RAB-5 (A), ARL-8::GFP and mCherry::RAB-7 (B), or ARL-8::GFP and NUC-1::mCherry (C). Bars, 2.5 μm.
Because ARL-8::GFP was observed in both lysosomes and phagosomes, we investigated whether phagosomal localization of ARL-8 was simply a consequence of phagosome–lysosome fusion. Time-lapse analysis using transgenic animals expressing both ARL-8::GFP and NUC-1::mCherry showed that ARL-8::GFP was localized to phagosomes in which no apparent incorporation of NUC-1::mCherry was observed (0 min in Figure 4C and Supplemental Movie S1). ARL-8::GFP-labeled phagosomes then gradually fused with NUC-1::mCherry-labeled lysosomes, leading to the formation of phagolysosomes in which NUC-1::mCherry accumulated (24 min in Figure 4C). Given that ARL-8::GFP was detected in NUC-1::mCherry-negative compartments (Figure 3A), phagosomal localization of ARL-8::GFP may be archived not only via phagosome–lysosome fusion but also via fusion of these compartments with phagosomes before phagolysosome formation.
Localization of ARL-8 on phagosomes requires RAB-7 and the HOPS-complex subunits
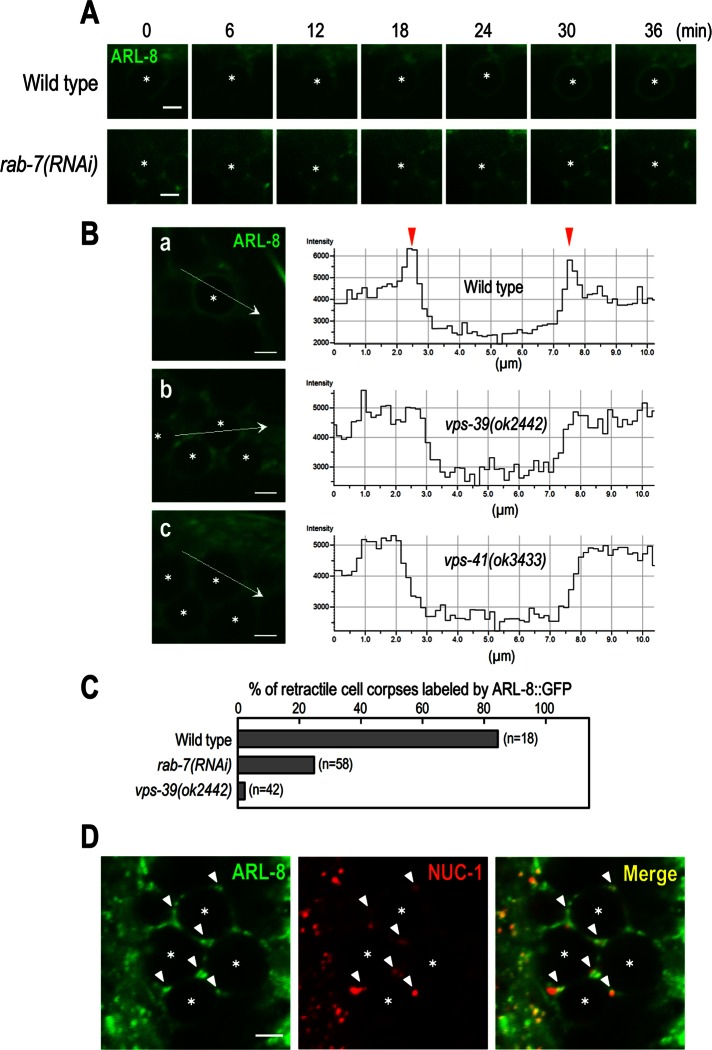
Because ARL-8 colocalized with RAB-7 at phagosomal membranes, we investigated whether phagosomal localization of ARL-8 requires RAB-7. We found that ARL-8::GFP failed to localize to phagosomes in rab-7(RNAi) mutants and persisted as punctate fluorescence around phagosomes (Figure 5A). This result indicates that rab-7 is required for the phagosomal localization of ARL-8. Studies suggested that the HOPS complex functions downstream of rab-7 during phagosomal maturation: loss of function of the HOPS-complex component VPS-39 leads to the accumulation of arrested phagosomes at the RAB-7–positive stage (Kinchen et al., 2008; Kinchen and Ravichandran, 2010; Akbar et al., 2011). To determine whether the HOPS complex is involved in the phagosomal localization of ARL-8, we analyzed the ARL-8::GFP localization in the deletion mutants of HOPS-complex subunits VPS-39 and VPS-41. Whereas localization of ARL-8::GFP on the phagosomal membranes in wild-type animals was observed as two fluorescence peaks along the line scan (arrowheads in Figure 5B), no apparent phagosomal localization of ARL-8::GFP was observed in vps-39(ok2422) and vps-41(ok3433) animals (Figure 5B). These results indicate that both rab-7 and the HOPS complex are required for ARL-8 localization to phagosomes. In contrast, ARL-8::GFP colocalized with NUC-1::mCherry in vps-39(ok2422) animals to a similar extent as in wild-type animals (Figure 5D), suggesting that ARL-8::GFP localizes to lysosomes in a HOPS complex–independent manner.
FIGURE 5:
Analysis of ARL-8::GFP localization in rab-7 and the HOPS-complex mutants. (A) Time-lapse images of ARL-8::GFP signal in the gonad of wild-type and rab-7(RNAi) animals. Here 0 min represents the point at which mCherry::RAB-5 (not shown) was first detected on phagosomal membranes. Asterisks indicate germ cell corpses. Bars, 2.5 μm. (B) Fluorescence images of the gonad of wild-type (a), vps-39(ok2442) (b), or vps-41(ok3433) (c) animals expressing ARL-8::GFP and line-scan quantitation of the fluorescence signal at the position indicated by the white arrow in a–c. Asterisks indicate germ cell corpses. Bars, 2.5 μm. (C) The ratio of cell corpses labeled by ARL-8::GFP in the indicated animals. n, number of cell corpses scored. (D) Fluorescence images of the gonad arm of vps-39(ok2442) coexpressing ARL-8::GFP and NUC-1::mCherry. ARL-8::GFP-labeled punctate structures colocalized with NUC-1::mCherry (arrowheads). Asterisks indicate germ cell corpses. Bar, 2.5 μm.
ARL-8 physically interacts with VPS-41 and is required for phagosome–lysosome fusion
We investigated whether ARL-8 can physically interact with the HOPS-complex components. Purified recombinant Flag-tagged ARL-8 proteins prebound to either GTPγS or GDP were incubated with HEK293T cell lysates containing Myc-tagged VPS-41 and then immunoprecipitated with anti-Flag M2 agarose. We found that GTPγS-bound Flag-ARL-8 preferentially associated with Myc-VPS-41 (Figure 6A), which is consistent with a report showing that GTP-bound active Arl8b binds to VPS41 in mammalian cells (Garg et al., 2011). These results suggest that VPS-41 may function as an effector protein of ARL-8 in the phagocytic pathway. Finally, we investigated whether arl-8 is required for phagolysosome formation. In wild-type animals, NUC-1::mCherry-labeled lysosomes were observed on the surface of phagosomes, and NUC-1::mCherry fluorescence then gradually increased inside the phagosomes, which likely represents phagolysosome formation via phagosome–lysosome fusion. We observed severe defects in the incorporation of NUC-1:mCherry into phagosomes of the arl-8 mutants: NUC-1:mCherry-labeled punctate structures were observed near the phagosomes of the arl-8 mutants, but the fluorescence failed to be incorporated into the phagosomes (Figure 6, B and C). These results suggest that phagosome–lysosome fusion is impaired in arl-8 mutants. We tested whether loss of arl-8 affects the morphology of lysosomes, since Arl8 and rab7 have recently been shown to be necessary for lysosome tubulation in mammalian macrophages (Mrakovic et al., 2012). We found that NUC-1::mCherry-labeled tubular structures were impaired in arl-8 mutants (Supplemental Figure S1), suggesting that arl-8 may be involved in the formation of normal lysosomes.
FIGURE 6:
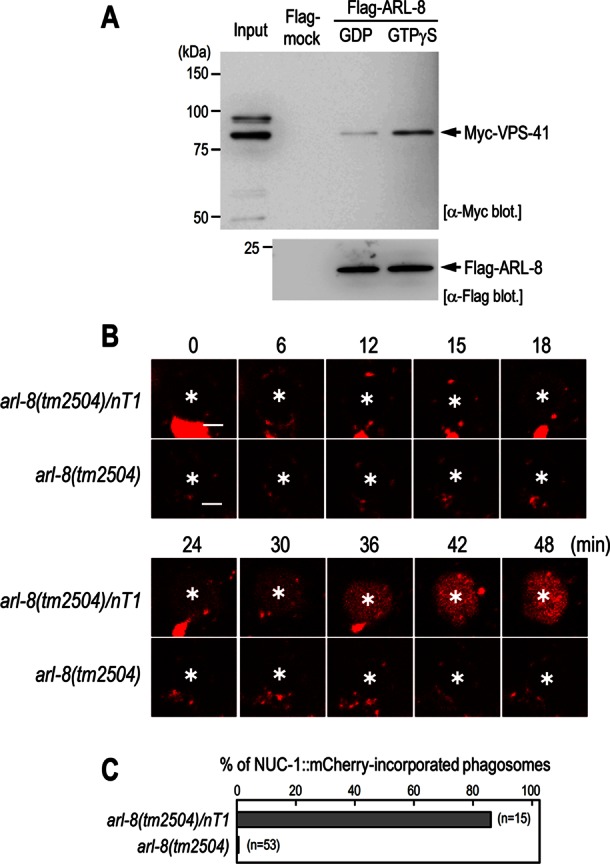
ARL-8 physically interacts with VPS-41 and is required for phagosome–lysosome fusion. (A) Pull-down assay using Flag-ARL-8 and Myc-VPS-41. Purified Flag-ARL-8 proteins preloaded with either GDP or GTPγS were incubated with HEK293T cell lysates containing Myc-VPS-41 and immunoprecipitated using anti-Flag M2 agarose. The precipitated fractions were then subjected to Western blot analysis using the indicated antibodies. (B) Time-lapse images of NUC-1::mCherry signal in the gonad of arl-8(tm2504)/nT1 and arl-8(tm2504) animals. Asterisks indicate germ cell corpses. Here 0 min represents the point at which GFP::RAB-5 (not shown) was first detected on phagosomal membranes. Bars, 2.5 μm. (C) Quantitation of fusion events in the indicated animals. Data are expressed as the ratio of cell corpses that incorporated NUC-1::mCherry within 1 h after their appearance. n, number of cell corpses scored.
DISCUSSION
The engulfment and clearance of apoptotic cells by neighboring cells or professional phagocytes is crucial to tissue homeostasis and the regulation of immune responses. In the present study, we identified arl-8 as a novel factor essential for the efficient degradation of apoptotic cells in C. elegans. Deletion of arl-8 resulted in the accumulation of apoptotic germ cells. The cell corpses were engulfed by phagocytic sheath cells in arl-8 mutants; however, the phagosomes containing cell corpses arrested in RAB-7–positive stages and exhibited impaired cell corpse degradation. ARL-8 localized primarily to lysosomes and physically interacted with the HOPS-complex component VPS-41. Finally, the loss of arl-8 caused defective phagosome–lysosome fusion. These results suggest that ARL-8 mediates phagolysosome formation to promote the degradation of apoptotic cells in vivo.
Phagosomes are dynamic, membrane-bound organelles that degrade a variety of particles, including potentially pathogenic microorganisms and apoptotic cells. After particle internalization, the nascent phagosome undergoes a complex maturation process involving sequential recruitment of endosomal compartments (Vieira et al., 2002; Kinchen and Ravichandran, 2008). Rab5 is transiently recruited to and subsequently removed from nascent phagosomes and is required for phagosomal maturation into a later Rab7-positive stage (Duclos et al., 2000; Vieira et al., 2003; Roberts et al., 2006; Kinchen and Ravichandran, 2010). Although the duration of RAB-5 at phagosomes was slightly longer in arl-8 mutants than in wild type, RAB-7 was recruited to the phagosomes, indicating that phagosomes undergo the transition from early- to late-stage maturation in arl-8 mutants. Consistent with this, the phagosomes of arl-8 mutants were stained with acid-tropic dyes (AO and LysoTracker Red), which preferentially stain late-stage, internalized apoptotic cells. These results suggest that phagosomes are arrested at a RAB-7–positive late stage of maturation in arl-8 mutants.
We previously showed that ARL-8 localizes primarily to lysosomes in the macrophage-like coelomocytes (Nakae et al., 2010). Consistent with this, most ARL-8::GFP-labeled structures were positive for the lysosomal enzyme marker NUC-1::mCherry in sheath cells. Note that a fraction of ARL-8::GFP-labeled structures were negative for NUC-1::mCherry. ARL-8 may thus be localized also to other membrane compartments besides lysosomes (e.g., endosomes). Given that early/late endosomal components were recruited to phagosomes during phagosomal maturation (Desjardins et al., 1994; Henry et al., 2004) and that ARL-8::GFP was observed at phagosomes before the formation of phagolysosomes (Figure 4C), ARL-8-positive endosomes may fuse with phagosomes to form ARL-8–labeled phagosomes, which subsequently fuse with lysosomes to form phagolysosomes. Further analysis is required to determine whether the preceding association of ARL-8 with phagosomes is involved in the subsequent phagosome–lysosome fusion.
Previous studies showed that RAB-7–positive phagosomes finally fuse with lysosomes to form the phagolysosomes in which cell corpses are degraded. The molecular mechanism underlying this process is poorly understood. Studies suggested that the HOPS complex functions downstream of RAB-7 to mediate phagosome–lysosome fusion (Kinchen et al., 2008; Akbar et al., 2011). Given that the yeast HOPS complex is recruited to Ypt7p/Rab7 on vacuolar membranes and interacts with SNAREs required for membrane fusion, the HOPS complex may be recruited to phagosomes via RAB-7 in sheath cells and involved in phagosome–lysosome fusion. Of interest, a recent report showed that human Arl8b interacts with the HOPS-complex subunit VPS41 and is involved in the recruitment of the HOPS components to Arl8b-positive compartments (Garg et al., 2011). Given that ARL-8 localized primarily to lysosomes and physically interacted with VPS-41, the HOPS complex may be recruited to both RAB-7–positive phagosomes and ARL-8–positive lysosomes, thereby promoting fusion between phagosomes and lysosomes. Alternatively, given that phagosomal localization of ARL-8 preceded phagolysosome formation and required the HOPS-complex components, the HOPS complex may be involved in the fusion of ARL-8–positive endosomal compartments with phagosomes to promote phagosomal maturation required for the subsequent fusion with lysosomes. To test these possibilities, we tried to determine the subcellular localization of the HOPS complex in sheath cells; however, various attempts to create transgenic animals that express sufficient levels of GFP- or mCherry-fusion proteins of the HOPS components have been unsuccessful. Future studies are thus needed to clarify the functional relationships among ARL-8, RAB-7, and the HOPS complex in the phagocytic pathway.
We previously reported that ARL-8 is required for endocytic traffic to lysosomes in macrophage-like coelomocytes in C. elegans (Nakae et al., 2010). The present study suggests that ARL-8 plays an important role in the delivery of phagocytosed cargo to lysosomes. Consistent with these findings, mammalian Arl8b was shown to be involved in the transport of various cargoes to lysosomes in macrophages (Garg et al., 2011). In contrast, Arl8b was reported to be dispensable for the trafficking of endocytosed BSA to lysosomes in HeLa cells (Kaniuk et al., 2011). The requirement of Arl8b for cargo trafficking to lysosomes may thus depend on cell type and/or cellular status. Uncovering the lysosomal delivery regulatory mechanism mediated by Arl8/ARL-8 may help to elucidate the spatiotemporal control of degradation of cargo molecules in lysosomes.
MATERIALS AND METHODS
Strains and genetics
Worm cultures and genetic crosses were typically performed according to standard protocols (Brenner, 1974). The N2 Bristol strain was used as the wild-type strain. Other strains used in this study are as follows: CB3203 ced-1(e1735) I, YB513 arl-8(tm2504) IV/nT1[qIs51](IV;V), VC2542 vps-39(ok2442) V/nT1[qIs51](IV;V), VC2784 +/szT1[lon-2(e678)] I; vps-41(ok3433)/szT1 X, YB1439 arl-8(tm2504) IV/nT1[qIs51](IV;V); qxIs58[Pced-1::lmp-1::Cherry]; tdEx955 [Pced-1::arl-8::venus, rol(su1006)], YB2187 arl-8(tm2504) IV/nT1[qIs51](IV;V); tdEx1504[Pced-1::gfp::rab-5, Pced-1::nuc-1::mCherry, rol-6(su1006)], YB1468 arl-8(tm2504) IV/nT1[qIs51](IV;V); qxIs58[Pced-1::lmp-1::Cherry]; tdEx974 [Pced-1::gfp::rab-5, rol-6(su1006)], YB1471 arl-8(tm2504) IV/nT1[qIs51](IV;V); qxIs58[Pced-1::lmp-1::Cherry]; tdEx977 [Pced-1::gfp::rab-7, rol-6(su1006)], YB2034 arl-8(tm2504) IV/nT1[qIs51](IV;V); tdEx1385[Pced-1::nuc-1::mCherry, rol-6(su1006)], YB2003 tdEx1361[Pced-1::nuc-1::mCherry, rol-6(su1006)], YB2036 tdIs27[Parl-8::arl-8::gfp, rol-6(su1006)]; tdEx1387[Pced-1::nuc-1::mCherry], YB1966 tdIs27[Parl-8::arl-8::gfp, rol-6(su1006)]; tdEx1332[Pced-1::mCherry::rab-5], YB2068 tdIs27[Parl-8::arl-8::gfp, rol-6(su1006)]; tdEx1418[Pced-1::mCherry::rab-7], YB2098 vps-39(ok2442) V/nT1[qIs51](IV;V); tdIs27[Parl-8::arl-8::gfp, rol-6(su1006)], YB2270 +/szT1[lon-2(e678)] I; vps-41(ok3433)/szT1 X; tdIs27[Parl-8::arl-8::gfp, rol-6(su1006)], YB2221 vps-39(ok2442) V/nT1[qIs51](IV;V); tdIs27[Parl-8::arl-8::gfp, rol-6(su1006)]; tdEx1534[Pced-1::nuc-1::mCherry]. The strains were maintained at 15 or 20ºC. tdIs27[Parl-8::arl-8::gfp, rol-6(su1006)] was isolated by ultraviolet irradiation methods (Mitani, 1995). qxIs58[Pced-1::lmp-1::Cherry] was kindly provided by X. Wang (National Institute of Biological Sciences, Beijing, China). Germline transformation experiments were conducted by injecting various constructs with coinjection markers (Mello et al., 1991).
Plasmid constructs
To generate Pced-1::arl-8::venus, a 5-kb fragment of the upstream promoter region of the ced-1 gene was amplified from genomic DNA by PCR using the primers 5′-GCGGCCGCTTGCGGCTGCAAAAAAACAGG-3′ and 5′-GGATCCCTGGATTAGTCATACCTCCTG-3′ and subsequently cloned into the venus-fusion expression vector pFX- arl-8::venus, which contains the entire coding region of the arl-8 gene. To generate Pced-1::gfp::rab-7, the entire coding region of the rab-7 gene was amplified from genomic DNA by PCR using the primers 5′-CGGGATCCAATGTCGGGAACCAGAAAGAAG-3′ and 5′-CGGGATCCTTAACAATTGCATCCCGAATTCTG-3′ and subsequently cloned into the GFP-fusion expression vector pKK04; this insertion was followed by the insertion of the genomic DNA fragment containing the promoter region of the ced-1 gene. A similar strategy was used to generate Pced-1::gfp::rab-5 using the primer sets 5′-GCGGATCCTATGGCCGCCCGAAACG-3′ and 5′-CGGGATCCTTATTTACAGCATGAACCCTTTTGTTGC-3′). To generate Pced-1::nuc-1::mCherry, the entire coding region of the nuc-1 gene was amplified from genomic DNA by PCR using the primers 5′-GCGGCCGCATGGGCTTGTCTCCTGCC-3′ and 5′-GCGGCCGCATGCACAATTATTTTGGGTTGCAA-3′ and subsequently cloned into the mCherry-fusion expression vector pFX-Pced-1::mCherry, which contains the promoter region of the ced-1 gene. To generate Pced-1::mCherry::rab-5 and Pced-1::mCherry::rab-7, the entire coding regions of rab-5 and rab-7 were prepared by BamHI digestion of Pced-1::gfp::rab-5 and Pced-1::gfp::rab-7, respectively, and the resultant DNA fragments were subcloned into the BamHI site of the mCherry-fusion expression vector pKK06, which contains the promoter region of the ced-1 gene.
Quantification of cell corpses
Germ cell corpses were scored using Nomarski optics. Cell corpses in the germline meiotic region of one gonad arm were scored 12, 24, 36, and 48 h after the L4 stage. To record the duration of the cell corpses, adult animals (48 h post L4 stage) were mounted on 4% agarose pads with 2 mM levamisole in M9 medium. The gonad arm was observed under Nomarski optics manually every 5 min for 180 min. The temperature was maintained at 18–22ºC during the scoring.
Acridine orange and LysoTracker staining
AO staining was performed as described previously, with minor modifications (Xiao et al., 2009). Adult animals were incubated in 250 μl of M9 medium containing AO (0.14 mg/ml) and OP50 bacteria for 90 min in the dark. The stained animals were transferred to growth medium plates to recover for 1 h and then examined under fluorescence microscopy to score the AO-positive germ cell corpses. LysoTracker staining was performed as described previously, with slight modifications (Guo et al., 2010). Briefly, adult animals were dissected in staining buffer (20 mM Na 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4, 60 mM NaCl, 32 mM KCl, 3 mM Na2HPO4, 5 mM MgCl2, 2 mM levamisole, 0.1% tricaine, 1 μM LysoTracker Red DND-99) and examined by fluorescence microscopy.
Microscopy
Nomarski and fluorescence images were obtained with a Zeiss Axio Imager M1 microscope system equipped with AxioVision software (Zeiss, Jena, Germany). Confocal images were acquired with a Nikon ECLIPSE TE2000-E (Nikon, Melville, NY) equipped with a CSU10 (Yokogawa, Tokyo, Japan)/iXon DV887 (Andor, Belfast, United Kingdom) confocal scanner unit and processed with Nikon NIS-Elements AR3.2, ImageJ (National Institutes of Health, Bethesda, MD), and Photoshop (Adobe, San Jose, CA) software. For quantification of the fluorescence intensity of GFP on phagosomes, the area of the limiting phagosomal membrane in each image was selected manually using ImageJ software, and the fluorescence intensity in the selected area was measured.
RNA interference experiments
The C. elegans open reading frame–RNA interference (RNAi) library, version 1.1, was purchased from Open Biosystems (Huntsville, AL). The feeding RNAi protocol was performed as described previously (Kamath et al., 2001; Poteryaev et al., 2007). Briefly, adult animals (P0) were placed on RNAi plates, and F1 adult animals were observed. The animals were maintained at 20ºC during RNAi treatment.
In vitro pull-down assay
Flag-tagged ARL-8 (Flag-ARL-8) was produced in the Escherichia coli strain BL21-CodonPlus (DE3)-RIL (Agilent Technologies, Santa Clara, CA) using pCold TF expression vector (Takara Bio, Otsu, Japan). cDNA encoding vps-41 (isoform a) was subcloned into pMyc-CMV5 vector, and the expression vector was transfected into HEK293T cells using Lipofectamine2000 (Life Technologies, Carlsbad, CA). The transfected cells were homogenized 24 h after transfection in buffer A (10 mM triethanolamine-acetic acid, pH 7.6, 250 mM sucrose and 1 mM EDTA) and centrifuged (200,000 × g, 30 min, 4ºC). The supernatant was mixed with purified Flag-ARL-8 proteins that had been preloaded with GDP or GTPγS and bound to anti–Flag M2 agarose (Sigma-Aldrich, St. Louis, MO), and the mixture was incubated for 1.5 h at 4ºC in buffer B (37 mM Tris-HCl, pH 7.5, 2.6 mM triethanol amine-acetic acid, pH 7.6, 74 mM NaCl, 65 mM sucrose, 3.7 mM MgCl2, 0.26 mM EDTA, 0.74 mM dithiothreitol [DTT], 2.22 mM dimyristoylphosphatidyl choline [DMPC], 0.37 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride, and 3.7 μM GDP or GTPγS). Anti-Flag M2 agarose in the mixture was then washed three times with buffer C (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 5 mM MgCl2, 1 mM DTT, 3 mM DMPC, and 5 μM GDP or GTPγS), followed by incubation with Flag peptide (200 μg/ml; Sigma-Aldrich) for 30 min at 4ºC. The Flag-peptide eluted fraction was subjected to Western blot analysis using anti-Myc and anti-Flag antibodies.
Supplementary Material
Acknowledgments
We gratefully acknowledge R. Y. Tsien for the mCherry plasmids, X. Wang for qxIs58[Pced-1::lmp-1::Cherry], and present and former members of the Katada laboratory for useful discussions. Some strains were provided by the CGC, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). This work was supported in part by research grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan and the Japan Society for the Promotion of Science (in both cases to K.K., K.S., M.F., T.K., K.G.-A., and S.M.) and the Japan Science and Technology Agency (to K.G.-A. and S.M.)
Abbreviations used:
- AO
acridine orange
- DIC
differential interference contrast
- GFP
green fluorescent protein
- RNAi
RNA interference
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-08-0628) on March 13, 2013.
*Present address: Saitama University Brain Science Institute, 255 Shimo-Okubo, Sakura-ku, Saitama 338-8570, Japan.
REFERENCES
- Akbar MA, Tracy C, Kahr WH, Kramer H. The full-of-bacteria gene is required for phagosome maturation during immune defense in Drosophila. J Cell Biol. 2011;192:383–390. doi: 10.1083/jcb.201008119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagshaw RD, Callahan JW, Mahuran DJ. The Arf-family protein, Arl8b, is involved in the spatial distribution of lysosomes. Biochem Biophys Res Commun. 2006;344:1186–1191. doi: 10.1016/j.bbrc.2006.03.221. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins M, Huber LA, Parton RG, Griffiths G. Biogenesis of phagolysosomes proceeds through a sequential series of interactions with the endocytic apparatus. J Cell Biol. 1994;124:677–688. doi: 10.1083/jcb.124.5.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclos S, Diez R, Garin J, Papadopoulou B, Descoteaux A, Stenmark H, Desjardins M. Rab5 regulates the kiss and run fusion between phagosomes and endosomes and the acquisition of phagosome leishmanicidal properties in RAW 264.7 macrophages. J Cell Sci. 2000;113:3531–3541. doi: 10.1242/jcs.113.19.3531. [DOI] [PubMed] [Google Scholar]
- Elliott MR, Ravichandran KS. Clearance of apoptotic cells: implications in health and disease. J Cell Biol. 2010;189:1059–1070. doi: 10.1083/jcb.201004096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epp N, Rethmeier R, Kramer L, Ungermann C. Membrane dynamics and fusion at late endosomes and vacuoles—Rab regulation, multisubunit tethering complexes and SNAREs. Eur J Cell Biol. 2011;90:779–785. doi: 10.1016/j.ejcb.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Fadeel B. Programmed cell clearance. Cell Mol Life Sci. 2003;60:2575–2585. doi: 10.1007/s00018-003-3145-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadok VA, Bratton DL, Rose DM, Pearson A, Ezekewitz RA, Henson PM. A receptor for phosphatidylserine-specific clearance of apoptotic cells. Nature. 2000;405:85–90. doi: 10.1038/35011084. [DOI] [PubMed] [Google Scholar]
- Garg S, Sharma M, Ung C, Tuli A, Barral DC, Hava DL, Veerapen N, Besra GS, Hacohen N, Brenner MB. Lysosomal trafficking, antigen presentation, and microbial killing are controlled by the Arf-like GTPase Arl8b. Immunity. 2011;35:182–193. doi: 10.1016/j.immuni.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumienny TL, Lambie E, Hartwieg E, Horvitz HR, Hengartner MO. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development. 1999;126:1011–1022. doi: 10.1242/dev.126.5.1011. [DOI] [PubMed] [Google Scholar]
- Guo P, Hu T, Zhang J, Jiang S, Wang X. Sequential action of Caenorhabditis elegans Rab GTPases regulates phagolysosome formation during apoptotic cell degradation. Proc Natl Acad Sci USA. 2010;107:18016–18021. doi: 10.1073/pnas.1008946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry RM, Hoppe AD, Joshi N, Swanson JA. The uniformity of phagosome maturation in macrophages. J Cell Biol. 2004;164:185–194. doi: 10.1083/jcb.200307080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann I, Munro S. An N-terminally acetylated Arf-like GTPase is localised to lysosomes and affects their motility. J Cell Sci. 2006;119:1494–1503. doi: 10.1242/jcs.02958. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2:RESEARCH0002. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniuk NA, et al. Salmonella exploits Arl8B-directed kinesin activity to promote endosome tubulation and cell-to-cell transfer. Cell Microbiol. 2011;13:1812–1823. doi: 10.1111/j.1462-5822.2011.01663.x. [DOI] [PubMed] [Google Scholar]
- Kinchen JM, Cabello J, Klingele D, Wong K, Feichtinger R, Schnabel H, Schnabel R, Hengartner MO. Two pathways converge at CED-10 to mediate actin rearrangement and corpse removal in C. elegans. Nature. 2005;434:93–99. doi: 10.1038/nature03263. [DOI] [PubMed] [Google Scholar]
- Kinchen JM, Doukoumetzidis K, Almendinger J, Stergiou L, Tosello-Trampont A, Sifri CD, Hengartner MO, Ravichandran KS. A pathway for phagosome maturation during engulfment of apoptotic cells. Nat Cell Biol. 2008;10:556–566. doi: 10.1038/ncb1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinchen JM, Ravichandran KS. Phagosome maturation: going through the acid test. Nat Rev Mol Cell Biol. 2008;9:781–795. doi: 10.1038/nrm2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinchen JM, Ravichandran KS. Identification of two evolutionarily conserved genes regulating processing of engulfed apoptotic cells. Nature. 2010;464:778–782. doi: 10.1038/nature08853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen MP, et al. An Arf-like small G protein, ARL-8, promotes the axonal transport of presynaptic cargoes by suppressing vesicle aggregation. Neuron. 2010;66:710–723. doi: 10.1016/j.neuron.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettre G, Hengartner MO. Developmental apoptosis in C. elegans: a complex CEDnario. Nat Rev Mol Cell Biol. 2006;7:97–108. doi: 10.1038/nrm1836. [DOI] [PubMed] [Google Scholar]
- Liu QA, Hengartner MO. Candidate adaptor protein CED-6 promotes the engulfment of apoptotic cells in C. elegans. Cell. 1998;93:961–972. doi: 10.1016/s0092-8674(00)81202-7. [DOI] [PubMed] [Google Scholar]
- Lu Q, Zhang Y, Hu T, Guo P, Li W, Wang X. C. elegans Rab GTPase 2 is required for the degradation of apoptotic cells. Development. 2008;135:1069–1080. doi: 10.1242/dev.016063. [DOI] [PubMed] [Google Scholar]
- Mangahas PM, Yu X, Miller KG, Zhou Z. The small GTPase Rab2 functions in the removal of apoptotic cells in Caenorhabditis elegans. J Cell Biol. 2008;180:357–373. doi: 10.1083/jcb.200708130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitani S. Genetic regulation of mec-3 gene expression implicated in the specification of the mechanosensory neuron cell types in Caenorhabditis elegans. Develop Growth Differ. 1995;37:551–557. doi: 10.1046/j.1440-169X.1995.t01-4-00010.x. [DOI] [PubMed] [Google Scholar]
- Mrakovic A, Kay JG, Furuya W, Brumell JH, Botelho RJ. Rab7 and Arl8 GTPases are necessary for lysosome tubulation in macrophages. Traffic. 2012;13:1667–1679. doi: 10.1111/tra.12003. [DOI] [PubMed] [Google Scholar]
- Nakae I, Fujino T, Kobayashi T, Sasaki A, Kikko Y, Fukuyama M, Gengyo-Ando K, Mitani S, Kontani K, Katada T. The Arf-like GTPase Arl8 mediates delivery of endocytosed macromolecules to lysosomes in Caenorhabditis elegans. Mol Biol Cell. 2010;21:2434–2442. doi: 10.1091/mbc.E09-12-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson DP, Brett CL, Merz AJ. Vps-C complexes: gatekeepers of endolysosomal traffic. Curr Opin Cell Biol. 2009;21:543–551. doi: 10.1016/j.ceb.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikiori M, Mori M, Dohi K, Okamura H, Katoh E, Naito S, Meshi T, Ishikawa M. A host small GTP-binding protein ARL8 plays crucial roles in tobamovirus RNA replication. PLoS Pathog. 2011;7:e1002409. doi: 10.1371/journal.ppat.1002409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteryaev D, Fares H, Bowerman B, Spang A. Caenorhabditis elegans SAND-1 is essential for RAB-7 function in endosomal traffic. EMBO J. 2007;26:301–312. doi: 10.1038/sj.emboj.7601498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Horvitz HR. The engulfment process of programmed cell death in Caenorhabditis elegans. Annu Rev Cell Dev Biol. 2004;20:193–221. doi: 10.1146/annurev.cellbio.20.022003.114619. [DOI] [PubMed] [Google Scholar]
- Roberts EA, Chua J, Kyei GB, Deretic V. Higher order Rab programming in phagolysosome biogenesis. J Cell Biol. 2006;174:923–929. doi: 10.1083/jcb.200603026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa-Ferreira C, Munro S. Arl8 and SKIP act together to link lysosomes to kinesin-1. Dev Cell. 2011;21:1171–1178. doi: 10.1016/j.devcel.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto S, Tsujimura K, Koide Y. Rab GTPases regulating phagosome maturation are differentially recruited to mycobacterial phagosomes. Traffic. 2011;12:407–420. doi: 10.1111/j.1600-0854.2011.01165.x. [DOI] [PubMed] [Google Scholar]
- Vieira OV, Botelho RJ, Grinstein S. Phagosome maturation: aging gracefully. Biochem J. 2002;366:689–704. doi: 10.1042/BJ20020691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira OV, Bucci C, Harrison RE, Trimble WS, Lanzetti L, Gruenberg J, Schreiber AD, Stahl PD, Grinstein S. Modulation of Rab5 and Rab7 recruitment to phagosomes by phosphatidylinositol 3-kinase. Mol Cell Biol. 2003;23:2501–2514. doi: 10.1128/MCB.23.7.2501-2514.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, et al. Cell corpse engulfment mediated by C. elegans phosphatidylserine receptor through CED-5 and CED-12. Science. 2003;302:1563–1566. doi: 10.1126/science.1087641. [DOI] [PubMed] [Google Scholar]
- Wickner W. Membrane fusion: five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu Rev Cell Dev Biol. 2010;26:115–136. doi: 10.1146/annurev-cellbio-100109-104131. [DOI] [PubMed] [Google Scholar]
- Wu YC, Horvitz HR. The C. elegans cell corpse engulfment gene ced-7 encodes a protein similar to ABC transporters. Cell. 1998;93:951–960. doi: 10.1016/s0092-8674(00)81201-5. [DOI] [PubMed] [Google Scholar]
- Wu YC, Stanfield GM, Horvitz HR. NUC-1, a Caenorhabditis elegans DNase II homolog, functions in an intermediate step of DNA degradation during apoptosis. Genes Dev. 2000;14:536–548. [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Chen D, Fang Z, Xu J, Sun X, Song S, Liu J, Yang C. Lysosome biogenesis mediated by vps-18 affects apoptotic cell degradation in Caenorhabditis elegans. Mol Biol Cell. 2009;20:21–32. doi: 10.1091/mbc.E08-04-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Lu N, Zhou Z. Phagocytic receptor CED-1 initiates a signaling pathway for degrading engulfed apoptotic cells. PLoS Biol. 2008;6:e61. doi: 10.1371/journal.pbio.0060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Hartwieg E, Horvitz HR. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 2001;104:43–56. doi: 10.1016/s0092-8674(01)00190-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.