The partitioning membrane of dividing plant cells is made by homotypic fusion of trans-Golgi network–derived membrane vesicles delivered to the division plane. The cytokinesis-specific syntaxin of Arabidopsis forms two different types of SNARE complexes, which can functionally replace each other in membrane fusion during cytokinesis.
Abstract
Membrane fusion is mediated by soluble N-ethylmaleimide–sensitive factor attachment protein receptor (SNARE) complexes. Although membrane fusion is required for separating daughter cells in eukaryotic cytokinesis, the SNARE complexes involved are not known. In plants, membrane vesicles targeted to the cell division plane fuse with one another to form the partitioning membrane, progressing from the center to the periphery of the cell. In Arabidopsis, the cytokinesis-specific Qa-SNARE KNOLLE interacts with two other Q-SNAREs, SNAP33 and novel plant-specific SNARE 11 (NPSN11), whose roles in cytokinesis are not clear. Here we show by coimmunoprecipitation that KNOLLE forms two SNARE complexes that differ in composition. One complex is modeled on the trimeric plasma membrane type of SNARE complex and includes, in addition to KNOLLE, the promiscuous Qb,c-SNARE SNAP33 and the R-SNARE vesicle-associated membrane protein (VAMP) 721,722, also involved in innate immunity. In contrast, the other KNOLLE-containing complex is tetrameric and includes Qb-SNARE NPSN11, Qc-SNARE SYP71, and VAMP721,722. Elimination of only one or the other type of KNOLLE complex by mutation, including the double mutant npsn11 syp71, causes a mild or no cytokinesis defect. In contrast, the two double mutants snap33 npsn11 and snap33 syp71 eliminate both types of KNOLLE complexes and display knolle-like cytokinesis defects. Thus the two distinct types of KNOLLE complexes appear to jointly mediate membrane fusion in Arabidopsis cytokinesis.
INTRODUCTION
Intracellular membrane fusion is necessary for the maintenance of eukaryotic membrane compartments and mediates the delivery of membrane-associated proteins to their destinations along the secretory or endocytic pathways (Jahn and Scheller, 2006; Lipka et al., 2007; Südhof and Rothman, 2009). In heterotypic fusion, membrane vesicles originate from one membrane compartment and fuse with a different target membrane, whereas homotypic fusion involves membranes of similar composition, for example, fusion of vacuoles in yeast (Wickner, 2010). Plant cytokinesis is a special case of homotypic fusion, in that trans-Golgi network–derived membrane vesicles are targeted along the microtubules of the phragmoplast to the plane of cell division, where they fuse with one another to form a novel membrane compartment: the cell plate (Segui-Simarro et al., 2004). Reorganization of the phragmoplast microtubules into an expanding open cylinder by repeated microtubule depolymerization in the center and polymerization of additional microtubules at the outer face of the phragmoplast results in delivery of later-arriving membrane vesicles to the margin of the cell plate. As a consequence of this dynamics, the cell plate expands from the center to the periphery of the cell and fuses with the parental plasma membrane at a predefined cortical division site, followed by its maturation into a stretch of plasma membrane separating the daughter cells (Jürgens, 2005).
Membrane fusion is mediated by the interaction of soluble N-ethylmaleimide–sensitive factor attachment receptor (SNARE) proteins through their highly conserved SNARE domains (Jahn and Scheller, 2006). Based on a glutamine (Q) or arginine (R) residue in the so-called 0-layer of the SNARE domain, SNAREs can be classified as Q- or R-SNAREs. Q-SNAREs can be further divided into three subgroups: Qa-, Qb-, and Qc-SNAREs. In analogy to the first described Qa-SNARE, the syntaxin 1 of mammals, Qa-SNAREs are often referred to as syntaxins (Bennett et al., 1992). Qa-SNAREs have a distinct domain organization: the N-terminal half comprises three α-helices, which are separated by a linker from the SNARE domain, followed by a C-terminal hydrophobic anchor (“tail anchor”) that mediates insertion into the endoplasmic reticulum (ER) membrane via the GET pathway (Denic, 2012). The α-helices can fold back on to the SNARE domain, which represents the closed, fusion-incompetent conformation of the syntaxin, or be away from the SNARE domain, which represents the open, fusion-competent conformation of the syntaxin (Südhof and Rothman, 2009). Qb-SNAREs and Qc-SNAREs both carry a unique N-terminal helical domain, followed by its particular SNARE domain and a C-terminal hydrophobic anchor. The SNAP25-like SNARE proteins harbor two SNARE domains, Qb and Qc, separated by a flexible linker and thus are referred to as Qb,c-SNAREs. Unlike the well-studied animal R-SNAREs known as synaptobrevins, plant (and most other) R-SNAREs carry an N-terminal longin domain that might play targeting and regulatory roles, in addition to the SNARE domain and a C-terminal hydrophobic anchor (Rossi et al., 2004; Uemura et al., 2005; Lipka et al., 2007).
The fusion of vesicles with the plasma membrane is generally mediated by the formation of a trimeric SNARE complex, consisting of a Qa-SNARE/syntaxin, a SNAP25-like SNARE (Qb,c-SNARE), and an R-SNARE/vesicle-associated membrane protein (VAMP). The formation of tetrameric SNARE complexes, assembled by Qa-, Qb-, Qc-, and R-SNARE domains on four different proteins, accomplishes membrane fusion in the endomembrane system (Fukuda et al., 2000). A functional SNARE complex, normally consisting of 3Q- and 1R-SNARE domains, is only formed by the interaction of cognate SNARE partners. After a vesicle has been tethered to the target compartment, the vesicle-localized R-SNARE/VAMP interacts with a preformed target (t)-SNARE complex, consisting of Qa-, Qb,- and Qc-SNAREs (endosomal type) or Qa- and Qb,c-SNAREs (plasma membrane type) to form a trans-SNARE complex. The interaction of the four coiled-coil SNARE domains is believed to overcome the energy barrier for membrane fusion (Jahn and Scheller, 2006).
In Arabidopsis, cytokinesis requires a cytokinesis-specific syntaxin (Qa-SNARE) named KNOLLE (also known as SYP111), which is also plant specific and interacts with ubiquitously expressed Qb,c-SNARE SNAP33 and the Qb-SNARE novel plant-specific SNARE 11 (NPSN11; Lukowitz et al., 1996; Lauber et al., 1997; Heese et al., 2001; Zheng et al., 2002). Embryos lacking KNOLLE protein develop abnormally from early on, displaying strong cytokinesis defects such as unfused vesicles, cell-wall stubs, and enlarged multinucleate cells. These knolle-mutant embryos give rise to morphologically highly abnormal seedlings that do not develop further and eventually die (Lauber et al., 1997). Although snap33-knockout mutant seedlings show slight cytokinesis defects, they only die later for cytokinesis-unrelated reasons, which is consistent with the accumulation of SNAP33 at the plasma membrane in nondividing cells and at the cell plate during cytokinesis (Heese et al., 2001). The SNAP25 homologue, SNAP33, also interacts with plasma membrane–localized syntaxin PEN1 (also known as SYP121) and R-SNAREs VAMP721/722, forming a SNARE complex involved in fungal pathogen response (Collins et al., 2003; Kwon et al., 2008). NPSN11 is a Qb-SNARE that localizes to the cell plate and may have a cytokinesis-specific function, although npsn11-knockout plants show no cytokinesis defects and are viable (Zheng et al., 2002). Thus neither of the two known SNARE interactors of KNOLLE is essential for cytokinesis, suggesting some form of redundancy. SNAP33 is a member of a small protein family of which the other two, SNAP29 and SNAP30, are also able to interact with KNOLLE in a yeast two-hybrid assay (Heese et al., 2001). Similarly, NPSN11 has two close paralogues, NPSN12 and NPSN13, of unknown function (Lipka et al., 2007; Sanderfoot, 2007).
Only two complete SNARE complexes have been identified in Arabidopsis: the plasma membrane type (Qa/Qb,c/R) PEN1/SNAP33/VAMP721,722, involved in pathogen response, and the endosomal type (Qa/Qb/Qc/R) SYP22/VTI11/SYP51/VAMP727, involved in vacuolar trafficking (Ebine et al., 2008; Kwon et al., 2008). In being cytokinesis specific, KNOLLE is unique among the syntaxins, which raises the possibility that the cytokinetic SNARE complex may have an unusual composition, for example, comprising Q-SNAREs only. Alternatively, cytokinesis might involve more than one type of KNOLLE-containing SNARE complex. To address this problem, we identified SNARE interactors of the known cytokinetic SNAREs by coimmunoprecipitation analysis and characterized them functionally by subcellular localization and mutant analysis.
RESULTS
Lack of interaction between KNOLLE-interacting Q-SNAREs SNAP33 and NPSN11 suggests different types of KNOLLE-containing SNARE complexes in cytokinesis
To analyze the relationship between the three Q-SNAREs KNOLLE, SNAP33, and NPSN11 that have been localized to the cell plate in dividing cells (Lauber et al., 1997; Heese et al., 2001; Zheng et al., 2002; see also Supplemental Figure S1), we generated transgenic plants expressing functional Myc or XFP fusions of the three proteins, performed immunoprecipitation experiments with anti-Myc or anti-GFP beads, and probed each precipitate for the presence of the two other proteins (Figure 1). KNOLLE associated with the other two Q-SNARE proteins, although more strongly with SNAP33 than with NPSN11 (Figure 1A). In contrast, the other two Q-SNAREs only interacted with KNOLLE and not with one another (Figure 1, B and C). Because the reciprocal experiments yielded comparable results for the strength of protein interaction, KNOLLE seems to associate preferentially with SNAP33 and also with NPSN11. Thus the cytokinesis-specific syntaxin KNOLLE appears to form at least two distinct SNARE complexes.
FIGURE 1:
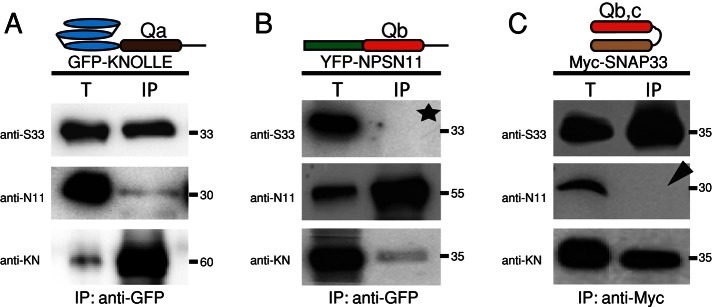
Interaction analysis of known Q-SNARE proteins involved in cytokinesis. GFP-KNOLLE (A), YFP-NPSN11 (B), and Myc-SNAP33 (C) were immunoprecipitated from total protein extracts of Arabidopsis inflorescences. (A) GFP-KNOLLE coprecipitated NPSN11 and SNAP33. (B) YFP-NPSN11 coprecipitated KNOLLE but not SNAP33 (asterisk). (C) Myc-SNAP33 coprecipitated KNOLLE but not NPSN11 (arrowhead). Protein was detected in total extract (T) and immunoprecipitate (IP) by Western blot using anti-SNAP33 (S33), anti-NPSN11 (N11), or anti-KNOLLE (KN) antiserum. Note that there is no interaction of endogenous NPSN11, KNOLLE, or SNAP33 with their YFP-, GFP-, or Myc-tagged fusion protein; the immunoblots show only GFP-KNOLLE, YFP-NPSN11, and Myc-SNAP33, as indicated by the molecular weight (in kilodaltons) on the right side.
Identification of R-SNAREs for the incomplete KNOLLE-containing SNARE complexes
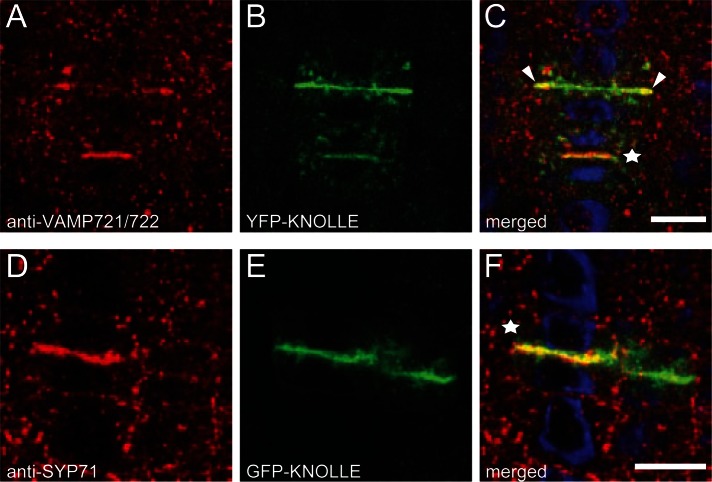
The two postulated KNOLLE-containing SNARE complexes were incomplete, both lacking the R-SNARE, whereas the NPSN11-containing complex also lacked a Qc-SNARE (see Table 1 for overview of SNAREs analyzed here). We attempted to identify KNOLLE-interacting R-SNAREs among candidate R-SNAREs of the VAMP721-727 family localized as green fluorescent protein (GFP) fusions to the plasma membrane by transient overexpression in Arabidopsis protoplasts, as has been KNOLLE itself (Uemura et al., 2004). Of these, VAMP721 and VAMP722 were particularly interesting candidates because they form complexes with Qb,c-SNARE SNAP33 and syntaxin (Qa-SNARE) PEN1 at the plasma membrane and the vamp721 vamp722 double mutant is lethal (Lipka et al., 2007; Kwon et al., 2008). We analyzed their subcellular localization by immunofluorescence with antiserum that detected the two closely related R-SNAREs VAMP721 and VAMP722, as revealed by the analysis of the individual knockout mutants (Supplemental Figure S2). Both proteins accumulated at the cell plate in dividing cells and did not give strong signals at the plasma membrane but instead displayed punctate patterns that coalesced into endosomal brefeldin A (BFA) compartments upon BFA treatment, which blocks post-Golgi trafficking, in interphase cells (Figure 2, A–C, and Supplemental Figure S2). Of interest, the VAMP721/722 signal colocalized almost completely with KNOLLE in early stages of cell-plate formation, whereas at later stages, the two signals mainly colocalized at the margin of the expanding cell plate, where the membrane vesicles are delivered along the microtubules of the phragmoplast (Figure 2, A–C, white arrowheads; Segui-Simarro et al., 2004; Jürgens, 2005). To examine whether VAMP721/722 might be interchangeable R-SNARE members of the two KNOLLE complexes, we performed reciprocal immunoprecipitation experiments with antisera precipitating KNOLLE, NPSN11, SNAP33, or VAMP721/722 (Figure 3). The results demonstrated interaction with all three Q-SNAREs with VAMP721/722, suggesting that the two distinct KNOLLE complexes share the same R-SNARE(s).
TABLE 1:
SNAREs involved in Arabidopsis cytokinesis.
| SNARE (AtGen #) | Category | Localization |
|---|---|---|
| KNOLLE (At1g08560) | Qa (syntaxin) | CP/TGN/MVB |
| SNAP33 (At5g61210) | Qb,c | CP/PM/E |
| NPSN11 (At2g35190) | Qb | CP/PM/E |
| SYP71 (At3g09740) | Qc | CP/PM/E/ER |
| VAMP721 (At1g04750) | R (VAMP) | CP/PM/E |
| VAMP722 (At2g33120) | R (VAMP) | CP/PM/E |
AtGen #, gene identifier (www.arabidopsis.org); CP, cell plate; E, endosome; MVB, multivesicular body; PM, plasma membrane.
FIGURE 2:
Cell-plate localization of R-SNAREs VAMP721 and VAMP722 and Qc-SNARE SYP71. (A–C) Immunofluorescence localization of VAMP721 and VAMP722 (red) at the cell plate (C, asterisk) and endosomal structures in root cells of YFP-KNOLLE (green) transgenic seedlings. Note that the VAMP721/722 signal is restricted to the periphery of the KN-labeled cell plate in late cytokinesis (arrowheads in C). (D–F) Immunofluorescence localization of SYP71 (red) in root cells of GFP-KNOLLE– (green) expressing seedlings. Note colocalization of SYP71 with GFP-KNOLLE at the cell plate (F, asterisk). DAPI-stained nuclei in merged images (C, F) are shown in blue. Scale bars, 5 μm.
FIGURE 3:
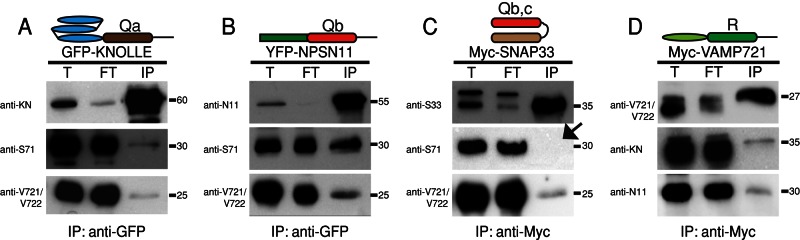
Interaction analysis of candidate SNARE proteins involved in cytokinesis. GFP-KNOLLE (A), YFP-NPSN11 (B), Myc-SNAP33 (C), and Myc-VAMP721 (D) were immunoprecipitated from total protein extracts of Arabidopsis inflorescences. (A) GFP-KNOLLE coprecipitated SYP71 and R-SNAREs VAMP721 and VAMP722. (B) YFP-NPSN11 coprecipitated SYP71 and R-SNAREs VAMP721 and VAMP722. (C) Myc-SNAP33 coprecipitated R-SNAREs VAMP721 and VAMP722 but not SYP71 (arrow). The upper band in (T) and (FT) represents a protein that cross-reacted with the SNAP33 antiserum and was not precipitated. (D) Myc-VAMP721 coprecipitated KNOLLE and NPSN11. Protein was detected in total extract (T), flowthrough (FT), and immunoprecipitate (IP) by Western blot using anti-SNAP33 (S33), anti-NPSN11 (N11), anti-KNOLLE (KN), anti-SYP71 (S71), or anti-VAMP721/722 (V721/722) antiserum. Note that there is no interaction of endogenous KNOLLE, NPSN11, SNAP33, or VAMP721/722 with their GFP-, YFP-, or Myc-tagged fusion protein; the immunoblots show only GFP-KNOLLE, YFP-NPSN11, Myc-SNAP33, and Myc-VAMP721/722, as indicated by the molecular weight (in kilodaltons) on the right side.
The single-knockout mutants of VAMP721 or VAMP722 have no obvious mutant phenotype, whereas the vamp721 vamp722 double mutant is lethal (Kwon et al., 2008). The double-mutant seedlings grew very slowly and eventually died, displaying cytokinesis defects such as incomplete cell walls or cell-wall stubs, as demonstrated for snap33-mutant seedlings (Heese et al., 2001; Supplemental Figure 3). The defective-cytokinesis phenotype of the vamp721 vamp722 double mutant was also described by Zhang et al. (2011) when this article was being revised. However, expression of the two R-SNAREs from the mitosis-specific KNOLLE promoter only partially rescued the double-mutant phenotype (Supplemental Figure S4; Lukowitz et al., 1996), supporting the idea that VAMP721 and VAMP722 are also involved in processes other than cytokinesis (Kwon et al., 2008). This result and the rather mild cytokinesis defects of the vamp721 vamp722 double mutant suggest that there might be some additional redundancy. Very recently, vamp721/VAMP721 vamp722/vamp722 seedling roots were shown to be more sensitive than wild type to growth-inhibiting ABA treatment, which suggests that VAMP721 and VAMP722 play a general role in secretory traffic to the plasma membrane (Yi et al., 2013) in addition to their role in cytokinesis.
Identification of a missing Qc-SNARE for the endomembrane type of KNOLLE-containing SNARE complex
The KNOLLE/SNAP33/VAMP721,722 complex appeared complete, conforming to the plasma membrane type of trimeric SNARE complex. However, the other KNOLLE complex still lacked a Qc-SNARE. A candidate was suggested from subcellular localization studies of the Qc-SNARE SYP71 fused to GFP, which accumulated at both ER and plasma membrane (Suwastika et al., 2008). A previous study involving transient overexpression of GFP-fusion proteins in Arabidopsis protoplasts did not reveal any Qc-SNARE localizing at the plasma membrane (Uemura et al., 2004). We confirmed the localization of GFP-SYP71 to ER and plasma membrane and also detected GFP-SYP71 at the cell plate (Supplemental Figure 5A). Using antiserum raised against SYP71, we observed accumulation of endogenous SYP71 at the cell plate, where it colocalized with KNOLLE (Figure 2, D–F). Whether SYP71 interacts with other cytokinetic SNARE proteins was tested by immunoprecipitation (Figure 3). SYP71 was coimmunoprecipitated with KNOLLE and NPSN11 but not SNAP33, indicating the existence of at least two very different KNOLLE complexes in cytokinesis. In addition to the trimeric plasma membrane type of SNARE complex containing the promiscuous Qb,c-SNARE SNAP33 and R-SNARE VAMP721 or VAMP722, there is a tetrameric endomembrane type of SNARE complex that comprises KNOLLE, Qb-SNARE NPSN11, Qc-SNARE SYP71, and R-SNARE VAMP721 or VAMP722.
Functional requirement and redundancy of KNOLLE-containing SNARE complexes in cytokinesis
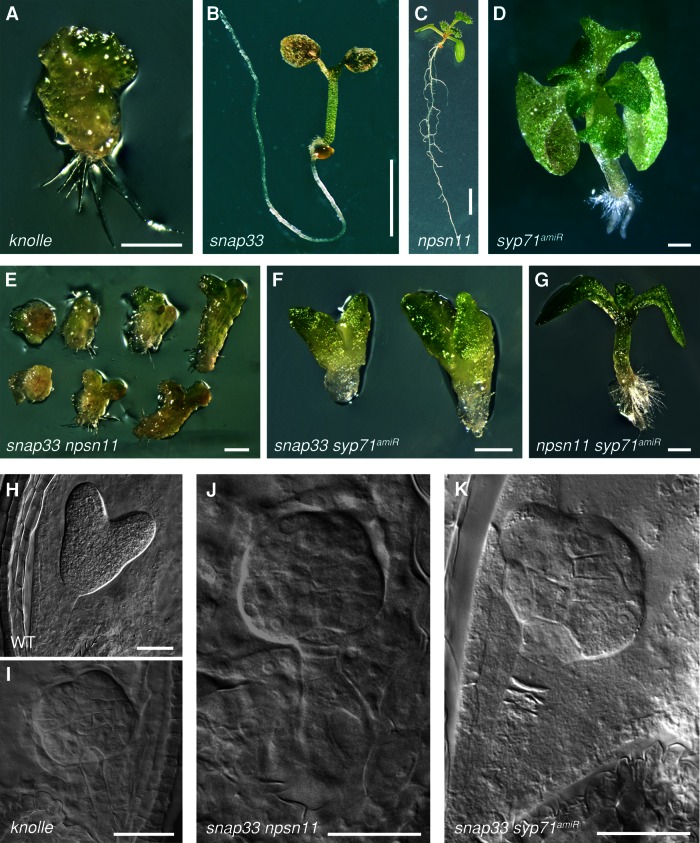
Of all the members of the two distinct cytokinetic SNARE complexes, only KNOLLE has been identified in a forward-genetic screen on the basis of its characteristic seedling phenotype and shown to display strong cytokinesis defects in embryogenesis, such as the accumulation of unfused membrane vesicles in the cell division plane (Mayer et al., 1991; Lukowitz et al., 1996; Lauber et al., 1997; Figure 4A). A knockout line for the KNOLLE-interacting Qb,c-SNARE SNAP33 shows mild cytokinesis defects only at the seedling stage, but the snap33 mutants die later because of cytokinesis-unrelated defects (Heese et al., 2001; Figure 4B). A knockout line for NPSN11 does not show obvious cytokinesis defects, and npsn11 homozygous plants are viable (Zheng et al., 2002; Figure 4C). A T-DNA insertion line (GABI-KAT 367A08) in which the SYP71 gene was disrupted carried two additional T-DNA insertions. Although this line segregated mutant embryos and seedlings displaying strong morphological abnormalities, the genetic basis of this phenotype was not clear (Supplemental Figure S5, B–D and F). Furthermore, the mutant phenotype did not resemble any known cytokinesis-defective phenotype. As an alternative approach, we generated artificial microRNA that specifically targeted SYP71 mRNA (amiRNA(SYP71)) but not the closely related paralogues SYP72 and SYP73 (Supplemental Figure S5, C, E, and F). Using the GAL4>>UAS two-component expression system in which the strong embryonic RPS5A promoter drives expression, we detected a slightly milder version of the abnormal-seedling phenotype of the genetically complex knockout line (Figure 4D). This correlated well with low-level accumulation of SYP71 mRNA, indicating that gene expression was strongly reduced but not abolished (Supplemental Figure S5, B, E, and F). We used this syp71-knockdown allele, named syp71amiR in the following, for further functional analyses.
FIGURE 4:
Q-SNARE double mutants display cytokinesis defects. (A–D) Seedling phenotypes of single mutants: (A) knolle knockout, (B) snap33 knockout, (C) npsn11 knockout, and (D) syp71 knockdown (syp71amiR). (E–G) Seedling phenotypes of double mutants: (E) snap33 npsn11– and (F) snap33 syp71amiR–mutant seedlings resemble knolle-mutant seedlings (compare with A); (G) npsn11 syp71amiR–mutant seedlings resemble syp71amiR-mutant seedlings (compare with D). (H–K) Embryo phenotypes of double mutants (chloral hydrate–cleared preparations of heart-stage embryos): (H) wild-type control, (I) knolle mutant, (J) snap33 npsn11, and (K) snap33 syp71amiR double mutants. Scale bars, 0.5 mm (A, D–G), 0.5 cm (B, C), 50 μm (H–K).
SNAP33, NPSN11, and SYP71 are members of small protein families, which might explain why their absence does not result in a strong or any cytokinesis-defective phenotype, respectively (Sanderfoot, 2007). Alternatively, if the two distinct KNOLLE-containing SNARE complexes were functionally related, mediating membrane fusion in cytokinesis, one might expect a strong cytokinesis defect in a double mutant that eliminates at least one component from each complex. Thus the phenotype of snap33 npsn11 and snap33 syp71 double mutants might resemble the knolle-mutant phenotype, whereas the npsn11 syp71 double mutant might not display a cytokinesis-defective phenotype. Because npsn11 is homozygous viable, we analyzed the progeny of snap33/SNAP33 npsn11/npsn11 plants. The snap33 npsn11 double-mutant seedlings resembled knolle seedlings morphologically (Figure 4, E, compare with A). Similarly, snap33 npsn11 double-mutant embryos resembled knolle embryos morphologically (Figure 4, J, compare with I). Furthermore, nearly 25% of the embryo progeny (60 of 251 analyzed) displayed very specific, severe cytokinesis defects at the ultrastructural level, such as incomplete cell walls or cell-wall stubs and cells with supernumerary nuclei (Figure 5, A–D). This figure is close to the expected proportion of 25% doubly homozygous mutant embryos. In addition, bands of unfused vesicles were detected in the former cell division plane between two nuclei, closely resembling the knolle subcellular phenotype (Figure 5, E and F). Of interest, this snap33 npsn11 double-mutant phenotype was completely rescued by expression of yellow fluorescent protein (YFP)–NPSN11 from the cytokinesis-specific KNOLLE promoter. Specifically, 18% of the seedlings (119 of 653 analyzed) that were produced by self-pollination of snap33/SNAP33 npsn11/npsn11 KN::YFP-NPSN11/- plants clearly displayed the snap33 single-mutant phenotype and were genotyped by PCR as snap33 npsn11 double mutants containing the KN::YFP-NPSN11 transgene, demonstrating that the expression of NPSN11 during cytokinesis is sufficient to rescue the cell division phenotype. Thus eliminating both NPSN11 and SNAP33 corresponds to the absence of KNOLLE, which strongly suggests that these two distinct KNOLLE complexes of different composition jointly promote membrane fusion in plant cytokinesis.
FIGURE 5:
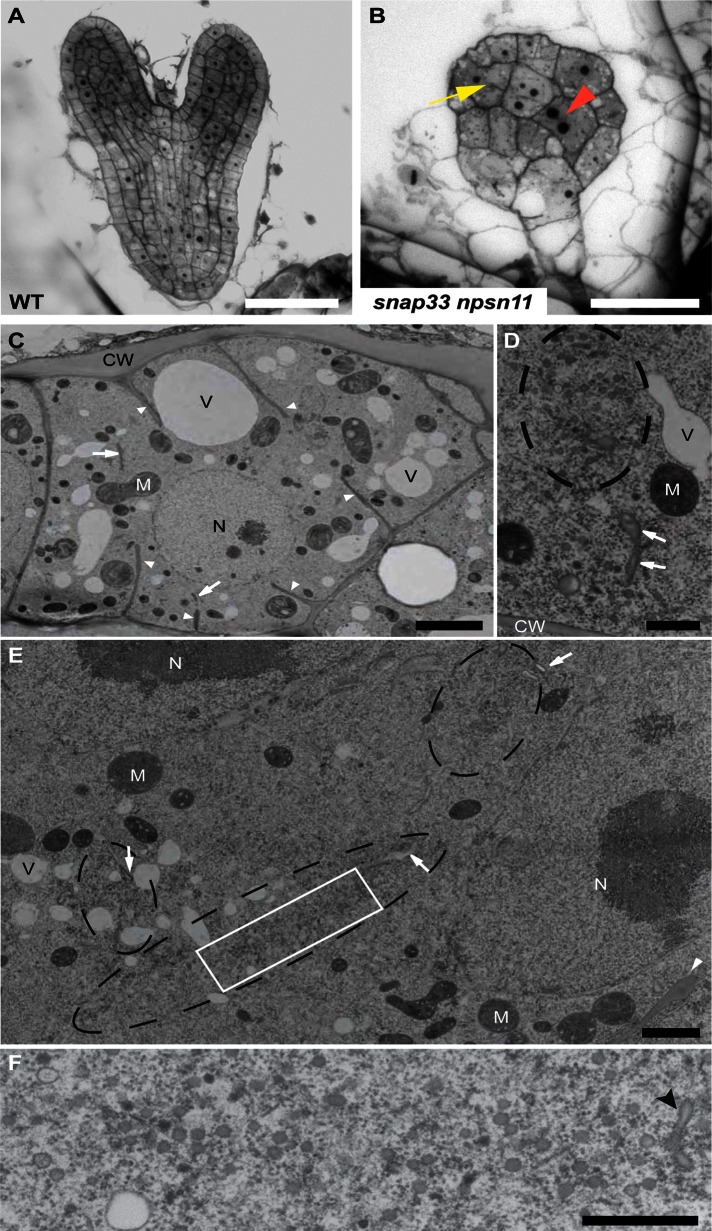
Cell-plate formation is severely affected in snap33 npsn11–mutant embryos. (A, B) Semithin sections of Spurr-embedded ovules from wild-type (A) and snap33 +/− npsn11 −/− (B) plants stained with toluidine blue. Cells of snap33 npsn11 (B) mutant embryos are enlarged, multinucleate (red arrowhead), and impaired in cell-plate formation, as indicated by the appearance of cell-wall stubs (yellow arrow). This was never seen in wild-type embryos (A). (C–F) Transmission electron microscopy images of ultrathin sections of snap33 npsn11–mutant embryos after high-pressure freezing, freeze substitution, and embedding in epoxy resin. (C) Overview of an interphase cell. Note internal cell-wall fragments (arrows) and cell-wall stubs (arrowheads). (D) Detailed view of a cluster of unfused vesicles (dashed circle) next to the parental cell wall (CW). Cell plate fragments (arrows) are indicated. (E) Vesicle bands (dashed circles) aligned in the plane of cell division are visible between two nuclei (N). Arrows indicate cell-plate fragments originated by the fusion of some vesicles. A cell-wall stub (arrowhead) is visible in the lower right corner. (F) Magnification of boxed area in E. A number of unfused electron-dense vesicles and a tubulovesicular structure (arrowhead) aggregated in the plane of cell division. CW, cell wall; M, mitochondrion; N, nucleus; V, vacuole. Scale bars, 50 μm (A, B), 3 μm (C), 0.5 μm (D, F), 1.5 μm (E).
As an independent test for this conclusion, we also generated double mutants of syp71amiR with snap33 and with npsn11 and compared the phenotypes of these two with the phenotype of the snap33 npsn11 double mutant (Figure 4, F and G, compare with E). Self-pollination of snap33/SNAP33 npsn11/NPSN11 doubly heterozygous plants gave 6% knolle-like seedling progeny (199 of 3467; Supplemental Table S1). Similarly, snap33 syp71amiR seedlings displayed knolle-like defects (6% of 498 seedlings analyzed), although this double-mutant phenotype was slightly weaker than the npsn11 snap33 phenotype, which might reflect the incomplete knockdown of SYP71 by amiRNA expression (Figure 4F and Supplemental Table S1). Furthermore, snap33 syp71amiR embryos resembled both snap33 npsn11 and knolle embryos (Figure 4, K, compare with I and J). In contrast, the double mutant npsn11 syp71amiR did not display any additional phenotype other than the one described earlier for syp71amiR among 506 seedlings analyzed (Figure 4, G, compare with D; and Supplemental Table S1). Thus these data support the conclusion that the two different types of KNOLLE-containing SNARE complexes are functionally redundant, mediating cytokinesis in Arabidopsis.
DISCUSSION
The mode of cytokinesis is very different between plants and nonplant eukaryotes (Barr and Gruneberg, 2007). Nonetheless, the abscission of the midbody during the late stage of animal cytokinesis superficially resembles the formation of the plant cell plate, in that both processes involve targeting of membrane vesicles to the plane of cell division, where they fuse with one another and with the parental plasma membrane (Albertson et al., 2005; Jürgens, 2005). SNAREs that act in abscission of the midbody in animal cytokinesis are regular plasma membrane SNAREs, such as syntaxin 2 and endobrevin/VAMP-8 (Low et al., 2003). Although the missing Q-SNARE partner(s) have not been reported, a SNARE complex of syntaxin 2, SNAP-23, and VAMP-8 appears to be involved in zymogen granule exocytosis in pancreatic cells (Pickett et al., 2007). Thus a plasma membrane type of SNARE complex might have been recruited to mediate membrane fusion during the final stage of cytokinesis in animals.
Here we identified the first complete SNARE complex(es) acting in eukaryotic cytokinesis. Surprisingly, and presumably in contrast to animals, plant cytokinesis involves two SNARE complexes of different composition. The cytokinesis-specific syntaxin KNOLLE forms not only a trimeric, plasma membrane type of complex (Qa/Qb,c/R) with promiscuous SNARE partners but also a tetrameric, endomembrane type of SNARE complex (Qa/Qb/Qc/R) that contains separate Qb-SNARE NPSN11 and Qc-SNARE SYP71. Of interest, the trimeric SNARE complex involved in cytokinesis differs from the previously described SNARE complex involved in innate immunity at the plasma membrane only by its syntaxin or Qa-SNARE—KNOLLE (also known as SYP111) versus PEN1 (also known as SYP121)—whereas Qb,c-SNARE SNAP33 and R-SNAREs VAMP721,722 are promiscuous (Kwon et al., 2008). This would suggest that the syntaxin alone confers specificity of SNARE action in membrane fusion. Indeed, when expressed from the KNOLLE promoter, PEN1 accumulated strongly at the cell plate but was unable to rescue the knolle mutant, and this difference in activity between the two syntaxins was attributed to the SNARE domain (Reichardt et al., 2011). How the SNARE domain could account for the functional difference between KNOLLE and PEN1 is not obvious, especially because the two SNARE domains can interact with the same SNARE partners. However, KNOLLE function in cytokinesis also depends on its interaction with the SM protein KEULE, which plays an essential role in cytokinesis (Park et al., 2012).
Promiscuous interactions between plant syntaxins and other SNAREs were previously reported. The vacuolar syntaxin SYP22 interacts with the two R-SNAREs VAMP727 and VAMP713 in different tetrameric complexes that also include Q-SNAREs VTI11 and SYP51 (Ebine et al., 2008, 2011). Similarly, SYP121/PEN1 interacts with both VAMP721/722 and VAMP727 but not with VAMP713 (Kwon et al., 2008; Ebine et al., 2011). Whereas the composition of the plasma membrane–localized trimeric SNARE complex has been identified as PEN1-SNAP33-VAMP721/722, it is not known whether the PEN1-VAMP727 complex also includes SNAP33. Although these studies revealed promiscuous interactions among plant SNAREs, KNOLLE appears to be unique in that this cytokinesis-specific syntaxin can form both tetrameric and trimeric SNARE complexes.
The biological significance of two different SNARE complexes mediating the same process—membrane fusion in cytokinesis—is not clear. The probably low antigen density on small vesicles (40–60 nm in diameter) does not afford distinction between adjacent vesicles carrying different SNARE proteins. However, there is no functional distinction between the two SNARE complexes in cytokinesis, and both might well reside on the same vesicle destined to fuse with other vesicles during cell plate formation. There is no evidence for one or the other KNOLLE complex mediating delivery of unique cargo to the forming cell plate. The relative contributions of the two KNOLLE complexes to cytokinesis are difficult to assess, although KNOLLE appears to interact more often or more strongly with SNAP33 than with NPSN11 and SYP71. However, it is not known whether the signal intensity of coimmunoprecipitated material might reflect the actual amount of interacting protein. Finally, it should be noted that the coimmunoprecipitation experiments were done with extracts from inflorescences, which might not be representative for all developmental stages, although there is no major difference between the two complexes in regard to the expression profile of their components. Because KNOLLE has only been reported from flowering plants, it will be interesting to examine whether the existence of the different KNOLLE-containing SNARE complexes is a conserved feature of flowering plant species (Sanderfoot, 2007).
Materials and Methods
Plant material and generation of transgenic lines
Arabidopsis thaliana genotypes used were wild type (Col-0), knolleX37-2 (Ler/Nd; Lukowitz et al., 1996), and snap33 (Ws; Heese et al., 2001). In addition, T-DNA insertion lines from the Arabidopsis Biological Resource Center, the Nottingham Arabidopsis Stock Center, and GABI-Kat were analyzed by PCR and DNA sequencing to identify homozygous vamp721 (At1g04750; SALK_037273, GABI_052G12), vamp722 (At2g33120; SALK_119149), npsn11 (At2g35190; SALK_068094), and syp71 (At3g09740; GABI_367A08) mutants.
The following rescue genotypes were generated by crossing or transformation: snap33 npsn11 plants expressing YFP-NPSN11 and vamp721 vamp722 plants expressing Myc-VAMP721 or YFP-VAMP722. Transgenic plants were generated by Agrobacterium tumefaciens–mediated transformation (Clough and Bent, 1998).
Plants were grown either on soil or on vertically oriented agar plates with 2.15 g/l Murashige and Skoog (1/2 MS) medium containing 1% sucrose in growth chambers at 20–24°C with a 16-h photoperiod or in continuous light.
Molecular cloning and transcriptional analysis
Cloning of artificial microRNA (amiRNA) for SYP71 was done as described (Schwab et al., 2006). For the two-component expression system the amiRNA was cloned under the GAL4-responsive UAS element, and these reporter lines were crossed with the RPS5A::GAL4 activator lines (Weijers et al., 2003). For subcellular localization studies, the DNA sequence encoding Myc epitope, GFP, or YFP was fused in-frame to the ATG codon of the respective coding sequence and downstream of the KNOLLE 5′ regulatory sequences (Müller et al., 2003).
For transcriptional analysis, total RNA was extracted from seedlings or one to two rosette leaves by the TRIZOL RNA extraction method (Chomczynski, 1993). After DNA digestion (DNase I; Fermentas, Glen Burnie, MD), first-strand cDNA was synthesized with RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas) using an oligo dT-primer. The resulting cDNA was used for PCR.
Primers used for cloning, genotyping, and reverse transcription (RT)-PCR are listed in Supplemental Table S2.
Immunoprecipitation
The immunoprecipitation procedure was modified from Karlova et al. (2006). Total protein extracts were prepared from 1–3 g of inflorescences. We added 30 μl of agarose-conjugated lama anti-GFP (GFP-trap; ChromoTek, Martinsried, Germany) or mouse anti-Myc (clone 4A6; Millipore, Billerica, MA) antibodies to 2 ml of protein extract and incubated them at 4°C for 1–3 h on a rolling incubator. All immunoprecipitation experiments were repeated at least twice. Immunoblot analysis was done as described previously (Lauber et al., 1997). Antibody dilutions were as follows: rabbit anti-KNOLLE (1:5000; Lauber et al., 1997), rabbit anti-SNAP33 (1:5000; Heese et al., 2001), rabbit anti-NPSN11 (1:1500; Zheng et al., 2002), rabbit anti-SYP71 (1:1000; Sanderfoot et al., 2001), rabbit anti-VAMP721/722 (1:1500; (Kwon et al., 2008), and sheep anti-rabbit immunoglobulin G–POD polyclonal antibody (1:1000; Millipore).
Immunofluorescence microscopy and live imaging
Live imaging, whole-mount immunofluorescence (IF), and 4′,6-diamidino-2-phenylindole (DAPI) staining of seedlings were performed as described (Lauber et al., 1997; Völker et al., 2001). Confocal laser scanning microscopy was done with a Leica TCS-SP2 microscope (Leica, Wetzlar, Germany). Images were obtained with a 63× water-immersion objective and Leica software and processed using Photoshop CS3 (Adobe, San Jose, CA) and Illustrator CS3 (Adobe) software (Lauber et al., 1997; Völker et al., 2001). Antibodies and their dilutions used for IF were rabbit anti-KNOLLE (1:2000; Lauber et al., 1997), mouse anti-Myc 9E10 (1:600; Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-NPSN11 (1:600; Zheng et al., 2002), rabbit anti-SYP71 (1:200; Sanderfoot et al., 2001), rabbit anti-VAMP721 (1:1000; Kwon et al., 2008), fluorescein isothiocyanate–conjugated goat anti-rabbit (1:600; Dianova, Hamburg, Germany), and Cy3-conjugated goat anti-mouse (1:600; Dianova).
Electron microscopy
Ultrastructural analysis of embryos was carried out after high-pressure freezing, freeze substitution (2.5% osmium tetroxide in acetone), and embedding in epoxy resin (Epon; Roth, Karlsruhe, Germany; Reichardt et al., 2007; Stierhof and El Kasmi, 2010).
Phenotypic analysis
Seedlings and whole-mount chloral hydrate preparations of embryos were analyzed using a Leica MZ125 or Leica MZFLIII binocular or a Zeiss Axiophot microscope (Zeiss, Jena, Germany). Images were taken with a COOLPIX 990 digital camera (Nikon, Melville, NY), a Leica DC200 camera, or an AxioCam, using AxioVision 4.8.1 software. Images were processed using Photoshop CS3 and CS5. Sectioning was done as described (Heese et al., 2001).
Supplementary Material
Acknowledgments
We thank Barbara Maier, Carola Nill, and Franziska Birkhold for technical assistance, Paul Schulze-Lefert (Max Planck Institute, Cologne, Germany) for kindly providing the anti-VAMP721,722 antiserum, Tomohiro Uemura (University of Tokyo, Tokyo, Japan) for kindly providing the SYP71::GFP-SYP71 plant line, the Arabidopsis Biological Resource Center and Nottingham Arabidopsis Stock Center for plant material, and Rita Gross-Hardt, Martin Bayer, and members of our lab for critical reading of the manuscript. This work was supported by Grant Ju 179/12-1 from the Deutsche Forschungsgemeinschaft.
Abbreviations used:
- ABA
abscisic acid
- amiRNA
artificial microRNA
- BFA
brefeldin A
- CP
cell plate
- CW
cell wall
- DAPI
4′,6-diamidino-2-phenylindole
- E
endosome
- ER
endoplasmic reticulum
- FT
flowthrough
- GFP
green fluorescent protein
- IF
immunofluorescence
- IP
immunoprecipitate
- KN
KNOLLE
- M
mitochondrion
- MVB
multivesicular body
- N
nucleus
- N11
NPSN11
- NPSN
novel plant SNARE
- PM
plasma membrane
- RT
reverse transcription
- S33
SNAP33
- S71
SYP71
- SNAP
synaptosomal-associated protein
- SNARE
soluble N-ethylmaleimide–sensitive factor attachment protein receptor
- SYP
syntaxin of plants
- T
total extract
- V
vacuole
- V721/722
VAMP721/722
- VAMP
vesicle-associated membrane protein
- XFP
X fluorescent protein
- YFP
yellow fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-02-0074) on March 20, 2013.
Present addresses: *Department of Biology, University of North Carolina, Chapel Hill, NC 27599;
‡Institute of Molecular Biotechnology, 1030 Vienna, Austria.
†These authors contributed equally.
REFERENCES
- Albertson R, Riggs B, Sullivan W. Membrane traffic: a driving force in cytokinesis. Trends Cell Biol. 2005;15:92–101. doi: 10.1016/j.tcb.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Barr FA, Gruneberg U. Cytokinesis: placing and making the final cut. Cell. 2007;131:847–860. doi: 10.1016/j.cell.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Bennett MK, Calakos N, Scheller RH. Syntaxin: a synaptic protein implicated in docking of synaptic vesicles at presynaptic active zones. Science. 1992;257:255–259. doi: 10.1126/science.1321498. [DOI] [PubMed] [Google Scholar]
- Chomczynski P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques. 1993;15:532–534. 536–537. [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998);16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Collins NC, et al. SNARE-protein-mediated disease resistance at the plant cell wall. Nature. 2003;425:973–977. doi: 10.1038/nature02076. [DOI] [PubMed] [Google Scholar]
- Denic V. A portrait of the GET pathway as a surprisingly complicated young man. Trends Biochem Sci. 2012;37:411–417. doi: 10.1016/j.tibs.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebine K, et al. A SNARE complex unique to seed plants is required for protein storage vacuole biogenesis and seed development of Arabidopsis thaliana. Plant Cell. 2008;20:3006–3021. doi: 10.1105/tpc.107.057711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebine K, et al. A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat Cell Biol. 2011;13:853–859. doi: 10.1038/ncb2270. [DOI] [PubMed] [Google Scholar]
- Fukuda R, McNew JA, Weber T, Parlati F, Engel T, Nickel W, Rothman JE, Söllner TH. Functional architecture of an intracellular membrane t-SNARE. Nature. 2000;407:198–201. doi: 10.1038/35025084. [DOI] [PubMed] [Google Scholar]
- Heese M, Gansel X, Sticher L, Wick P, Grebe M, Granier F, Jürgens G. Functional characterization of the KNOLLE-interacting t-SNARE AtSNAP33 and its role in plant cytokinesis. J Cell Biol. 2001;155:239–249. doi: 10.1083/jcb.200107126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R, Scheller RH. SNAREs—engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- Jürgens G. Plant cytokinesis: fission by fusion. Trends Cell Biol. 2005;15:277–283. doi: 10.1016/j.tcb.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Karlova R, Boeren S, Russinova E, Aker J, Vervoort J, de Vries S. The Arabidopsis somatic embryogenesis receptor-like kinase 1 protein complex includes brassinosteroid-insensitive 1. Plant Cell. 2006;18:626–638. doi: 10.1105/tpc.105.039412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C, et al. Co-option of a default secretory pathway for plant immune responses. Nature. 2008;451:835–840. doi: 10.1038/nature06545. [DOI] [PubMed] [Google Scholar]
- Lauber MH, Waizenegger I, Steinmann T, Schwarz H, Mayer U, Hwang I, Lukowitz W, Jürgens G. The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J Cell Biol. 1997;139:1485–1493. doi: 10.1083/jcb.139.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka V, Kwon C, Panstruga R. SNARE-ware: the role of SNARE-domain proteins in plant biology. Annu Rev Cell Dev Biol. 2007;23:147–174. doi: 10.1146/annurev.cellbio.23.090506.123529. [DOI] [PubMed] [Google Scholar]
- Low SH, Li X, Miura M, Kudo N, Quinones B, Weimbs T. Syntaxin 2 and endobrevin are required for the terminal step of cytokinesis in mammalian cells. Dev Cell. 2003;4:753–759. doi: 10.1016/s1534-5807(03)00122-9. [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Mayer U, Jürgens G. Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell. 1996;84:61–71. doi: 10.1016/s0092-8674(00)80993-9. [DOI] [PubMed] [Google Scholar]
- Mayer U, Torres Ruiz RA, Berleth T, Miséra S, Jürgens G. Mutations affecting body organization in the Arabidopsis embryo. Nature. 1991;353:402–407. [Google Scholar]
- Müller I, Wagner W, Völker A, Schellmann S, Nacry P, Küttner F, Schwarz-Sommer Z, Mayer U, Jürgens G. Syntaxin specificity of cytokinesis in Arabidopsis. Nat Cell Biol. 2003;5:531–534. doi: 10.1038/ncb991. [DOI] [PubMed] [Google Scholar]
- Park M, Touihri S, Müller I, Mayer U, Jürgens G. Sec1/Munc18 protein stabilizes fusion-competent syntaxin for membrane fusion in Arabidopsis cytokinesis. Dev Cell. 2012;22:989–1000. doi: 10.1016/j.devcel.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Pickett JA, Campos-Toimil M, Thomas P, Edwardson JM. Identification of SNAREs that mediate zymogen granule exocytosis. Biochem Biophys Res Commun. 2007;359:599–603. doi: 10.1016/j.bbrc.2007.05.128. [DOI] [PubMed] [Google Scholar]
- Reichardt I, Stierhof Y-D, Mayer U, Richter S, Schwarz H, Schumacher K, Jürgens G. Plant cytokinesis requires de novo secretory trafficking but not endocytosis. Curr Biol. 2007;17:2047–2053. doi: 10.1016/j.cub.2007.10.040. [DOI] [PubMed] [Google Scholar]
- Reichardt I, Slane D, El Kasmi F, Knöll C, Fuchs R, Mayer U, Lipka V, Jürgens G. Mechanisms of functional specificity among plasma-membrane syntaxins in Arabidopsis. Traffic. 2011;12:1269–1280. doi: 10.1111/j.1600-0854.2011.01222.x. [DOI] [PubMed] [Google Scholar]
- Rossi V, Banfield DK, Vacca M, Dietrich LE, Ungermann C, D'Esposito M, Galli T, Filippini F. Longins and their longin domains: regulated SNAREs and multifunctional SNARE regulators. Trends Biochem Sci. 2004;12:682–688. doi: 10.1016/j.tibs.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Sanderfoot A. Increases in the number of SNARE genes parallels the rise of multicellularity among the green plants. Plant Physiol. 2007;144:6–17. doi: 10.1104/pp.106.092973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot AA, Kovaleva V, Bassham DC, Raikhel NV. Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell. Mol Biol Cell. 2001;12:3733–3743. doi: 10.1091/mbc.12.12.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segui-Simarro JM, Austin JR 2nd, White EA, Staehelin LA. Electron tomographic analysis of somatic cell plate formation in meristematic cells of Arabidopsis preserved by high-pressure freezing. Plant Cell. 2004;16:836–856. doi: 10.1105/tpc.017749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stierhof Y-D, El Kasmi F. Strategies to improve the antigenicity, ultrastructure preservation and visibility of trafficking compartments in Arabidopsis tissue. Eur J Cell Biol. 2010;89:285–297. doi: 10.1016/j.ejcb.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Südhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwastika IN, Uemura T, Shiina T, Sato MH, Takeyasu K. SYP71, a plant-specific Qc-SNARE protein, reveals dual localization to the plasma membrane and the endoplasmic reticulum in Arabidopsis. Cell Struct Funct. 2008;33:185–192. doi: 10.1247/csf.08024. [DOI] [PubMed] [Google Scholar]
- Uemura T, Sato MH, Takeyasu K. The longin domain regulates subcellular targeting of VAMP7 in Arabidopsis thaliana. FEBS Lett. 2005;13:2842–2846. doi: 10.1016/j.febslet.2005.04.022. [DOI] [PubMed] [Google Scholar]
- Uemura T, Ueda T, Ohniwa RL, Nakano A, Takeyasu K, Sato MH. Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct Funct. 2004;29:49–65. doi: 10.1247/csf.29.49. [DOI] [PubMed] [Google Scholar]
- Völker A, Stierhof Y-D, Jürgens G. Cell cycle-independent expression of the Arabidopsis cytokinesis-specific syntaxin KNOLLE results in mistargeting to the plasma membrane and is not sufficient for cytokinesis. J Cell Sci. 2001;114:3001–3012. doi: 10.1242/jcs.114.16.3001. [DOI] [PubMed] [Google Scholar]
- Weijers D, Van Hamburg JP, Van Rijn E, Hooykaas PJ, Offringa R. Diphtheria toxin-mediated cell ablation reveals interregional communication during Arabidopsis seed development. Plant Physiol. 2003;133:1882–1892. doi: 10.1104/pp.103.030692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W. Membrane fusion: five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu Rev Cell Dev Biol. 2010;26:115–136. doi: 10.1146/annurev-cellbio-100109-104131. [DOI] [PubMed] [Google Scholar]
- Yi C, Park S, Yun HS, Kwon C. Vesicle-associated membrane proteins 721 and 722 are required for unimpeded growth of Arabidopsis under ABA application. J Plant Physiol. 2013;170:529–533. doi: 10.1016/j.jplph.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhang H, Liu P, Hao H, Jin JB, Lin J. Arabidopsis R-SNARE proteins VAMP721 and VAMP722 are required for cell plate formation. PLoS One. 2011;6:e26129. doi: 10.1371/journal.pone.0026129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Bednarek SY, Sanderfoot AA, Alonso J, Ecker JR, Raikhel NV. NPSN11 is a cell plate-associated SNARE protein that interacts with the syntaxin KNOLLE. Plant Physiol. 2002;129:530–539. doi: 10.1104/pp.003970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.