Abstract
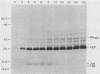
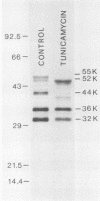
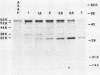
Processing and secretion of the alkaline extracellular protease (AEP) from the yeast Yarrowia lipolytica was studied by pulse-chase and immunoprecipitation experiments. Over half of newly synthesized AEP was secreted by 6 min. Over 99% of AEP activity which was external to the cytoplasmic membrane was located in the supernatant medium. Polypeptides of 55, 52, 44, 36, and 32 kilodaltons (55K, 52K, 44K, 36K, and 32K polypeptides) were immunoprecipitated from [3H]leucine-labeled cell extracts by rabbit antibodies raised against mature, secreted AEP (32K polypeptide). Experiments with tunicamycin and endoglycosidase H indicated that the 55K, 52K, and 44K polypeptides contained about 2 kilodaltons of N-linked oligosaccharide and that the 36K and 32K polypeptides contained none. Results of pulse-chase experiments did not fit a simple precursor-product relationship of 55K----52K----44K----36K----32K. In fact, maximum labeling intensity of the 52K polypeptide occurred later than for the 44K and 36K polypeptides. Secretion of polypeptides of 19 and 20 kilodaltons derived from the proregion of AEP indicated that one major processing pathway was 55K----52K----32K. The gene coding for AEP (XPR2) was cloned and sequenced. The sequence and the immunoprecipitation results suggest that AEP is originally synthesized with an additional preproI-proII-proIII amino-terminal region. Processing definitely involves cleavage(s) after pairs of basic amino acids and the addition of one N-linked oligosaccharide. Signal peptidase cleavage, dipeptidyl aminopeptidase cleavages, and at least one additional proteolytic cleavage may also be involved.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ammerer G., Hunter C. P., Rothman J. H., Saari G. C., Valls L. A., Stevens T. H. PEP4 gene of Saccharomyces cerevisiae encodes proteinase A, a vacuolar enzyme required for processing of vacuolar precursors. Mol Cell Biol. 1986 Jul;6(7):2490–2499. doi: 10.1128/mcb.6.7.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbers G. A., Hoekman W. A., Bloemendal H., de Jong W. W., Kleinschmidt T., Braunitzer G. Proline- and alanine-rich N-terminal extension of the basic bovine beta-crystallin B1 chains. FEBS Lett. 1983 Sep 19;161(2):225–229. doi: 10.1016/0014-5793(83)81013-8. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner W. M. Use of fluorography for sensitive isotope detection in polyacrylamide gel electrophoresis and related techniques. Methods Enzymol. 1983;96:215–222. doi: 10.1016/s0076-6879(83)96019-6. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Cheng S. C., Ogrydziak D. M. Extracellular RNase produced by Yarrowia lipolytica. J Bacteriol. 1986 Nov;168(2):581–589. doi: 10.1128/jb.168.2.581-589.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S. C., Ogrydziak D. M. Processing and secretion of the Yarrowia lipolytica RNase. J Bacteriol. 1987 Apr;169(4):1433–1440. doi: 10.1128/jb.169.4.1433-1440.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirico W. J., Waters M. G., Blobel G. 70K heat shock related proteins stimulate protein translocation into microsomes. Nature. 1988 Apr 28;332(6167):805–810. doi: 10.1038/332805a0. [DOI] [PubMed] [Google Scholar]
- Chvatchko Y., Howald I., Riezman H. Two yeast mutants defective in endocytosis are defective in pheromone response. Cell. 1986 Aug 1;46(3):355–364. doi: 10.1016/0092-8674(86)90656-2. [DOI] [PubMed] [Google Scholar]
- Ciejek E., Thorner J. Recovery of S. cerevisiae a cells from G1 arrest by alpha factor pheromone requires endopeptidase action. Cell. 1979 Nov;18(3):623–635. doi: 10.1016/0092-8674(79)90117-x. [DOI] [PubMed] [Google Scholar]
- Clare J. J., Davidow L. S., Gardner D. C., Oliver S. G. Cloning and characterisation of the ribosomal RNA genes of the dimorphic yeast, Yarrowia lipolytica. Curr Genet. 1986;10(6):449–452. doi: 10.1007/BF00419872. [DOI] [PubMed] [Google Scholar]
- Cleveland D. W., Fischer S. G., Kirschner M. W., Laemmli U. K. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem. 1977 Feb 10;252(3):1102–1106. [PubMed] [Google Scholar]
- Cryer D. R., Eccleshall R., Marmur J. Isolation of yeast DNA. Methods Cell Biol. 1975;12:39–44. doi: 10.1016/s0091-679x(08)60950-4. [DOI] [PubMed] [Google Scholar]
- Davidow L. S., Kaczmarek F. S., DeZeeuw J. R., Conlon S. W., Lauth M. R., Pereira D. A., Franke A. E. The Yarrowia lipolytica LEU2 gene. Curr Genet. 1987;11(5):377–383. doi: 10.1007/BF00378180. [DOI] [PubMed] [Google Scholar]
- Davidow L. S., O'Donnell M. M., Kaczmarek F. S., Pereira D. A., DeZeeuw J. R., Franke A. E. Cloning and sequencing of the alkaline extracellular protease gene of Yarrowia lipolytica. J Bacteriol. 1987 Oct;169(10):4621–4629. doi: 10.1128/jb.169.10.4621-4629.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R. J., Koch B. D., Werner-Washburne M., Craig E. A., Schekman R. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature. 1988 Apr 28;332(6167):800–805. doi: 10.1038/332800a0. [DOI] [PubMed] [Google Scholar]
- Efimov V. A., Buryakova A. A., Reverdatto S. V., Chakhmakhcheva O. G., Ovchinnikov YuA Rapid synthesis of long-chain deoxyribooligonucleotides by the N-methylimidazolide phosphotriester method. Nucleic Acids Res. 1983 Dec 10;11(23):8369–8387. doi: 10.1093/nar/11.23.8369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmon B., Novick P., Schekman R. Compartmentalized assembly of oligosaccharides on exported glycoproteins in yeast. Cell. 1981 Aug;25(2):451–460. doi: 10.1016/0092-8674(81)90063-5. [DOI] [PubMed] [Google Scholar]
- Fournier P., Gaillardin C., Persuy M. A., Klootwijk J., van Heerikhuizen H. Heterogeneity in the ribosomal family of the yeast Yarrowia lipolytica: genomic organization and segregation studies. Gene. 1986;42(3):273–282. doi: 10.1016/0378-1119(86)90231-3. [DOI] [PubMed] [Google Scholar]
- Gaillardin C., Ribet A. M. LEU2 directed expression of beta-galactosidase activity and phleomycin resistance in Yarrowia lipolytica. Curr Genet. 1987;11(5):369–375. doi: 10.1007/BF00378179. [DOI] [PubMed] [Google Scholar]
- Hubbard S. C., Ivatt R. J. Synthesis and processing of asparagine-linked oligosaccharides. Annu Rev Biochem. 1981;50:555–583. doi: 10.1146/annurev.bi.50.070181.003011. [DOI] [PubMed] [Google Scholar]
- Johnson L. M., Bankaitis V. A., Emr S. D. Distinct sequence determinants direct intracellular sorting and modification of a yeast vacuolar protease. Cell. 1987 Mar 13;48(5):875–885. doi: 10.1016/0092-8674(87)90084-5. [DOI] [PubMed] [Google Scholar]
- Julius D., Blair L., Brake A., Sprague G., Thorner J. Yeast alpha factor is processed from a larger precursor polypeptide: the essential role of a membrane-bound dipeptidyl aminopeptidase. Cell. 1983 Mar;32(3):839–852. doi: 10.1016/0092-8674(83)90070-3. [DOI] [PubMed] [Google Scholar]
- Julius D., Brake A., Blair L., Kunisawa R., Thorner J. Isolation of the putative structural gene for the lysine-arginine-cleaving endopeptidase required for processing of yeast prepro-alpha-factor. Cell. 1984 Jul;37(3):1075–1089. doi: 10.1016/0092-8674(84)90442-2. [DOI] [PubMed] [Google Scholar]
- Julius D., Schekman R., Thorner J. Glycosylation and processing of prepro-alpha-factor through the yeast secretory pathway. Cell. 1984 Feb;36(2):309–318. doi: 10.1016/0092-8674(84)90224-1. [DOI] [PubMed] [Google Scholar]
- Kaiser C. A., Preuss D., Grisafi P., Botstein D. Many random sequences functionally replace the secretion signal sequence of yeast invertase. Science. 1987 Jan 16;235(4786):312–317. doi: 10.1126/science.3541205. [DOI] [PubMed] [Google Scholar]
- Kreil G., Haiml L., Suchanek G. Stepwise cleavage of the pro part of promelittin by dipeptidylpeptidase IV. Evidence for a new type of precursor--product conversion. Eur J Biochem. 1980 Oct;111(1):49–58. doi: 10.1111/j.1432-1033.1980.tb06073.x. [DOI] [PubMed] [Google Scholar]
- Kukuruzinska M. A., Bergh M. L., Jackson B. J. Protein glycosylation in yeast. Annu Rev Biochem. 1987;56:915–944. doi: 10.1146/annurev.bi.56.070187.004411. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lehle L. Biosynthesis of the core region of yeast mannoproteins. Formation of a glucosylated dolichol-bound oligosaccharide precursor, its transfer to protein and subsequent modification. Eur J Biochem. 1980 Aug;109(2):589–601. doi: 10.1111/j.1432-1033.1980.tb04832.x. [DOI] [PubMed] [Google Scholar]
- Mahoney W. C., Duksin D. Biological activities of the two major components of tunicamycin. J Biol Chem. 1979 Jul 25;254(14):6572–6576. [PubMed] [Google Scholar]
- Mellman I., Fuchs R., Helenius A. Acidification of the endocytic and exocytic pathways. Annu Rev Biochem. 1986;55:663–700. doi: 10.1146/annurev.bi.55.070186.003311. [DOI] [PubMed] [Google Scholar]
- Moehle C. M., Tizard R., Lemmon S. K., Smart J., Jones E. W. Protease B of the lysosomelike vacuole of the yeast Saccharomyces cerevisiae is homologous to the subtilisin family of serine proteases. Mol Cell Biol. 1987 Dec;7(12):4390–4399. doi: 10.1128/mcb.7.12.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P., Ferro S., Schekman R. Order of events in the yeast secretory pathway. Cell. 1981 Aug;25(2):461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- Novick P., Schekman R. Export of major cell surface proteins is blocked in yeast secretory mutants. J Cell Biol. 1983 Feb;96(2):541–547. doi: 10.1083/jcb.96.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogrydziak D. M., Scharf S. J. Alkaline extracellular protease produced by Saccharomycopsis lipolytica CX161-1B. J Gen Microbiol. 1982 Jun;128(6):1225–1234. doi: 10.1099/00221287-128-6-1225. [DOI] [PubMed] [Google Scholar]
- Poritz M. A., Siegel V., Hansen W., Walter P. Small ribonucleoproteins in Schizosaccharomyces pombe and Yarrowia lipolytica homologous to signal recognition particle. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4315–4319. doi: 10.1073/pnas.85.12.4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers S., Michaelis S., Broek D., Santa Anna S., Field J., Herskowitz I., Wigler M. RAM, a gene of yeast required for a functional modification of RAS proteins and for production of mating pheromone a-factor. Cell. 1986 Nov 7;47(3):413–422. doi: 10.1016/0092-8674(86)90598-2. [DOI] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roffman S., Sanocka U., Troll W. Sensitive proteolytic enzyme assay using differential solubilities of radioactive substrates and products in biphasic systems. Anal Biochem. 1970 Jul;36(1):11–17. doi: 10.1016/0003-2697(70)90326-x. [DOI] [PubMed] [Google Scholar]
- Rothman J. H., Stevens T. H. Protein sorting in yeast: mutants defective in vacuole biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cell. 1986 Dec 26;47(6):1041–1051. doi: 10.1016/0092-8674(86)90819-6. [DOI] [PubMed] [Google Scholar]
- Salminen A., Novick P. J. A ras-like protein is required for a post-Golgi event in yeast secretion. Cell. 1987 May 22;49(4):527–538. doi: 10.1016/0092-8674(87)90455-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schekman R. Protein localization and membrane traffic in yeast. Annu Rev Cell Biol. 1985;1:115–143. doi: 10.1146/annurev.cb.01.110185.000555. [DOI] [PubMed] [Google Scholar]
- Simms P. C., Ogrydziak D. M. Structural gene for the alkaline extracellular protease of Saccharomycopsis lipolytica. J Bacteriol. 1981 Jan;145(1):404–409. doi: 10.1128/jb.145.1.404-409.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens T., Esmon B., Schekman R. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell. 1982 Sep;30(2):439–448. doi: 10.1016/0092-8674(82)90241-0. [DOI] [PubMed] [Google Scholar]
- Tarentino A. L., Plummer T. H., Jr, Maley F. The release of intact oligosaccharides from specific glycoproteins by endo-beta-N-acetylglucosaminidase H. J Biol Chem. 1974 Feb 10;249(3):818–824. [PubMed] [Google Scholar]
- Trimble R. B., Maley F. Subunit structure of external invertase from Saccharomyces cerevisiae. J Biol Chem. 1977 Jun 25;252(12):4409–4412. [PubMed] [Google Scholar]
- Valls L. A., Hunter C. P., Rothman J. H., Stevens T. H. Protein sorting in yeast: the localization determinant of yeast vacuolar carboxypeptidase Y resides in the propeptide. Cell. 1987 Mar 13;48(5):887–897. doi: 10.1016/0092-8674(87)90085-7. [DOI] [PubMed] [Google Scholar]
- Vasantha N., Thompson L. D., Rhodes C., Banner C., Nagle J., Filpula D. Genes for alkaline protease and neutral protease from Bacillus amyloliquefaciens contain a large open reading frame between the regions coding for signal sequence and mature protein. J Bacteriol. 1984 Sep;159(3):811–819. doi: 10.1128/jb.159.3.811-819.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J. A., Ferrari E., Henner D. J., Estell D. A., Chen E. Y. Cloning, sequencing, and secretion of Bacillus amyloliquefaciens subtilisin in Bacillus subtilis. Nucleic Acids Res. 1983 Nov 25;11(22):7911–7925. doi: 10.1093/nar/11.22.7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford C. A., Daniels L. B., Park F. J., Jones E. W., Van Arsdell J. N., Innis M. A. The PEP4 gene encodes an aspartyl protease implicated in the posttranslational regulation of Saccharomyces cerevisiae vacuolar hydrolases. Mol Cell Biol. 1986 Jul;6(7):2500–2510. doi: 10.1128/mcb.6.7.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Ogrydziak D. M. Extracellular acid proteases produced by Saccharomycopsis lipolytica. J Bacteriol. 1983 Apr;154(1):23–31. doi: 10.1128/jb.154.1.23-31.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- van Heerikhuizen H., Ykema A., Klootwijk J., Gaillardin C., Ballas C., Fournier P. Heterogeneity in the ribosomal RNA genes of the yeast Yarrowia lipolytica; cloning and analysis of two size classes of repeats. Gene. 1985;39(2-3):213–222. doi: 10.1016/0378-1119(85)90315-4. [DOI] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986 Jun 11;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985 Jul 5;184(1):99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]