Summary
Recent exon sequencing studies have revealed that over 19% of human tumors have mutations in subunits of mSWI/SNF (BAF) complexes. To investigate the underlying mechanism we studied human synovial sarcoma (SS), in which transformation results from the translocation of exactly 78 amino acids of SSX to the SS18 subunit of BAF complexes. We demonstrate that the SS18-SSX fusion protein competes for assembly with wildtype SS18, forming an altered complex lacking the tumor suppressor BAF47 (hSNF5). The altered complex binds the Sox2 locus and reverses polycomb-mediated repression, resulting in Sox2 activation. Sox2 is uniformly expressed in SS tumors and is essential for proliferation. Increasing the concentration of wildtype SS18 leads to reassembly of wildtype complexes retargeted away from the Sox2 locus, polycomb-mediated repression of Sox2 and cessation of proliferation. This mechanism of transformation depends on only two amino acids of SSX providing a potential foundation for therapeutic intervention.
Introduction
Exon sequencing in human malignancy has provided paradigm-changing insights into pathogenesis (Lander, 2011) but is often limited by the fact that mutation frequencies are correlative, leaving open the possibility that other primary events are responsible for tumor initiation. This correlative aspect has emerged particularly from recent exon sequencing studies of human cancers, which have defined frequent mutations in chromatin regulators(Dawson and Kouzarides, 2012). By contrast, precise chromosomal translocations which define cancer subsets provide strong support for an initiating role. Chromatin regulation has often been thought to play supportive roles and hence a potential instructive or initiating function for chromatin regulators in human cancer is less clear.
Chromatin regulation is essential for appropriate and timely gene expression. This process is achieved by several mechanisms including DNA-methylation, histone modifications and ATP-dependent chromatin remodeling. One of the most well characterized chromatin remodeling complexes studied to date is the SWI/SNF (BAF) complex, which was discovered in yeast (Peterson and Herskowitz, 1992) and plays a general role in gene activation through nucleosome remodeling, thereby allowing accessibility of transcription factors to their recognition sites. In flies, the Swi2/Snf2 ATPase homologue, Brahma was discovered in screens for trithroax genes (Tamkun et al., 1992) and opposes polycomb function. Mammalian complexes have two SWI2-like ATPases (Brg1 and Brm) and a second ATPase, β-actin, and are combinatorially assembled from gene families that encode the 15 subunits. Fewer than half of the subunits are related to yeast SWI/SNF, others are related to RSC and SWR1 subunits (Cairns et al., 1996; Krogan et al., 2003; Mizuguchi et al., 2004) and hence the name BAF (Brg/Brm-associated complexes) is commonly used. The complexes appear to have undergone evolutionary changes in response to the emergence of multicellularity, polycomb mediated repression, DNA methylation, and a larger genome size (Wu et al., 2009). The role of combinatorial assembly is seen most clearly in the mammalian nervous system in which neurons have a family of highly specialized neuron-specific complexes involved in dendritic morphogenesis (Lessard et al., 2007; Wu et al., 2007; Yoo et al., 2009). Recent genetic studies in flies have suggested that the fly homologue of the neural specific BAF (nBAF) subunit, BAF53b has an instructive role in targeting dendritic trees to their correct termini (Tea and Luo, 2011). Instructive roles are also suggested from studies demonstrating that forcing the formation of nBAF complexes leads to the conversion of fibroblasts to neurons (Yoo et al., 2011). Specialized complexes are also found in pluripotent cells (esBAF complexes) (Ho et al., 2009) and recreating the esBAF complex subunit composition in fibroblasts facilitates iPS cell formation (Singhal et al., 2010). These recent studies suggest an instructive role for these ATP-dependent chromatin regulators that was not anticipated from earlier studies.
Recent exome sequencing studies of primary, early human cancers have repeatedly discovered mutations to subunits of polymorphic BAF complexes. Indeed, analysis of the 44 exome sequencing studies published to date indicate that 19.6% of all human cancers have mutations in at least one subunit (Kadoch et al., submitted). For example, BAF250a is mutated in 57% of clear cell ovarian cancers, BAF180 (polybromo) is mutated in 41% of renal cancers (Varela et al., 2011), and medulloblastomas have frequent mutations in Brg, BAF53a or BAF60b (Jones et al., 2012). The significance of perturbation to ATP-dependent chromatin remodeling complexes in tumorigenesis has been most strongly demonstrated in studies focusing on a particular class of tumors, malignant rhabdoid tumors (MRTs), in which the subunit BAF47 (hSnf5) is biallelically inactivated in nearly 100% of cases reported (Versteege et al., 1998). Patients often have inherited a defective SNF5 allele and the remaining wild-type allele is lost in the tumors, implicating them as tumor suppressors. Conditional biallelic inactivation of Snf5 in mouse models results in a fully penetrant phenotype with median onset to tumor development at only 11 weeks (Roberts et al., 2002). The preference for mutation of specific subunits in specific malignancies suggests that different combinatorial assemblies have roles in tissue-specific oncogenic processes, consistent with roles for specialized BAF complexes in neurogenesis and other biologic processes (de la Serna et al., 2001; Lickert et al., 2004; Wu et al., 2009). Because of the possibility that the frequent BAF subunit mutations might be playing a relatively non-specific role in oncogenesis, we initiated studies on a cancer type, human synovial sarcoma (SS) where nearly all tumors have a precise translocation involving a specific subunit, indicating that the translocation is the initiating oncogenic event.
Human synovial sarcoma accounts for 8-10% of all soft tissue malignancies and most commonly arises in the extremities of young adults (Weiss, 2001). A recurrent chromosomal translocation, t(X;18)(p11.2;q11.2) fuses the SS18 gene on chromosome 18 to one of three closely related genes on the X chromosome, SSX1, SSX2 and rarely SSX4, resulting in an in-frame fusion protein in which the eight C-terminal amino acids of SS18 are replaced with 78 amino acids from the SSX C-terminus(Clark et al., 1994; de Leeuw et al., 1995; Skytting et al., 1999). This remarkably precise translocation is present in greater than 95% of cases and has been established as pathognomonic for the disease with clinical diagnosis confirmed by karyotyping and RT-PCR for SS18-SSX transcripts (Hiraga et al., 1998; Sandberg and Bridge, 2002). The presence of this translocation is the defining feature of synovial sarcomas and is often the only cytogenetic abnormality (dos Santos et al., 1997; Limon et al., 1991); hence, this is very likely to be the driving oncogenic event in the development of these tumors. However, the mechanism of SS18-SSX transformation has been unclear.
Both SS18 and SSX proteins lack known DNA binding motifs, yet they appear to be acting through transcriptional regulatory mechanisms. SS18 is a nuclear protein which has been suggested to interact with chromatin remodeling factors such as Brg/Brm containing complexes (Nagai et al., 2001; Thaete et al., 1999), and the transformation potential of the SS18-SSX fusion has been shown to require Brg/Brm(Nagai et al., 2001). Fusion partners SSX1, 2, and 4 are members of a family of nine human SSX genes which encode highly similar proteins with 73-92% homology (cDNA homology 87-96%) (Smith and McNeel, 2010) and conserved intron/exon junctions. SSX3 and SSX5 have not been found as fusion partners in tumors although they are highly similar to the oncogenic fusion partners. mRNA expression of SSX genes are restricted to the testes and have been detected at low levels in the thyroid.
Here we demonstrate that SS18 is a dedicated, highly stable subunit of BAF complexes. We find that the fusion of SS18 with SSX produces a protein that binds to the complex and evicts both the wild-type SS18 and the tumor suppressor BAF47 (hSNF5). This altered complex then binds to Sox2, relieving H3K27me3 repression thereby activating Sox2, which we find is required for proliferation. Importantly, SS18-SSX-driven complex disruption is determined by a 2 amino acid (aa) hydrophilic region of SSX. Assembly of wild type complexes and proliferative quiescence can be produced by increasing the concentration of the wild type SS18, making this region an excellent drug target.
Results
SS18 is a subunit of mammalian SWI/SNF-like BAF complexes
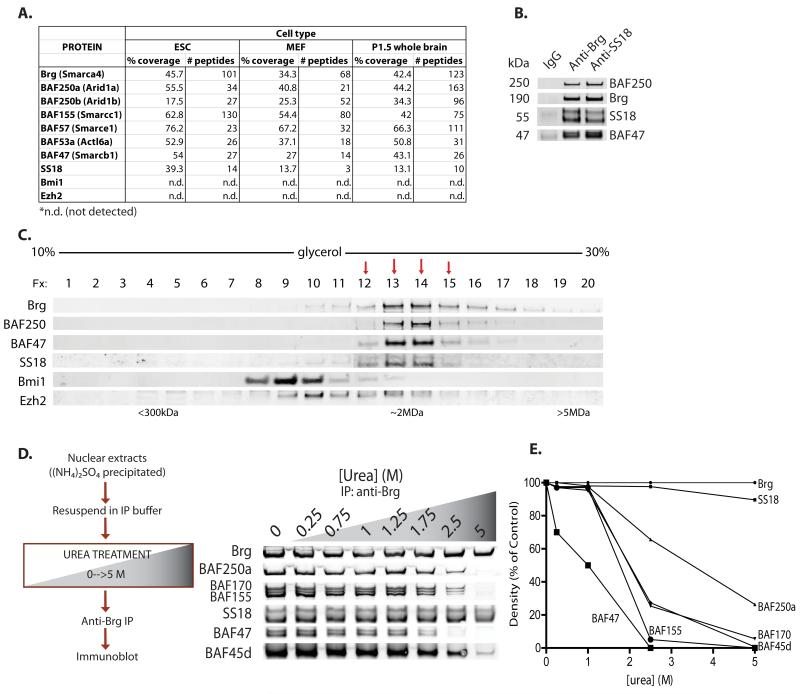
To better understand the composition of BAF complexes, we used a rapid biochemical/affinity purification approach to isolate endogenous complexes from non-transformed cells. Ammonium sulfate fractionation was followed by rapid affinity purification using a highly specific antibody to a genetically non-essential epitope in the Brg/Brm ATPase subunits (Ho et al., 2009). SS18 (synovial sarcoma translocation, chromosome 18) peptides were found in highly pure, endogenous BAF complexes in all tissue types examined, with the exception of post-mitotic adult neurons. Numbers of peptides and percent coverage for the protein SS18 were comparable to those of established BAF complex subunits, suggesting it is a subunit of BAF complexes (Figure 1A). Immunoprecipitation studies using anti-Brg as well as antibodies specific to other established mSWI/SNF complex components including BAF250a, BAF155 and BAF47 confirmed the association of SS18 with native BAF complexes; similarly, reciprocal immunoprecipitation using an antibody to SS18 revealed known components of BAF complexes(Figure 1B, Figure S1A). Two bands are detected for human SS18 due to alternative splicing(Figure S1B). Purification of complexes using anti-Brg and anti-SS18 antibodies revealed similar banding patterns upon silver stain analyses (Figure S1C). In order to determine whether SS18 was dedicated exclusively to BAF complexes, we performed glycerol gradient sedimentation analyses, which demonstrated the presence of SS18 only in fractions containing Brg and other BAF complex subunits (fractions 12-15). SS18 did not associate with polycomb repressor complexes PRC1 or PRC2, as indicated by Bmi1 or Ezh2 immunoblots, respectively, or as a free monomer in earlier fractions of the gradient (Figure 1C). Results were comparable in several cell types assayed including cell lines ES E14, Raji, 293T, and CCRF-CEM as well as primary human fibroblasts. Using urea-based denaturation studies, we determined that SS18 was remarkably stably bound to the complex, to a greater extent than most other subunits including BAF47, BAF155 and BAF 170, requiring denaturing conditions of greater than 5M urea to dissociate (Figure 1D, Figure 1E), similar to ribosomal subunits. The observation that SS18 remains bound when other subunits have dissociated indicates that SS18 binds directly to a stable core complex of Brg, BAF53a, and beta-actin (Zhao et al., 1998). These results demonstrate that SS18 is a dedicated subunit of mSWI/SNF or BAF complexes with binding characteristics similar to those of ribosomal subunits.
Figure 1. SS18 is a dedicated, stable subunit of mSWI/SNF (BAF) complexes.
(A) Composition of BAF complexes isolated from ES cells, MEFs and brain as determined by mass spectrometric analysis. See also Figure S1C.
(B) Immunoprecipitation (IP) using anti-Brg and anti-SS18 antibodies in 293T nuclear extracts (NE). See also Figure 1A,B.
(C) Glycerol gradient (10-30%) analysis on ES cell NE.
(D) Left, Schematic for urea-based denaturation analyses; right, anti-Brg IPs on 293T NE preps treated with 0-5M urea.
(E) Quantitative densitometry on urea denaturation immunoblots.
SS18-SSX integrates into BAF complexes and alters complex composition
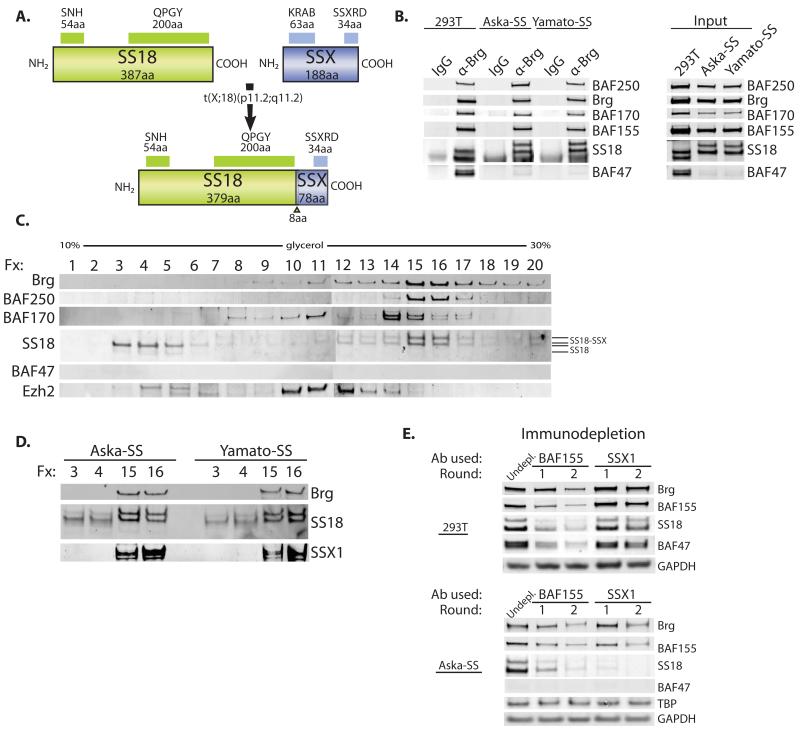
The invariant molecular feature of human synovial sarcoma is the SS18-SSX fusion protein in which the C-terminal 78 amino acids of SSX are fused in frame with amino acids 1-379 of the SS18 subunit (Figure 2A). To investigate the oncogenic mechanism we used two biphasic synovial sarcoma (SS) lines, Aska-SS and Yamato-SS, both of which bear the SS18-SSX1 chromosomal translocation (Naka et al., 2010). Anti-Brg immunoprecipitation studies performed on nuclear extracts isolated from synovial sarcoma cell lines, as compared to control 293T cells (and various other cell types), demonstrated that when SS18 was fused to its translocation partner SSX, the SS18-SSX1 fusion protein was indeed bound to BAF complexes, as reflected by an appropriate upshift in molecular weight of SS18 from 55kDa to 66 kDa upon immunoblot analysis (Figure 2B, left). Remarkably, we observed that both synovial sarcoma lines, as compared to several other cell types assayed, exhibited lower to absent total protein levels of the tumor suppressor subunit BAF47 (hSNF5 or INI1) (Figure 2B, right; Figure S2A, left), while transcripts were largely comparable (Figure S2A, right). Immunoprecipitated BAF complexes containing the SS18-SSX1 fusion protein showed nearly absent levels of wild-type SS18 on the complex. Input protein levels of the wild-type sized SS18 protein were also lowered, as were mRNA levels, suggesting reduced transcription (Figure S2B), and consistent with previously reported findings (Brodin et al., 2001). In addition, a prominent Brg peak is located at the promoter and in an intronic region of the SS18 gene as determined by ChIP-seq analysis in murine ES cells (Figure S2C) (Ho et al., 2011), suggesting auto-regulation of this locus. Density sedimentation analyses performed on nuclear extracts isolated from Aska-SS and Yamato-SS lines revealed disruption of BAF complex composition, in that wild-type SS18 protein no longer associated with the BAF complex fractions (fractions 15,16), and rather existed in fractions 3 and 4 suggesting its presence as a monomer (Figure 2C, Figure S2D). Quantitative densitometry of an anti-SS18 immunoblot of the glycerol gradient, revealed that only a small percentage (2-8%) of BAF complexes contains the wild-type SS18 protein in these cells (Figure S2E). Side-by-side molecular weight comparisons indicated that the SS18-SSX fusion protein, in both SS lines, was almost entirely associated with the BAF complex (denoted by Brg peaks in fractions 15,16) and the wild-type SS18 protein was present, albeit at lower protein levels, in the monomeric fractions of the gradient (fractions 3,4) (Figure 2D). This was further confirmed by immunoblotting using an anti-SSX1 antibody, which demonstrated the presence of SSX1 only in fractions containing Brg. As shown above, in SS lines containing the SS18-SSX fusion, BAF47 no longer associated with BAF complexes and was nearly absent from nuclear extracts indicative of degradation. This is particularly interesting given that BAF47 is a known tumor suppressor; loss of this subunit from the complex as a result of the integration of SS18-SSX might produce functional consequences similar to those of SNF5 inactivation. In order to further assess the degree of dedication of SS18 and SS18-SSX to the BAF complex, we performed depletion studies using two rounds of immunoprecipitation with polyclonal antibodies specific to a known complex subunit, BAF155, as well as to SS18’s fusion partner, SSX1 (Figure 2E). In 293T cells, BAF155 antibodies depleted SS18 protein from the nuclear extracts; SSX1 antibody did not deplete the lysate, as expected, in the wild-type setting. In the Aska-SS synovial sarcoma cell line, immunodepletion using the SSX1 antibody significantly depleted complex subunits Brg, BAF155, and SS18-SSX proteins from nuclear extracts to comparable levels as with anti-BAF155 antibody. These results collectively demonstrate that both wild-type SS18 and in synovial sarcoma, the SS18-SSX1 fusion protein, are dedicated to BAF complexes, but that the fusion protein alters subunit composition.
Figure 2. mSWI/SNF (BAF) complexes are disrupted in synovial sarcoma cells bearing the SS18-SSX1 fusion protein.
(A) Diagram of SS18-SSX fusion protein resulting from t(x;18) translocation, hallmark to synovial sarcoma.
(B) Anti-Brg IP (left) and total protein (right), in 293T cells as compared to synovial sarcoma (SS) cell lines, Aska-SS and Yamato-SS. See also Figure S2A-C.
(C) Glycerol gradient (10-30%, fractions 1-20) analysis on Aska-SS cell NE. See also Figure S2D,E.
(D) Side-by-side comparison of fractions 3,4 and 15,16 of Aska-SS glycerol gradient analysis.
(E) Immunodepletion studies performed on 293T and Aska-SS cells using anti-BAF155 and anti-SSX1 antibodies. (undepleted= antibody not added)
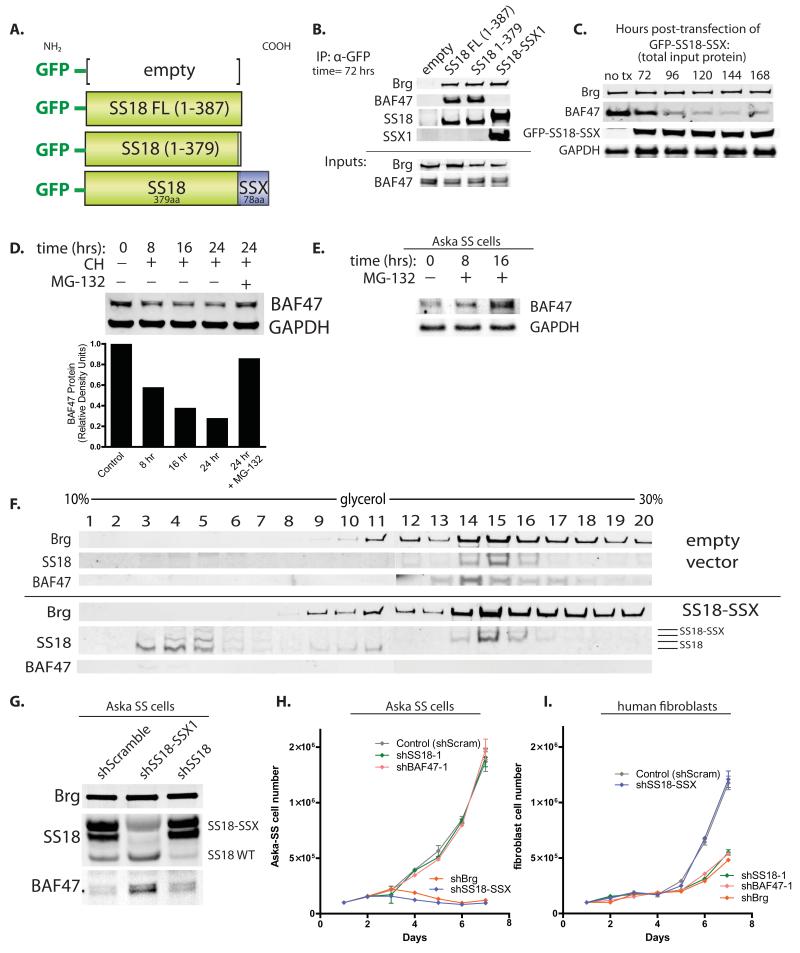
To understand how incorporation of SS18-SSX alters the biochemical subunit composition of BAF complexes, we produced N-terminally GFP-tagged constructs of SS18 FL (full length, aa1-387), SS18 aa1-379 (lacking the last C-terminal 8 aa which are lost in the fusion), and SS18-SSX using a pEGFP-based expression system (Figure 3A). Previous studies have established that the N-terminal SNH domain of SS18 is responsible for its BAF complex association (Nagai et al., 2001). Anti-GFP immunoprecipitations were performed to isolate BAF complexes which had incorporated the exogenously introduced SS18 or SS18-SSX variants. Intriguingly, we noted that expressing the SS18-SSX fusion protein resulted in the loss of BAF47 from the complex at 72 hours post-transfection (Figure 3B). Wild-type SS18 FL or SS18 1-379 both incorporated into BAF complexes but did not alter BAF47 binding to the complex. Input levels of BAF47 at this time point (72 hours) following introduction of SS18-SSX (and all variants tested) were comparable to those of untreated cells. Immunoblot analysis performed on total input protein harvested at 96 hours post-transfection with SS18-SSX indicated a marked decrease in BAF47 levels, with mRNA levels held stable, suggesting that BAF47 is first lost from the complex upon integration of SS18-SSX and subsequently degraded (Figure 3C, Figure S3). To understand the means by which BAF47 is degraded under normal conditions, we performed cyclohexamide (CH) chase experiments over 24 hours, plus and minus proteasome inhibitor treatment using MG-132 at the 24 hour time point. The protein half-life of BAF47 was approximately 10 hours after the addition of CH; BAF47 levels could be rescued from CH treatment with MG-132 to >85% of control levels, indicative of proteasome-mediated degradation (Figure 3D). Treatment of Aska-SS cells with MG-132 resulted in a substantial increase in BAF47 total protein levels (Figure 3E). Upon infection of SS18-SSX1 into 293T fibroblasts, wild-type SS18-containing complexes were readily replaced by SS18-SSX-containing complexes (fractions 14,15,16) and BAF47 levels were reduced as determined by glycerol gradient analyses (Figure 3F). Wild-type SS18 was observed in free, monomeric fractions of the glycerol gradient, as well as in transient lower molecular weight, Brg-associated fractions 9-11. These studies indicate that the SS18-SSX fusion incorporates into BAF complexes, replacing wild-type SS18, and ejecting and destabilizing BAF47.
Figure 3. SS18-SSX1 ejects BAF47 and wild-type SS18 to recapitulate BAF complex phenotype in synovial sarcoma cells.
(A) N-terminal GFP-tagged constructs of SS18 FL, SS18 1-379 (-8aa), and SS18-SSX.
(B) Anti-GFP IP of BAF complexes 72 hrs post-transfection with various pEGFP constructs in 293T fibroblasts. See also Figure S3.
(C) Immunoblot analysis on total protein isolated from transfected 293T cells at time t=0 hrs to t=168 hrs post- transfection with GFP-SS18-SSX.
(D) Top, Cyclohexamide (CH) chase treatment of 293T cells, t=0 to t=24 hours, +/− MG-132 proteosome inhibitor. Bottom, quantitative densitometry of BAF47 protein levels on immunoblot.
(E) Immunoblot analysis for BAF47 protein in Aska-SS cells treated with MG-132 proteosome inhibitor for t=8 and t=16 hours.
(F) Glycerol gradient analyses on 293T cells infected with lentivirus (LV) containing either empty vector (top half) or SS18-SSX (bottom half).
(G) shRNA-mediated knock-down (KD) of SS18-SSX and wild-type SS18 in Aska-SS cells.
(H) Cell proliferation analyses of Aska-SS cells infected with shScramble control vector or delivery constructs containing shRNA KD to BAF subunits. Cells plated in triplicate at 10^5 cells/well per condition. Error bars, s.d. of n=3 experiments.
(I) Cell proliferation analyses of human primary neonatal foreskin fibroblasts infected with shScramble control vector or shRNA KD to BAF complex subunits. Cells plated in triplicate at 10^5 cells/well per condition. Error bars, s.d. of n=3 experiments.
To understand whether low protein levels of BAF47 results specifically from the presence of the SS18-SSX1 fusion in SS cells, we generated shRNA-based knock down (KD) constructs specific for the 3′ UTR of SSX (based on (Takenaka et al., 2010)) to exclusively target SS18-SSX, but not wild-type SS18. Remarkably, we noted a substantial increase in BAF47 total protein levels upon KD of the SS18-SSX oncogenic fusion (Figure 3G). In addition, wild-type SS18 protein levels increased, suggesting relieved repression of SS18 upon KD of the SS18-SSX fusion. We assessed the effect of SS18-SSX KD on proliferation of both synovial sarcoma cell lines. Importantly, KD of the SS18-SSX fusion and of Brg, to which the SS18-SSX fusion was bound, resulted in a profound decrease in proliferation of synovial sarcoma cells (Figure 3H). By contrast, KD of wild-type SS18 and BAF47, subunits not contained in the SS18-SSX-containing BAF complexes, wild-type SS18 and BAF47, had little to no effect on synovial sarcoma cell proliferation (Figure 3H), suggesting that the aberrant residual complex is responsible for driving and maintaining cell proliferation. In human primary fibroblasts with wild-type complexes, KD of Brg, SS18 and BAF47 reduced proliferation; KD of SS18-SSX1 did not alter proliferation as compared to control hairpin (Figure 3I). These studies indicate that the eviction of BAF47 inactivates it and that it is no longer required for proliferation of the SS cell lines. Hence, the free BAF47 protein does not acquire a new function enabling transformation.
Synovial sarcoma cell gene expression features recapitulated: SS18-SSX induces Sox2 expression
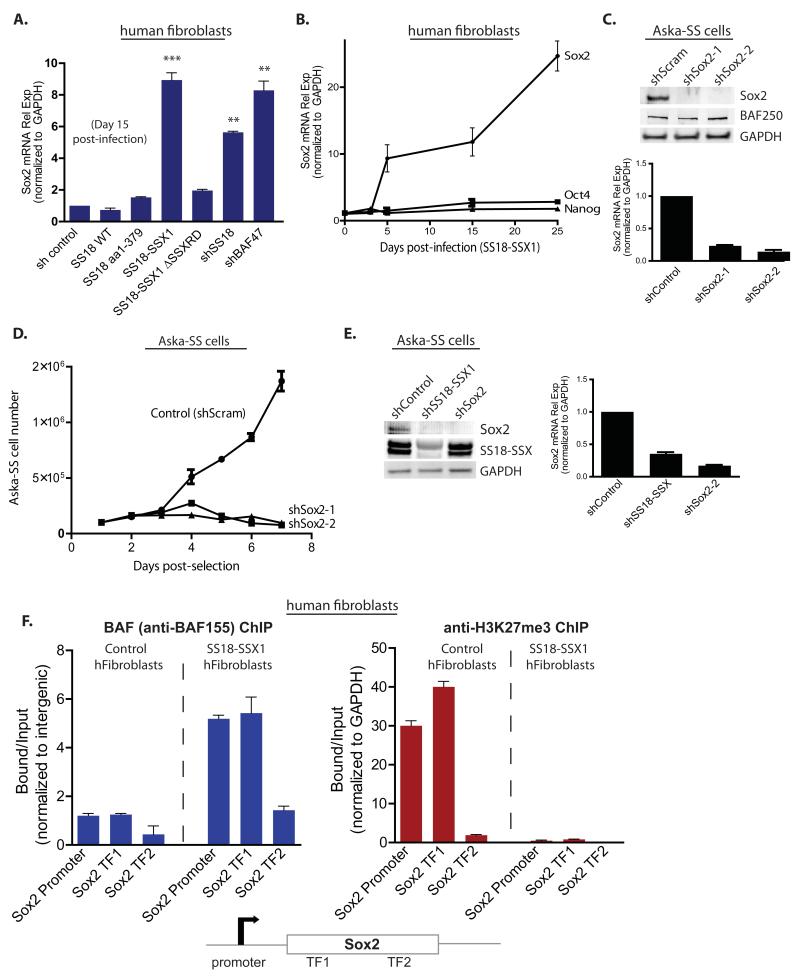
Several studies have demonstrated that SS cells harbor stem-cell like gene expression profiles (Garcia et al., 2012) (Naka et al., 2010). Moreover, Roberts and colleagues observed that tumors lacking the BAF47 tumor suppressor subunit also express stem cell-like signatures (Wilson et al., 2010). Naka and colleagues demonstrated that Aska-SS and Yamato-SS lines as well as 15/15 human tumor specimens of synovial sarcoma tested express mRNA transcripts of pluripotency factors Sox2, Oct4 and Nanog (Naka et al., 2010). We focused on Sox2 because of its role in oncogenesis (Bass et al., 2009). Introduction of SS18-SSX dramatically induced Sox2 mRNA in primary, untransformed human neonatal foreskin fibroblasts by 15 days post-infection and selection (Figure 4A). This induction was specific to the full SS18-SSX1 fusion and did not occur when the C-terminal 34 aa of the conserved SSXRD domain was removed from SSX1. To determine if Sox2 mRNA induction was driven by the partially formed complexes, we tested the effect of shRNA-mediated KD of SS18 and BAF47 in fibroblasts on Sox2 mRNA induction. Intriguingly, KD of SS18 and BAF47 both resulted in a statistically significant increase in Sox2 mRNA to levels nearly comparable to those resulting from overexpression of SS18-SSX (Figure 4A). At the protein level, BAF47 and SS18 appear to reciprocally regulate one another’s stability in fibroblasts as determined by KD of BAF47 and SS18 and immunoblot analysis for protein levels of each (Figure S4A). KD of Brg alone resulted in >70% reduction in protein levels, but did not induce Sox2. Collectively, these data suggest that the activity of aberrant complexes, which lack BAF47 and wild-type SS18, are responsible for Sox2 mRNA induction. Sox2 mRNA levels increased 23-fold by day 25 post-infection with SS18-SSX1 as compared to control (Figure 4B). Oct4 and Nanog mRNA were not induced significantly.
Figure 4. SS18-SSX1 induces Sox2 mRNA expression which drives SS cell proliferation.
(A) Sox2 mRNA levels at day 10 post-infection with LV containing either SS18/SS18-SSX variants or shRNAs to BAF complex subunits. (Normalized to GAPDH; ***p<0.005, **p<0.01). See also Figure S4A.
(B) Time course of Sox2, Oct4, and Nanog mRNA levels post-infection with SS18-SSX-containing LV. (Normalized to GAPDH; error bars reflect s.d. in n=5 separate experiments)
(C) shRNA-mediated knock-down of Sox2 in Aska-SS cells: top, immunoblot analysis; bottom, Sox2 mRNA levels.
(D) Proliferative analysis of Aska-SS cells treated with Sox2 shRNA KD LV. Control, shScramble.
(E) Left, Immunoblot analysis on Aska-SS cells treated with shControl or with either shSS18-SSX1 or shSox2-1; Right, Sox2 mRNA relative expression (normalized to GAPDH).
(F) Left, anti-BAF155 ChIP on human primary fibroblasts treated with either empty vector or SS18-SSX1, with subsequent qPCR for regions at the human Sox2 promoter and two Sox2 transcription factor (TF) binding sites within the exon. Right, anti-H3K27me3 ChIP. Error bars, s.d. of n=3 experiments. See also Figure S4B,C.
We sought to determine whether Sox2 was important for synovial sarcoma cell proliferation. To this end, we generated lentivirus containing two different shRNA hairpins to Sox2 which both effectively reduced Sox2 mRNA and protein in Aska SS cells (Figure 4C) and assessed proliferative capacity in vitro. shRNA-mediated KD of Sox2 profoundly reduced proliferation of Aska-SS cells as compared to scrambled shRNA control (Figure 4D). Intriguingly, Sox2 mRNA and protein levels were reduced in Aska-SS cells upon KD of the SS18-SSX1 fusion to levels comparable to those of cells treated with Sox2 shRNA itself (Figure 4E), indicating elevated levels of Sox2 were specifically due to the presence of SS18-SSX fusion.
To understand the mechanism of Sox2 induction by SS18-SSX we assessed BAF complex occupancy at the Sox2 promoter as well as two clusters of transcription factor (TF) binding sites within the Sox2 exonic region using our affinity-purified BAF155 polyclonal antibody. Intergenic regions were selected as normalization controls. SS18-SSX1-infected primary human fibroblasts demonstrated a significant increase in BAF complex occupancy at all three sites within the human Sox2 locus as compared to control fibroblasts (Figure 4F). In MEFs, there is a prominent H3K27me3 peak over the Sox2 locus as shown by MEF ChIP-seq studies (Mikkelsen et al., 2007), consistent with absent Sox2 expression in these cells (Figure S4B). Lentiviral introduction of SS18-SSX1 into primary human fibroblasts resulted in a striking decrease in H3K27me3 enrichment at all three sites tested within the Sox2 locus (Figure 4F).
To determine if the 78 aa tail of SSX was itself responsible for the targeting of BAF complexes to the Sox2 locus (perhaps by binding a transcription factor) we infected human fibroblasts with V5-tagged SSX78aa (as well as SS18FL and SS18-SSX). However, we did not find that the 78aa SSX fragment localized to the Sox2 locus (Figure S4C). These studies indicate that the SS18-SSX fusion functioning within the altered BAF complexes binds to and activates the Sox2 locus in fibroblasts by disrupting H3K27me3-mediated repression, which is likely directed by the actions of PRC2, the only complex known to place this mark(Chamberlain et al., 2008).
Molecular requirements of SS18-SSX for BAF47 ejection from BAF complexes
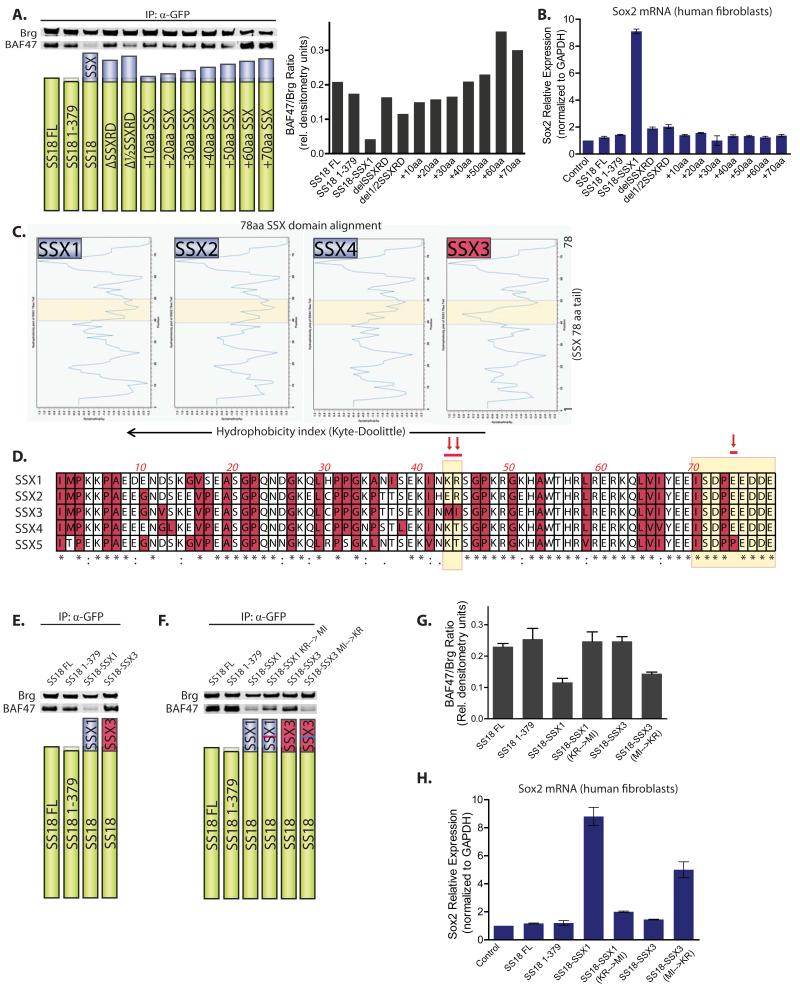
Because expression of SS18-SSX1 resulted in the ejection and subsequent degradation of the BAF47 subunit, we aimed to understand the features of the 78 amino acid SSX tail that could be responsible for this. We generated a series of truncation mutants: deleting the conserved SSXRD domain of 34 aa, deleting ½ of the SSXRD domain (17aa, hydrophobic) and adding amino acids in increments of 10 amino acids to the SS18 C-terminus(+10 through +70). We noted that SS18+10 through SS18+70 did not result in significant ejection of BAF47 from the complex as determined by immunoblot analysis and quantitative densitometry performed on immunoprecipitated complexes (Figure 5A). This implies that a region in the last 8 amino acids (SDPEEDDE) is required for BAF47 ejection. Deleting 1/2SSXRD resulted in slightly decreased levels of BAF47. Upon introduction of these variants into human fibroblasts, Sox2 mRNA induction was only observed with SS18-SSX1 (Figure 5B). Because none of these truncation mutants fully recapitulated the SS18-SSX1-induced BAF47 ejection and Sox2 mRNA induction phenotype, we turned to the fact that the only translocations that have been observed in human synovial sarcoma are SS18-SSX1, SS18-SSX2, and SS18-SSX4. SS18-SSX3 has never been observed in a human tumor. This family of 9 genes (SSX1-9) located at ch Xp11.2 is highly similar; protein homology among members ranges from 73-93% (Smith and McNeel, 2010). We used Kyte-Doolittle hydrophobicity analysis to compare the 78 C-terminal amino acids of SSX1,2,4 versus SSX3, which revealed a significant difference in hydrophobicity between amino acids 40-50 (Figure 5C), as highlighted. Upon peptide alignment of the 78 amino acids of SSX1- 4 it became clear that the amino acid composition at position 43, 44 was most discrepant between the SSX members observed in human SS tumors (1,2 and 4) and the non-oncogenic SSX3. SSX1,2, 4, contains lysine (K) and arginine (R), glutamic acid (E) and arginine (R), and lysine (K) and threonine (T), respectively at position 43,44, while SSX3 contains a methionine (M) and isoleucine (I) at these positions (Figure 5D, arrows). Given that SSX1 is a common fusion partner of SS18 in synovial sarcoma and SSX3 is not, we then sought to understand if SSX3 fused to SS18 could result in BAF47 ejection and Sox2 induction. To this end, we generated an SS18-SSX3 fusion protein (379 amino acids of SS18 fused to 78 amino acids of the SSX3 C-terminus). SS18-SSX3 was able to integrate into BAF complexes, as assessed by anti-GFP immunoprecipitation of BAF complexes, but failed to eject BAF47 (Figure 5E) from the complexes. Remarkably, replacement of amino acids 43,44 of SSX1 (KR) with those of SSX3 (MI) in the SS18-SSX1 fusion resulted in substantial loss of the ability to displace BAF47 (Figure 5F). Reciprocal amino acid substitution at position 43,44 in SS18-SSX3 (MI to KR) resulted in the gained ability of SS18-SSX3 to eject BAF47. Comparative densitometry accounting for the BAF47/Brg ratio is shown, representative of n=3 experiments (Figure 5G). Intriguingly, SS18-SSX1 as well as SS18-SSX3 (Δ43,44 MI→KR) significantly induced Sox2 mRNA; no other variant produced this phenotype (Figure 5H), lending further evidence that the loss of BAF47 (hSnf5) from mSWI/SNF complexes is necessary for the induction of Sox2 mRNA expression in synovial sarcoma. All three fusions reported in human synovial sarcomas (SSX1,2,4) produced BAF47 eviction, while SS18-SSX3 and SS18-SSX5 (which bears an amino acid change in the last 8 aa of SSX) fusions did not (Figure S5A,B).
Figure 5. Molecular requirements of the 78aa SSX peptide for BAF47 subunit ejection from mSWI/SNF-like BAF complexes.
(A) Left, Immunoblot analysis for BAF47 and Brg on anti-Brg IPs (Top) of 293T cells transfected with various SS18/SS18-SSX constructs (Bottom). Right, quantitative densitometry depicting BAF47/Brg protein ratios in IP studies.
(B) Sox2 mRNA levels in human fibroblasts day 15 post-infection with LV containing SS18 and SS18-SSX variants. Error bars= s.d.
(C) Hydrophobicity determination using Kyte-Doolittle algorithm for 78 C-terminal amino acids (aa) of SSX1-SSX4 proteins. Region of significant difference highlighted in yellow.
(D) Peptide alignment of SSX1- SSX5 C-terminal 78 aa. Pink arrows indicate aa of significant difference between SSX1/2/4 and SSX3 or SSX1/2/4 and SSX5; yellow highlight indicates regions determined to be critical for BAF47 ejection.
(E) Immunoblot analysis for BAF47 and Brg on anti-GFP IPs of 293T cells transfected with constructs as per above as well as SS18-SSX3; and (F) with SS18-SSX1, SS18-SSX1Δaa43,44 (KR→MI), SS18-SSX3, SS18-SSX3Δaa43,44 (MI→KR). See also Figure S5A.
(G) Quantitative densitometry depicting BAF47/Brg protein ratios in IP studies. Error bars= s.d. (H) Sox2 mRNA levels in human fibroblasts day 15 post-infection with LV containing various constructs. See also Figure S5B. Error bars=s.d.
Reversibility of BAF complex subunit composition and targeting in human synovial sarcoma
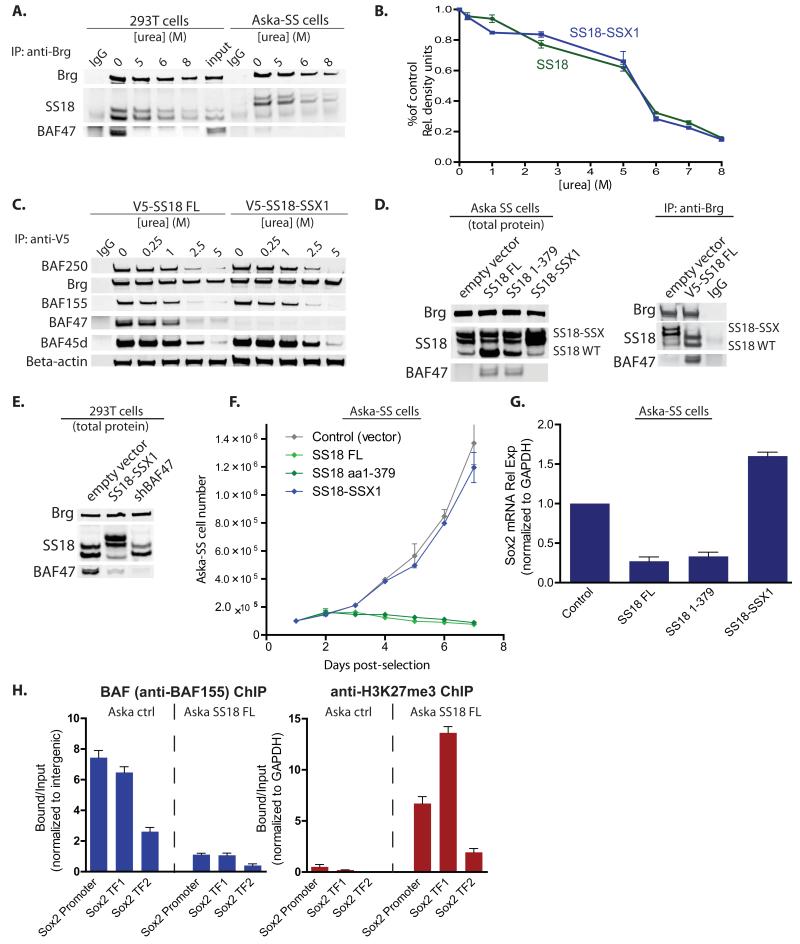
Our observation that SS18 was displaced or failed to assemble into BAF complexes in the presence of somewhat higher concentrations of the SS18-SSX fusion protein (Figures 2C, 3F) lead us to investigate the possibility that the transforming fusion protein and the wild-type protein might exist in a concentration-dependent equilibrium or could be competing for assembly into newly formed complexes. Urea-based denaturation experiments demonstrated that SS18 and SS18-SSX are both stably bound to BAF complexes and dissociate to comparable degrees from 0 to 8 M urea as shown by immunoblot and quantitative densitometry analyses (Figure 6A, 6B). BAF complex components dissociated at comparable levels across the urea denaturation series from V5-tagged SS18 and SS18-SSX, indicative of equal affinity binding of wild-type SS18 and SS18-SSX (Figure 6C). Moreover, Brg and β-actin remained bound to V5-SS18/SS18-SSX-purified complexes to >5M urea, suggesting that SS18/SS18-SSX is part of a highly stable core complex of Brg, BAF53a and β-actin.
Figure 6. Reversible integration, gene expression and occupancy by SS18 and SS18-SSX containing mSWI/SNF (BAF) complexes.
(A) Denaturation studies using 0-8M urea with subsequent immunoblot analysis for SS18 in 293T cells and SS18-SSX in Aska-SS cells. See also Figure S6.
(B) Quantitative densitometry of SS18 or SS18-SSX1 protein immunoblots from n=3 experimental replicates of urea denaturation 0<[urea]<8M. Y-axis: band quantitation/ untreated control. Error bars= s.d.
(C) IP using anti-V5 antibody in urea treated nuclear extracts isolated from 293T fibroblasts infected with either V5-SS18 or V5-SS18-SSX with immunoblotting for BAF complex components.
(D) Left, Immunoblot analysis on total protein isolated from Aska-SS cells with either SS18 or SS18 1-379 or SS18-SSX1 introduced via LV. Right, anti-Brg IP of complexes in either empty vector or V5-SS18FL treated conditions.
(E) Introduction of SS18-SSX1 and shBAF47 into 293T cells with subsequent immunoblot analysis on total protein.
(F) Cell proliferation analyses of Aska-SS cells infected with control vector, SS18, SS18 1-379, and SS18-SSX. Error bars= s.d.
(G) Sox2 mRNA relative expression (normalized to GAPDH) 10 days post infection with LV containing either control shScramble or overexpression of SS18, SS18 1-379, or SS18-SSX. Error bars= s.d.
(H) Left, anti-BAF155 ChIP on Aska-SS cells treated with either empty vector or SS18 FL, with subsequent qPCR for regions at the human Sox2 promoter and two Sox2 transcription factor (TF) binding sites within the exon. Right, anti-H3K27me3 ChIP at Sox2 locus in Aska-SS control treated and SS18 FL-treated cells. Error bars= s.d.
Given these findings and having observed that shRNA-mediated KD of the SS18-SSX1 fusion could restore BAF47 total protein levels (Figure 3G), we sought to determine whether overexpression of wild-type SS18 could also be sufficient to allow normal complexes to reform in synovial sarcoma cell lines and whether this could reverse the misassembly of synovial sarcoma BAF complexes and correct the gene expression phenotypes. Intriguingly, introduction of SS18 FL or SS18 1-379 resulted in a profound increase in BAF47 total protein levels by Day 10 post-infection (Figure 6D, left). Moreover, BAF complexes in Aska-SS cells infected with SS18 regained normal incorporation of wild-type SS18 and BAF47 subunits, suggesting concentration-driven re-integration of SS18 (Figure 6D, right). Introduction of SS18-SSX1 into 293T fibroblasts resulted in reduction of BAF47 total protein to a comparable degree as shRNA-mediated KD of BAF47 (Figure 6E). These studies indicate that the SS18-SSX fusion protein and the wild type SS18 protein compete for assembly into BAF complexes and that the transforming mutant protein can be displaced from BAF complexes to yield wild-type complexes by increasing the concentration of the wild-type protein.
Proliferation of SS cells was inhibited by introduction of wild-type SS18 and SS18 1-379, to a similar degree as in cells treated with shRNA-mediated KD of the SS18-SSX1 fusion (Figure 6F). In contrast, introduction of SS18-SSX into SS18-SSX-bearing synovial sarcoma Aska-SS cells had no appreciable effect on proliferation as compared to control. Sox2 mRNA expression levels in Aska-SS cells were reduced by 3- and 4-fold, upon overexpression of SS18FL and SS18 1-379, respectively (Figure 6G). In contrast, overexpression of SS18-SSX1 in these lines already bearing one translocated allele caused Sox2 mRNA levels to increase 1.7-fold above control levels relative to empty vector control indicating that the levels of Sox2 produced by the SS18-SSX fusion protein were not at maximum. Finally, Aska-SS synovial sarcoma cells infected with SS18FL to reverse the BAF complex phenotype exhibited a dramatically decreased occupancy of BAF complexes at the human Sox2 locus with a concomitant increase in H3K27me3 occupancy (Figure 6H). These studies indicate that normal BAF complexes can be reassembled in malignant cells by over expression of the wild type SS18 protein, leading to BAF complex removal from the Sox2 gene and resumption of normal repression of Sox2 by H3K27 trimethylation.
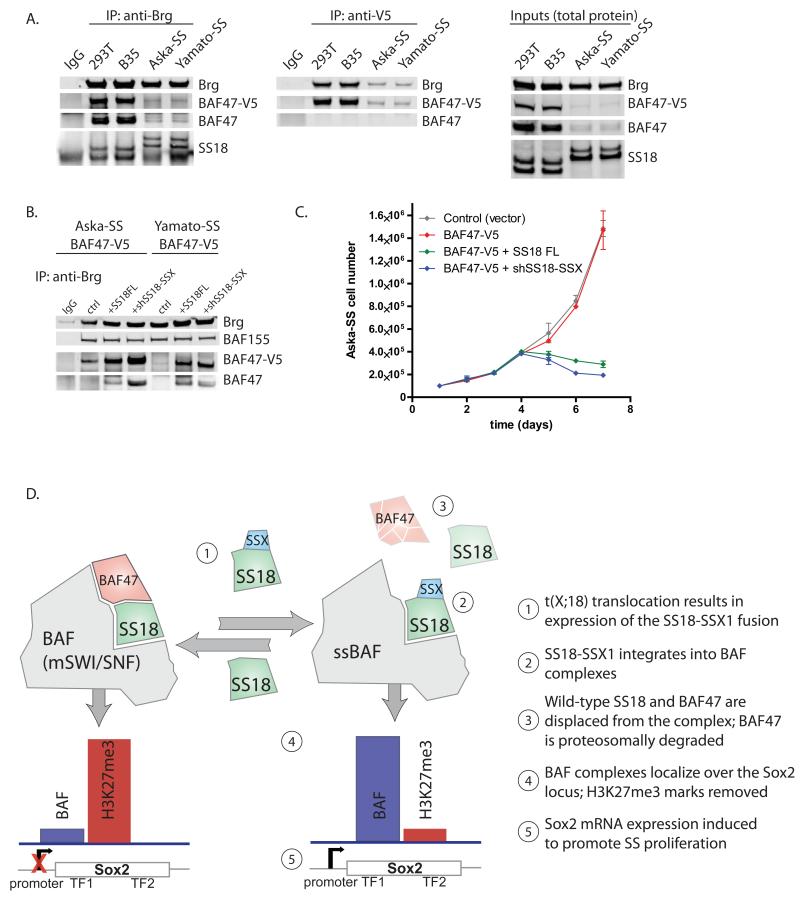
Finally, we aimed to test the potential for BAF47 overexpression to promote reassembly of wild-type BAF complexes containing BAF47 and SS18 in SS cells and its effect on proliferation. Notably, overexpressed V5-tagged BAF47 was unable to bind SS18-SSX containing complexes in both SS cell lines tested, as evidenced by low protein levels on complexes detected by anti-Brg and anti-V5 immunoprecipitations as well as by total protein immunoblots, suggestive of rapid degradation (Figure 7A). To test whether shifting aberrant complex assembly back to that of wild-type would allow for integration of the exogenous BAF47-V5 into complexes we infected SS cells containing BAF47-V5 with either SS18FL or shSS18-SSX. Indeed, in both lines, overexpression of SS18FL or KD of the SS18-SSX fusion resulted in increased incorporation and stabilization of BAF47-V5 as indicated by anti-Brg immunoprecipitation (Figure 7B). Intriguingly, BAF47 overxpression had no effect on SS cell proliferation in culture; however, proliferation was dramatically attenuated upon co-introduction of overexpressed SS18FL or KD of SS18-SSX, suggesting that BAF47 can only assemble into wild-type SS18-containing complexes and not complexes bearing the SS18-SSX fusion (Figure 7C).
Figure 7. A model for reversible transformation by the SS18-SSX1 oncogenic fusion.
(A) (left) Anti-Brg IPs on 293T, B35, Aska-SS and Yamato-SS cells bearing introduced BAF47-V5; (middle) anti-V5 IPs; (right) Total protein inputs.
(B) Anti-Brg IPs on nuclear extracts of Aska-SS and Yamato-SS cells with stably introduced BAF47-V5 and co-infection with either control vector, SS18FL, or shSS18-SSX.
(C) Proliferation analyses of Aska-SS cells infected with either control vector, BAF47-V5, or BAF47-V5+co-infected SS18FL, BAF47-V5+co-infected shSS18-SSX. Error bars= s.d.
(D) Model for reversible disruption of BAF complex composition and action upon SS18-SSX incorporation.
Discussion
Our studies demonstrate that in the two synovial sarcoma cell lines we have used, the fusion of SS18 with SSX, which is diagnostic of this tumor type, leads to assembly of aberrant BAF complexes that become targeted to the Sox2 locus, with loss of repressive H3K27me3 marks, driving Sox2 expression and proliferation of these cells (Figure 7D). The observation that Sox2 is activated in all SS studied ((Naka et al., 2010)) suggests this is a general mechanism of oncogenesis in these tumors. We find that the SS18-SSX fusion incorporates into BAF complexes and activates Sox2 expression, explaining the uniform activation of this gene in SS. But how do complexes containing the SS18-SSX fusion activate Sox2? BAF complexes containing the SS18-SSX fusion could be targeted by the interaction of SSX with a factor that binds the Sox2 locus. Alternatively, an incorrectly assembled complex could target the Sox 2 locus by changes to bromo-, chromo- and PHD domain presentation. We find that the 78aa of SSX alone is not targeted to the Sox2 locus when expressed in human fibroblasts (Figure S4B), indicating that it is the aberrantly assembled complex that targets the inactive Sox2 locus, reversing H3K27Me3-mediated repression, and leading to Sox2 activation.
Remarkably, the wild type SS18 protein is capable of replacing the SS18-SSX fusion in BAF complexes when expressed at somewhat higher levels than the fusion protein. The incorporation of wild-type and mutant proteins is unlikely to be due to direct binding competition. This conclusion arises from the fact that 8 M urea is required to remove either the wild-type SS18 protein from the wild type complexes or the SS18-SSX fusion from the malignant complexes. Hence, the two proteins most likely compete for assembly into complexes, with the product of the fusion allele winning in SS cells due to increased concentration.
The ability of SS18-SSX to disrupt BAF complexes maps to two regions of the SSX protein: The C-terminal 8 amino acids (SDPEEDDE) and a polar region of two amino acids present in the oncogenic members of the SSX family of proteins. Substitution of KR with MI, found in the non-transforming SSX3, restores normal complex assembly and gene regulation; substitution of MI with KR in SS18-SSX3 results in BAF47 ejection and increased Sox2 mRNA. In this regard, SSX5 is interesting in that it has KT at position 43, 44, combined with an amino acid substitution of P for E in the 8 terminal amino acids; SS18-SSX5 has not been found in translocations and does not eject BAF47, confirming the importance of both regions for oncogenicity. These two regions could interact to facilitate complex dissolution or form dimers in the malignant complexes. Structural studies will be necessary to define the precise mechanism. However, the ability of such a small region to lead to complex dissolution and the observation that the wild type and malignant protein are in a dynamic equilibrium indicates that the fusion containing the two amino acid essential region in the SSX tail (K43, R44) is an excellent target for developing therapeutics for this disease. A decoy molecule that causes SSX1 to resemble SSX3 would be expected to prevent eviction of BAF47, and thereby reverse the effects of the aberrant SS-BAF complex. This notion is consistent with the precision of the oncogenic translocation, in that all translocations discovered to date add exactly 78 amino acids of SSX1,2 or 4 to the SS18 protein at position 379.
In SS cells, the partially assembled complex gains the ability to bind the Sox2 gene, reversing H3K27Me3-mediated repression. Forcing correct assembly by expressing the wild-type SS18 causes the reassembly of wild-type complexes without the fusion thus reestablishing normal repression of Sox2 by polycomb. The fly Brahma protein was discovered from its ability to oppose polycomb and hence is known as a trithorax gene, however the underlying biochemical mechanisms have been controversial. In some studies, polycomb was found to prevent Brahma (BAP) complex binding, while in others it seemed that BAP or SWI/SNF directly recruited PolII, thereby opposing polycomb. Our studies suggest that somehow BAF complexes evict polycomb, however our temporal resolution is limited to the infection times (24-72 hrs) and hence we are unable to determine if the mechanism is direct physical eviction, or dilution of H3K27Me3 by nucleosome exchange with cell division since the measured rates of nucleosome turnover (Deal et al., 2010) are sufficient to remove most H3K27Me3 if methylation were prevented by the SS BAF complex. Evidence for BAF-polycomb opposition in malignancy has also been found with inactivation of BAF47 (hSNF5 or Ini1) in human malignant rhabdoid sarcoma (MRTs). In these tumors and in mouse models, polycomb was found to be removed from the INK4a locus upon introduction of BAF47 (hSNF5) (Kia et al., 2008). Understanding the underlying mechanism of polycomb opposition will require techniques that allow rapid recruitment of BAF complexes with a high degree of temporal and spatial control (Hathaway et al., 2012).
Synovial sarcoma is largely resistant to conventional, chemotherapy-based forms of treatment, underlining the need for an understanding of its pathogenesis. Disease-specific biologic agents which target SS18-SSX or its interactions have to date not been developed. Here we have shown that the SS18-SSX1 oncogenic fusion usurps SWI/SNF-like BAF complexes, resulting in activation of Sox2, which drives proliferation. Remarkably, the oncogenic fusion and wild-type SS18 bind to BAF complexes with comparable affinities, allowing directed assembly of oncogenic or wild-type complexes. Moreover, the composition of SS18-SSX-containing BAF complexes (lacking BAF47 and wild-type SS18) can be reversed by reducing the levels of SS18-SSX or by increasing levels of wild-type SS18. The observation that eviction of BAF47 from the complexes is dependent upon only 2 amino acids in SSX demonstrates an unusual mechanism of oncogenesis and opens a potential therapeutic avenue.
Experimental Procedures
Nuclear extract preparation and proteomic studies
Nuclear extract (NE) preparation and immunoprecipitation (IP) studies were performed as described in Ho et al, 2009 and Extended Experimental Procedures. Antibody specifications are presented in Table S1.
Transfection studies
Briefly, cells were plated in 6-well plates to 80% confluence prior to transfection using PEI (Poly(ethylenimine) in a 3:1 PEI:DNA ratio and were harvested at the appropriate time points thereafter.
Cell proliferation analyses
Cells were assessed for >95% viability prior to being plated at 10^5 cells/well in triplicate/condition in 12-well plates. Cell counts were determined using trypan blue exclusion-based methods.
Urea denaturation studies
NE (150 μg) were subjected to partial urea denaturation ranging from 0.25M to 8M urea (in IP buffer) for 15 min at RT prior to anti-Brg IP. The co-precipitated proteins were analyzed by immunoblot. Quantitative densitometry analyses were performed with the LiCor Oddessy Imaging System (Li-COR Biosciences).
Density sedimentation analyses
800 μg NE were resuspended in 300 μl of 0% glycerol HEMG buffer, and carefully overlaid on to a 10ml 10-30% glycerol (in HEMG buffer) gradient prepared in a 14×89 mm polyallomer centrifuge tube (Beckman # 331327). Tubes were centrifuged in an SW40 rotor at 4°C for 16 hrs at 40K RPM. 0.5 ml fractions were collected and used in analyses. See Extended Experimental Procedures.
Cyclohexamide/MG-132 studies
MG-132 (Calbiochem, #474790) (10mg/ml in DMSO) was used at 1:1000, cyclohexamide (Sigma, #C4859) (100mg/ml) at 1:100 in cell culture media. Briefly, cells were plated in 6-well plates and treated with the above agents for 0 to 24 hours and harvested with RIPA lysis buffer.
Gene Expression Profiling and Analysis
Total RNA was isolated using TRIzol® reagent (Invitrogen) and reverse transcribed into cDNA (SuperScript® III RT kit (Invitrogen)). Real-time PCR was performed using TaqMan Universal Master Mix with Taqman probes and/or SYBR green method with custom designed primers, normalized to GAPDH and/or 18S rRNA expression. All primers are listed in Supplemental Table S2.
shRNA-mediated knock down and lentiviral (LV) generation
shRNAs specific for human Brg1, BAF47, SS18, and Sox2 were purchased from Open Biosystems (Table S3). shRNA KD constructs for SS18-SSX and shScramble control were generated by annealed oligos (Table S3) and subsequent cloning into the pLK0.1 vector. LV was produced as described by Tiscornia et al., 2006. See Extended Experimental Procedures.
ChIP Analyses
Briefly, cells were crosslinked in formaldehyde, washed, and sonicated as described in Extended Experimental Procedures. ChIP antibodies: anti-BAF155 (in house generated), anti-H3K27me3 (Millipore, 07-449), V5 (Invitrogen, 46-0705). Primers used for real-time PCR listed in Table S2.
Supplementary Material
Highlights.
SS18 is a dedicated and stable subunit of mSWI/SNF (BAF) complexes
SS18-SSX fusion (the hallmark of synovial sarcomas) disrupts BAF complex composition
SS18-SSX1 induces Sox2 expression, necessary for SS cell proliferation
Synovial sarcoma cell proliferation is reversed via reassembly of normal complexes
Acknowledgements
This work was supported in part by NIH grants HD55391, RO1NS046789, and R01CA163195 (G.R.C.). G.R.C. is an Investigator of the Howard Hughes Medical Institute. C.K. is supported by the National Science Foundation (Graduate Research Fellowship Program). The authors are grateful to Kazuyuki Itoh, Norifume Naka, and Satoshi Takenaka (Osaka University, Japan) for kindly providing the Aska-SS and Yamato-SS synovial sarcoma cell lines and cDNA clone of the SS18-SSX fusion. We are also grateful to Clara Lee for technical assistance and N. Hathaway, O. Bell, A. Shalizi, and J. Ronan for helpful discussions and critical review of the manuscript.
Footnotes
Supplemental Information
Supplemental information includes 6 Supplemental Figures and 3 Supplemental Tables.
References
- Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, Kim SY, Wardwell L, Tamayo P, Gat-Viks I, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nature genetics. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin B, Haslam K, Yang K, Bartolazzi A, Xie Y, Starborg M, Lundeberg J, Larsson O. Cloning and characterization of spliced fusion transcript variants of synovial sarcoma: SYT/SSX4, SYT/SSX4v, and SYT/SSX2v. Possible regulatory role of the fusion gene product in wild type SYT expression. Gene. 2001;268:173–182. doi: 10.1016/s0378-1119(01)00412-7. [DOI] [PubMed] [Google Scholar]
- Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J, Rocques PJ, Crew AJ, Gill S, Shipley J, Chan AM, Gusterson BA, Cooper CS. Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nature genetics. 1994;7:502–508. doi: 10.1038/ng0894-502. [DOI] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- de la Serna IL, Carlson KA, Imbalzano AN. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat Genet. 2001;27:187–190. doi: 10.1038/84826. [DOI] [PubMed] [Google Scholar]
- de Leeuw B, Balemans M, Olde Weghuis D, Geurts van Kessel A. Identification of two alternative fusion genes, SYT-SSX1 and SYT-SSX2, in t(X;18)(p11.2;q11.2)-positive synovial sarcomas. Human molecular genetics. 1995;4:1097–1099. doi: 10.1093/hmg/4.6.1097. [DOI] [PubMed] [Google Scholar]
- Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328:1161–1164. doi: 10.1126/science.1186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos NR, de Bruijn DR, Balemans M, Janssen B, Gartner F, Lopes JM, de Leeuw B, Geurts van Kessel A. Nuclear localization of SYT, SSX and the synovial sarcoma-associated SYT-SSX fusion proteins. Hum Mol Genet. 1997;6:1549–1558. doi: 10.1093/hmg/6.9.1549. [DOI] [PubMed] [Google Scholar]
- Garcia CB, Shaffer CM, Alfaro MP, Smith AL, Sun J, Zhao Z, Young PP, VanSaun MN, Eid JE. Reprogramming of mesenchymal stem cells by the synovial sarcoma-associated oncogene SYT-SSX2. Oncogene. 2012;31:2323–2334. doi: 10.1038/onc.2011.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hathaway NA, Bell O, Hodges C, Miller EL, Neel DS, Crabtree GR. Dynamics and memory of heterochromatin in living cells. Cell. 2012;149:1447–1460. doi: 10.1016/j.cell.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga H, Nojima T, Abe S, Sawa H, Yamashiro K, Yamawaki S, Kaneda K, Nagashima K. Diagnosis of synovial sarcoma with the reverse transcriptase-polymerase chain reaction: analyses of 84 soft tissue and bone tumors. Diagn Mol Pathol. 1998;7:102–110. doi: 10.1097/00019606-199804000-00007. [DOI] [PubMed] [Google Scholar]
- Ho L, Miller EL, Ronan JL, Ho WQ, Jothi R, Crabtree GR. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat Cell Biol. 2011;13:903–913. doi: 10.1038/ncb2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A. 2009;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Jager N, Kool M, Zichner T, Hutter B, Sultan M, Cho YJ, Pugh TJ, Hovestadt V, Stutz AM, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol Cell Biol. 2008;28:3457–3464. doi: 10.1128/MCB.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470:187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- Limon J, Mrozek K, Mandahl N, Nedoszytko B, Verhest A, Rys J, Niezabitowski A, Babinska M, Nosek H, Ochalek T, et al. Cytogenetics of synovial sarcoma: presentation of ten new cases and review of the literature. Genes Chromosomes Cancer. 1991;3:338–345. doi: 10.1002/gcc.2870030504. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Nagai M, Tanaka S, Tsuda M, Endo S, Kato H, Sonobe H, Minami A, Hiraga H, Nishihara H, Sawa H, et al. Analysis of transforming activity of human synovial sarcoma-associated chimeric protein SYT-SSX1 bound to chromatin remodeling factor hBRM/hSNF2 alpha. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:3843–3848. doi: 10.1073/pnas.061036798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naka N, Takenaka S, Araki N, Miwa T, Hashimoto N, Yoshioka K, Joyama S, Hamada K, Tsukamoto Y, Tomita Y, et al. Synovial sarcoma is a stem cell malignancy. Stem cells (Dayton, Ohio) 2010;28:1119–1131. doi: 10.1002/stem.452. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- Roberts CW, Leroux MM, Fleming MD, Orkin SH. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumor suppressor gene Snf5. Cancer Cell. 2002;2:415–425. doi: 10.1016/s1535-6108(02)00185-x. [DOI] [PubMed] [Google Scholar]
- Sandberg AA, Bridge JA. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors. Synovial sarcoma. Cancer Genet Cytogenet. 2002;133:1–23. doi: 10.1016/s0165-4608(01)00626-4. [DOI] [PubMed] [Google Scholar]
- Singhal N, Graumann J, Wu G, Arauzo-Bravo MJ, Han DW, Greber B, Gentile L, Mann M, Scholer HR. Chromatin-Remodeling Components of the BAF Complex Facilitate Reprogramming. Cell. 2010;141:943–955. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- Skytting B, Nilsson G, Brodin B, Xie Y, Lundeberg J, Uhlen M, Larsson O. A novel fusion gene, SYT-SSX4, in synovial sarcoma. J Natl Cancer Inst. 1999;91:974–975. doi: 10.1093/jnci/91.11.974. [DOI] [PubMed] [Google Scholar]
- Smith HA, McNeel DG. The SSX family of cancer-testis antigens as target proteins for tumor therapy. Clin Dev Immunol. 2010;2010:150591. doi: 10.1155/2010/150591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka S, Naka N, Araki N, Hashimoto N, Ueda T, Yoshioka K, Yoshikawa H, Itoh K. Downregulation of SS18-SSX1 expression in synovial sarcoma by small interfering RNA enhances the focal adhesion pathway and inhibits anchorage-independent growth in vitro and tumor growth in vivo. International journal of oncology. 2010;36:823–831. doi: 10.3892/ijo_00000559. [DOI] [PubMed] [Google Scholar]
- Tamkun JW, Deuring R, Scott MP, Kissinger M, Pattatucci AM, Kaufman TC, Kennison JA. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992;68:561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- Tea JS, Luo L. The chromatin remodeling factor Bap55 functions through the TIP60 complex to regulate olfactory projection neuron dendrite targeting. Neural Dev. 2011;6:5. doi: 10.1186/1749-8104-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaete C, Brett D, Monaghan P, Whitehouse S, Rennie G, Rayner E, Cooper CS, Goodwin G. Functional domains of the SYT and SYT-SSX synovial sarcoma translocation proteins and co-localization with the SNF protein BRM in the nucleus. Hum Mol Genet. 1999;8:585–591. doi: 10.1093/hmg/8.4.585. [DOI] [PubMed] [Google Scholar]
- Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, Aurias A, Delattre O. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- Weiss SW, Goldblum JR. Enzinger and Weiss’s Soft Tissue Tumors. Fourth Edition edn Mosby, Inc.; St. Louis, MO: 2001. [Google Scholar]
- Wilson BG, Wang X, Shen X, McKenna ES, Lemieux ME, Cho YJ, Koellhoffer EC, Pomeroy SL, Orkin SH, Roberts CW. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell. 2010;18:316–328. doi: 10.1016/j.ccr.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136:200–206. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Olave IA, Qiu Z, Ghosh A, Graef IA, Crabtree GR. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.