Abstract
Enzymatic modification is a prevalent mechanism by which bacteria defeat the action of antibiotics. Aminoglycosides are often inactivated by aminoglycoside modifying enzymes encoded by genes present in the chromosome, plasmids, and other genetic elements. The AAC(6′)-Ib (aminoglycoside 6′-N-acetyltransferase type Ib) is an enzyme of clinical importance found in a wide variety of gram-negative pathogens. The AAC(6′)-Ib enzyme is of interest not only because of his ubiquity but also because of other characteristics, it presents significant microheterogeneity at the N-termini and the aac(6′)-Ib gene is often present in integrons, transposons, plasmids, genomic islands, and other genetic structures. Excluding the highly heterogeneous N-termini, there are 45 non-identical AAC(6′)-Ib related entries in the NCBI database, 32 of which have identical name in spite of not having identical amino acid sequence. While some variants conserved similar properties, others show dramatic differences in specificity, including the case of AAC(6′)-Ib-cr that mediates acetylation of ciprofloxacin representing a rare case where a resistance enzyme acquires the ability to utilize an antibiotic of a different class as substrate. Efforts to utilize antisense technologies to turn off expression of the gene or to identify enzymatic inhibitors to induce phenotypic conversion to susceptibility are under way.
Keywords: antibiotic resistance, aminoglycoside, inhibition, acetyltransferase, mobile elements, integron, transposon
Aminoglycosides and resistance
Aminoglycosides are bactericidal antibiotics that affect translation fidelity and, according to recent data, they may also stimulate the production of highly deleterious hydroxyl radicals (Vakulenko and Mobashery, 2003; Magnet and Blanchard, 2005; Jana and Deb, 2006; Kohanski et al., 2007; Majumder et al., 2007). Aminoglycosides are used to treat infections caused by gram-negative bacilli and, in combination with β-lactams or vancomycin, to treat some gram-positive pathogens, mainly staphylococci (Yao and Moellering, 2007). Since a step in the uptake process requires functional respiration, the spectrum of action of aminoglycosides is limited to aerobic bacteria (Muir et al., 1984). In addition to their most common uses, aminoglycosides can be utilized to treat diseases such as tuberculosis (Menzies et al., 2009; Brossier et al., 2010), plague, tularemia, brucellosis, endocarditis caused by enterococci, and others (Vakulenko and Mobashery, 2003; Yao and Moellering, 2007; Ramirez and Tolmasky, 2010). The fact that aminoglycosides also cause a decrease in eukaryotic translational fidelity permitted to initiate efforts to developed them as drugs to treat nonsense mutation related genetic disorders such as cystic fibrosis and Duchenne muscular dystrophy (Rich et al., 1990; Kellermayer, 2006; Hermann, 2007; Kondo et al., 2007; Zingman et al., 2007; Bidou et al., 2012; Kandasamy et al., 2012). A chemical labyrinthectomy using intratympanic injection of aminoglycosides is used when most treatments of Ménière's disease fail (Huon et al., 2012; Pacheu-Grau et al., 2012). Aminoglycoside-based drugs are also inhibitors of reproduction of the HIV virus, a property that could result in their utilization in the treatment of AIDS patients (Houghton et al., 2010).
The basic chemical structure of aminoglycosides is characterized by the presence of an aminocyclitol nucleus (streptamine, 2-deoxystreptamine, or streptidine) linked to amino sugars through glycosidic bonds. However, other compounds with different basic structures are also included within the aminoglycosides family, e.g., spectinomycin, an aminocyclitol not linked to amino sugars or compounds containing the aminocyclitol fortamine (Bryskier, 2005). They reach the cytoplasm of the bacterial cell in a three-step process, of which the first one is energy-independent and the following two are energy-dependent (Taber et al., 1987; Vakulenko and Mobashery, 2003; Ramirez and Tolmasky, 2010). At the molecular level, the action of aminoglycosides is characterized by interactions between the antibiotic molecule and the 16S rRNA. Although for all aminoglycosides the effect of this interaction is a change of conformation of the decoding A site producing one that resembles the closed state induced by interaction between cognate tRNA and mRNA, it must be noted that not all aminoglycosides seem to bind the same sites of the 16S rRNA. The consequence of the conformational change induced by the interaction 16S rRNA-aminoglycoside is the reduction of the proofreading capabilities of the ribosome, which in turns results in high levels of mistranslation (Bakker, 1992; Busse et al., 1992; Vakulenko and Mobashery, 2003; Vicens and Westhof, 2003; Magnet and Blanchard, 2005; Majumder et al., 2007; Zaher and Green, 2009; Ramirez and Tolmasky, 2010). Other molecular effects of some aminoglycosides have been described but it is not clear if some of them are not secondary to protein mistranslation. They include inhibition of 30S ribosomal subunit assembly, induction of RNA cleavage, or interference with the action of RNase P (Mikkelsen et al., 1999; Mehta and Champney, 2003; Belousoff et al., 2009).
Aminoglycosides are powerful tools against infections (Labaune et al., 2001; Avent et al., 2011) but unfortunately the levels of resistance are growing and in consequence failure of treatments with aminoglycosides is becoming more common (Galani et al., 2002; van ‘t Veen et al., 2005; Tolmasky, 2007a; Soler Bistue et al., 2008). Bacteria have developed numerous mechanisms to resist the action of aminoglycosides and cells can possess the genetic determinants for several of them enhancing the levels of resistance and making it very difficult to overcome all of them. Enzymatic inactivation by acetylation, adenylylation, or phosphorylation at different locations of the aminoglycoside molecule is among the most clinically relevant strategies bacteria use to resist the action of these antibiotics (Shaw et al., 1993; Vakulenko and Mobashery, 2003; Tolmasky, 2007a; Ramirez and Tolmasky, 2010; Chen et al., 2011; Chiang et al., 2013). The enzymes that catalyze these reactions are collectively known as aminoglycoside modifying enzymes. Other well studied mechanisms are: (1) mutation of the 16S rRNA or ribosomal proteins modify the target eliminating or reducing the interaction with the antibiotic molecule (O'Connor et al., 1991); (2) methylation of 16S rRNA, a mechanism found in most aminoglycoside-producing organisms and in clinical strains (Schmitt et al., 2009; Wachino and Arakawa, 2012); (3) reduced permeability to the antibiotic molecule by modification of the permeability of the outer membrane or diminished inner membrane transport (Hancock, 1981; Taber et al., 1987; Macleod et al., 2000; Over et al., 2001); (4) export outside the cell by active efflux pumps (Hocquet et al., 2003; Morita et al., 2012; Wachino and Arakawa, 2012); (5) sequestration by tight binding to a low active aminoglycoside acetyltransferase (Magnet et al., 2003); and (6) extracellular DNA shielding in biofilms (Chiang et al., 2013).
The general characteristics of all known aminoglycoside modifying enzymes have been recently reviewed (Ramirez and Tolmasky, 2010). This review will focus on the aminoglycoside 6′-N-acetyltransferase type Ib [AAC(6′)-Ib], which is of great clinical relevance and it is found in over 70% of AAC(6′)-I-producing gram-negative clinical isolates (Vakulenko and Mobashery, 2003), and has been the subject of numerous studies (Tolmasky, 2007a; Cambray and Mazel, 2008; Ramirez and Tolmasky, 2010).
The AAC(6′)-Ib protein
The aminoglycoside N-acetyltransferases (AAC) belong to the GCN5-related N-acetyltransferase superfamily, also known as GNAT. This is a large group of enzymes that includes about 10,000 proteins from all kinds of organisms that share the property to catalyze the acetylation of a primary amine in numerous acceptor molecules using acetyl CoA as donor substrate (Neuwald and Landsman, 1997; Dyda et al., 2000; Vetting et al., 2005). The AACs are subdivided in groups based on the position where the acetyl group is transferred in the acceptor aminoglycoside molecule. Known AACs catalyze acetylation at the 1 [AAC(1)], 3 [AAC(3)], 2′ [AAC(2′)], or 6′ [AAC(6′)] positions (Shaw et al., 1993; De Pascale and Wright, 2010; Ramirez and Tolmasky, 2010). AAC(6′) enzymes are the most numerous group of AACs, more than 40 have been described, and can be found in gram-negatives as well as gram-positives (Shaw et al., 1993; Miller et al., 1997; Wright, 1999; Tolmasky, 2007a; Ramirez and Tolmasky, 2010). AAC(6′) enzymes are subdivided in two groups, AAC(6′)-I and AAC(6′)-II, which are differentiated by the profile of the aminoglycosides inactivated. With a few exceptions, AAC(6′)-I enzymes specify resistance to several aminoglycosides plus amikacin and gentamicin C1a and C2 but not to gentamicin C1 (Shaw et al., 1993). On the other hand, AAC(6′)-II enzymes catalyze acetylation of all forms of gentamicin but not of amikacin (Rather et al., 1992). In addition, enzymes with extended spectrum that may merit addition of new subclasses of AAC(6′)-I enzymes have been recently described (Casin et al., 2003; Robicsek et al., 2006; Strahilevitz et al., 2009). Phylogenetic analyses divided the AAC(6′) enzymes into three clades. However, with the information available it is still not clear if all AAC(6′) enzymes evolved from a single origin or the three groups are less related and the 6′ acetylating activity has evolved independently at least three times (Salipante and Hall, 2003). According to the phylogenetic analyses recently communicated by Salipante and Hall (Salipante and Hall, 2003) the AAC(6′)-Ib is most closely related to AAC(6′)-IIa AAC(6′)-IIb, AAC(6′)-IIc, and AAC(6′)-IId.
There are numerous variants of AAC(6′)-Ib, many of them identified by modifications in the name of the enzyme such as the addition of subscripts or a prime symbol superscript (Cambray and Mazel, 2008; Ramirez and Tolmasky, 2010). However, a large number of versions of the protein, or predicted protein, have all been named AAC(6′)-Ib, which can be a source of confusion or indetermination. These variants mainly differ at the N-terminus, however one should be careful when considering these differences because not in all of them the N-terminus has been experimentally determined (Dery et al., 1997; Casin et al., 2003; Soler Bistue et al., 2006; Maurice et al., 2008). Table 1 shows a list of the aac(6′)-Ib gene versions found in different genetic environments and bacterial species. Some variants differing at the N-termini such as AAC(6′)-Ib3, AAC(6′)-Ib4, AAC(6′)-Ib6, and AAC(6′)-Ib7 have been compared and it was found that they have similar behavior (Casin et al., 1998) but variations as small as one or two amino acids at key positions proved to be of high relevance (Table 2). For example, the AAC(6′)-Ib11 found in S. Typhimurium has L and S residues at positions 118 and 119 as opposed to Q and L or Q and S, the amino acids present at these positions in all previously described enzymes, acquired an extended resistance spectrum that includes all three gentamicin forms (Casin et al., 2003). (Amino acid numbers throughout the text are based on the sequence corresponding to accession number AF479774.) Another example worth mentioning is the AAC(6′)-Ib', originally found in Pseudomonas fluorescens BM2687, but previously generated by site-directed mutagenesis in the laboratory (Table 1). This protein has a L to S substitution at amino acid 119 that confers the enzyme an AAC(6′)-II profile, i.e., the enzyme confers resistance to gentamicin but not amikacin (Rather et al., 1992; Lambert et al., 1994). A highly surprising effect occurred in the natural variant known as AAC(6′)-Ib-cr, which has the modifications W104R and N181Y (Tables 1, 2). The substrate spectrum was expanded to include quinolone antibiotics, crossing the barrier from the aminoglycosides (Robicsek et al., 2006). Since the first detection of the AAC(6′)-Ib-cr variant there have been numerous reports of its presence, and variants of it, across the world in different genetic environments suggesting an extraordinary ability to disseminate (Quiroga et al., 2007; Cattoir and Nordmann, 2009; Strahilevitz et al., 2009; Rodriguez-Martinez et al., 2011; Ruiz et al., 2012; De Toro et al., 2013). Furthermore, there have been cases where a strain was found to simultaneously include genes coding for AAC(6′)-Ib and AAC(6′)-Ib-cr (Kim et al., 2011). AAC(6′)-Ib is also found fused to the C-terminal end of AAC(3)-Ib protein within a class I integron found in a Pseudomonas aeruginosa strain (Dubois et al., 2002) and to the C-terminus of the AAC(6′)-30 also within a P. aeruginosa class I integron (Mendes et al., 2004).
Table 1.
AAC(6′)-Ib variants.
| Number | AAC(6′)-Ib enzyme | Gene allele | Genetic localization | Species | Reference |
|---|---|---|---|---|---|
| 1 | NP_608307 | aac(6′)-Ib | pJHCMW1::Tn1331, | K. pneumoniae, En. spp., | NP_608307, |
| pKPN4, | Pseudomonas putida, | YP_001338668, | |||
| pMET1::Tn1331.2, | Proteus mirabilis | YP_001928078, | |||
| pKlebpneu15S, | YP_001928081, | ||||
| pR23::Tn1331, | YP_002286819, | ||||
| pAAC154:: ΔTn1331, | YP_004455304, | ||||
| pColEST258, | YP_006958960, | ||||
| pJHC-MW1, | YP_006959190, | ||||
| Tn1332, | ZP_14492679 (contig), | ||||
| class 1 integron, | ZP_14498301 (contig), | ||||
| pRMH712::Tn1331, | ZP_14503930 (contig), | ||||
| SGI1-V::class 1 integron | ZP_14509538 (contig), | ||||
| ZP_14515174 (contig), | |||||
| ZP_14520767 (contig), | |||||
| ZP_14526392 (contig), | |||||
| ZP_14531781 (contig), | |||||
| ZP_14537604 (contig), | |||||
| ZP_14543183 (contig), | |||||
| ZP_14548764 (contig), | |||||
| ZP_14554292 (contig), | |||||
| ZP_14559831 (contig), | |||||
| ZP_14565429 (contig), | |||||
| ZP_14571055 (contig), | |||||
| ZP_14576504 (contig), | |||||
| ZP_14581777 (contig), | |||||
| ZP_14587733 (contig), | |||||
| ZP_14593028 (contig), | |||||
| ZP_14598930 (contig), | |||||
| ZP_19010829 (contig), | |||||
| AAC6_KLEPN, | |||||
| AF479774_5, AAA69747 | |||||
| AAA98404, ABA54975, | |||||
| ABR80438, ACB55476, | |||||
| ACB55479, ACI63081, | |||||
| ACL36604, ADK35766, | |||||
| AED98720, AED99555, | |||||
| AEG74535, AEW43367, | |||||
| EJJ31842 (contig), | |||||
| EJJ31842 (contig), | |||||
| EJJ31884 (contig), | |||||
| EJJ31888 (contig), | |||||
| EJJ48672 (contig), | |||||
| EJJ48714 (contig), | |||||
| EJJ49532 (contig), | |||||
| EJJ65671 (contig), | |||||
| EJJ65889 (contig), | |||||
| EJJ68295 (contig), | |||||
| EJJ79581 (contig), | |||||
| EJJ81436 (contig), | |||||
| EJJ85464 (contig), | |||||
| EJJ96302 (contig), | |||||
| EJJ96595 (contig), | |||||
| EJK03278 (contig), | |||||
| EJK13161 (contig), | |||||
| EJK16025 (contig), | |||||
| EJK18863 (contig), | |||||
| EJK30890 (contig), | |||||
| EJK33635 (contig), | |||||
| EKV58688 (contig) | |||||
| 2 | YP_002286969 | aac(6′)-Ib | p12::Tn1331 | K. pneumoniae, E. coli | YP_002286969, |
| ZP_16459764 (genomic scaffold), ZP_19016755 (contig), ACI63027, | |||||
| EGB78408 (contig), | |||||
| EKV58524 (contig) | |||||
| 3 | AAA26550 | aac(6′)-Ib | pAZ007 | Serratia marcescens | AAA26550 |
| 4 | AAR18814 | aac(6′)-Ib | pKP31::class 1 integron | K. pneumoniae | AAR18814 |
| 5 | CBI63199 | aac(6′)-Ib | Class 1 integron | P. aeruginosa | CBI63199 |
| 6 | CBI63201 | aac(6′)-Ib | Class 1 integron | P. aeruginosa | CBI63201 |
| 7 | CBI63203 | aac(6′)-Ib | Class 1 integron | P. aeruginosa | CBI63203 |
| 8 | ABG77519 | aac(6′)-Ib | Class 1 integron | P. aeruginosa | ABG77519 |
| 9 | CBL95252 | aac(6′)-Ib | Class 1 integron | P. aeruginosa | CBL95252 |
| 10 | CBL95256 | aac(6′)-Ib | Class 1 integron | P. aeruginosa | CBL95256 |
| 11 | CBI63204 | aac(6′)-Ib | Class 1 integron | P. aeruginosa | CBI63204 |
| 12 | CBI63202 | aac(6′)-Ib | Class 1 integron | P. aeruginosa | CBI63202 |
| 13 | YP_003937697 | aac(6′)-Ib | pETN48:: Δclass 1 integron | E. coli | YP_003937697, |
| CBX36023 | |||||
| 14 | ADC80806 | aac(6′)-Ib | pRYC103T24::class 1 integron In4-like, | E. coli, uncultured bacterium | ADC80806, AFR44153 |
| pKSP212::class 1 integron | |||||
| 15 | YP_005797131 | aac(6′)-Ib | Class 1 integron (Chromosome) | A. baumannii | YP_005797131, |
| AEN92376 | |||||
| 16 | YP_005525242 | aac(6′)-Ib | Class 1 integron (Chromosome) | A. baumannii | YP_005525242, |
| YP_006289231, | |||||
| YP_006848983, | |||||
| ZP_11603605 (contig), | |||||
| ZP_16142456 (contig), | |||||
| ZP_16146111 (contig), | |||||
| EGK45756 (seq0044), | |||||
| AEP05746, AFI94936, | |||||
| EKE64317 (contig), | |||||
| EKE64588 (contig), | |||||
| AFU38752 | |||||
| 17 | NP_863005 | aac(6′)-Ib | p1658/97::class 1 integron, class 1 integron (Chromosome), class 1 integron, plasmid In238a | E. coli, A. baumannii, | NP_863005, |
| K. pneumoniae, | YP_001844882, | ||||
| K. oxytoca, En. cloacae | AAO49600, ACZ55927, | ||||
| ACZ64698, AFS33307 | |||||
| 18 | ADC80825 | aac(6′)-Ib | pRYC103T24::class 1 integron | E. coli | ADC80825 |
| 19 | YP_002791392 | aac(6′)-Ib | pEC-IMP::class 1 integron, | En. cloacae, Salmonella enterica subsp. enterica serovar Bredeney, P. aeruginosa | YP_002791392, |
| pEC-IMPQ::class 1 integron, pb1004::class 1 integron, class 1 integron | YP_002791702, | ||||
| ACF59628, ACO54016, | |||||
| ACO54326, ADF47469 | |||||
| 20 | AEO50496 | aac(6′)-Ib | Class 1 integron | Se. marcescens | AEO50496 |
| 21 | ACB41759 | aac(6′)-Ib | Class 1 integron | E. coli | ACB41759 |
| 22 | BAL45797 | aac(6′)-Ib | pKPI-6::class 1 integron | K. pneumoniae | BAL45797 |
| 23 | AAC46343 | aac(6′)-Ib | Class 1 integron | P. aeruginosa | AAC46343 |
| 24 | AAD02244 | aac(6′)-Ib9 | Class 1 integron | P. aeruginosa | AAD02244 |
| 25 | YP_003108195 | aac(6′)-Ib-cr | pEK516, pEK499, | E. coli, K. pneumoniae | YP_003108195, |
| pEC_L8, pUUH239.2 | YP_003108338, | ||||
| YP_003829182, | |||||
| YP_005351453, | |||||
| ACQ41894, ACQ42045, | |||||
| ADL14076, AET17280 | |||||
| 26 | ZP_18354173 | aac(6′)-Ib-cr | K. pneumoniae | ZP_18354173 (genomic scaffold), EKF76226 | |
| 27 | ACD56150 | aac(6′)-Ib-cr | pHS1387::class 1 integron | Escherichia coli | ACD56150 |
| 28 | ADY02579 | aac(6′)-Ib-cr | Class 1 integron | Aeromonas media | ADY02579 |
| 29 | NP_957555 | aac(6′)-Ib-cr | pC15-1a, pKP96::class 1 | Escherichia coli, | NP_957555, |
| integron, pNDM-MAR, | K. pneumoniae, Kluyvera | YP_002332851, | |||
| pGUE-NDM, pKDO1, | ascorbata, mixed culture | YP_005352168, | |||
| pHe96, pKas96, | bacterium, K. oxytoca, | YP_006953881, | |||
| pECZ6-1::class 1 | Se. rubidaea, En. cloacae, | YP_006973732, | |||
| integron, Class 1 | Aeromonas | AAR25030, ABC17627, | |||
| integron, pLC108::class 1 | allosaccharophila, | ABM47029, ABY74389, | |||
| integron, pJIE101, | Providencia spp., Shigella spp., En. aerogenes | ACD03312, ACD03322, | |||
| ACM24788, ACT97328, | |||||
| ACT97332, ACT97345, | |||||
| ACT97681, ACV60575, | |||||
| ADA60222, ADE44336, | |||||
| ADP30789, ADU16107, | |||||
| ADU16118, ADY02556, | |||||
| AEC49701, AEC49704, | |||||
| AEL33522, AEO45791, | |||||
| AEO79936, AEO79967, | |||||
| AEP16466, AER36609, | |||||
| AEU10750, AEU10754, | |||||
| AFB82784, AFC38861, | |||||
| AFI72862, AFV52812, | |||||
| AFV70394 | |||||
| 30 | 1V0C_A | aac(6′)-Ib | Escherichia coli Chain A, Structure | 1V0C_A, 2BUE_A, | |
| 2VQY_A | |||||
| 31 | YP_006501621 | pKOX_R1::class 1 integron, class 1 integorn, | K. oxytoca, | YP_006501621, | |
| K. pneumoniae | AFM57748, AFN35014 | ||||
| 32 | ABC54722 | aac(6′)-Ib | pAS1::InVC117 | Vibrio cholerae | ABC54722 |
| 33 | BAE66666 | aac(6′)-Ib | Class 1 integron | Vibrio cholerae O1 | BAE66666 |
| 34 | YP_007232190 | aac(6′)-Ib | pPC9 | P. putida | YP_007232190 |
| 35 | ZP_16084267 | aac(6′)-Ib | Class 1 integron (Chromosome) | A. baumannii | ZP_16084267 (contig), |
| ZP_16086960 (contig), | |||||
| ZP_16140385 (contig), | |||||
| EKA73751 (contig), | |||||
| EKK08901 (contig), | |||||
| EKK18976 (contig) | |||||
| 36 | AFS51540 | aac(6′)-Ib9 | pKS208::class 1 integron | Uncultured bacterium | AFS51540 |
| 37 | YP_006957899 | aac(6′)-Ib-cr4 | pMdT | SS. enterica subsp. enterica serovar Typhimurium | YP_006957899, |
| AFU63391 | |||||
| 38 | ADZ96942 | aac(6′)-Ib-cr | Plasmid | K. pneumoniae | ADZ96942 |
| 39 | CAA42873 | aac(6′)-Ib3, aac(6′)-Ib5 | plasmid pCFF04 | P. aeruginosa | CAA42873 |
| 40 | AAB24284 | aac(6′)-Ib4 | pSP21::class 1 integron, | Se. spp., uncultured bacterium, En. cloacae | AAB24284, |
| pEl1573::class 1 integron | YP_006941442, | ||||
| YP_006965430 | |||||
| 41 | AAN41403 | aac(6′)-Ib11 | pSTI1::class 1 integron | SS. enterica subsp. enterica serovar Typhimurium | AAN41403 |
| 42 | YP_006903338 | aac(6′)-Ib | pNDM102337::class 1 integron, | Escherichia coli | YP_006903338, |
| pNDM10505::class 1 integron | YP_006953195 | ||||
| 43 | YP_006959139 | aac(6′)-Ib | pNDM10469::class 1 integron | K. pneumoniae | YP_006959139 |
| 44 | aac(6′)-Ib7 | Plasmid | En. cloacae, Citrobacter freundii | Not available | |
| 45 | aac(6′)-Ib8 | Plasmid | En. cloacae | Not available |
The pJHCMW1-encoded AAC(6′)-Ib variant (accession number NP_608307) was subjected to BLASTP and the identical proteins were identified. Those that were closely related but not identical were identified by numbers. The names given in the publications or GenBank entries are shown. Those that were named aacA4 were named aac(6′)-Ib here.
Table 2.
Phenotypes of representative mutants of AAC(6′)-Ib.
| Mutationa | AAC(6′)-Ib variant name | Phenotype | References |
|---|---|---|---|
| Y80C | S | Panaite and Tolmasky, 1998 | |
| D117A | S | Pourreza et al., 2005 | |
| L119S | AAC(6′)-Ib′, | Specificity, Gmr Aks | Rather et al., 1992; |
| AAC(6′)-Ib7, | Lambert et al., 1994; | ||
| AAC(6′)-Ibb8 | Casin et al., 2003 | ||
| Q118L, L119S | AAC(6′)-Ib11 | Specificity, Gmr Akr | Casin et al., 2003 |
| L120A | S | Pourreza et al., 2005 | |
| Y166A | Specificity, Aks Kmr | Shmara et al., 2001 | |
| E167A | S | Shmara et al., 2001 | |
| F171A | S | Shmara et al., 2001 | |
| F171L | Thermosensitive for Ak and Nm | Panaite and Tolmasky, 1998; Shmara et al., 2001 | |
| W104R, D181Y | AAC(6′)-Ib-cr | Expanded substrate spectrum including quinolones | Robicsek et al., 2006 |
S, susceptible.
Numbering from sequence in accession number AF479774.
The proteins differ at the amino terminus.
Subcellular localization studies of the AAC(6′)-Ib enzyme encoded by Tn1331 showed that the enzyme is homogeneously distributed in the cytoplasmic compartment (Dery et al., 2003). AAC(6′)-Ib was one of three aminoglycoside modifying enzymes used in a study consisting of molecular dynamics simulations of the enzymes and aminoglycoside ribosomal RNA binding site, unliganded, and complexed with an aminoglycoside, kanamycin A. These studies concluded that the enzymes efficiently mimic the nucleic acid environment of the ribosomal RNA binding cleft (Romanowska et al., 2013). Extensive studies using mutagenesis showed some interesting phenotypes such as modifications in specificity, enhanced activity, or selective thermosensitivity (Table 2) (Panaite and Tolmasky, 1998; Chavideh et al., 1999; Shmara et al., 2001; Casin et al., 2003; Pourreza et al., 2005; Kim et al., 2007; Maurice et al., 2008). In addition, alanine scanning showed that several amino acid substitutions by A had minor effects. These mutagenesis studies together with structural and enzymatic analyses led to a deep understanding of features and characteristics of AAC(6′)-Ib proteins (Rather et al., 1992; Vetting et al., 2004; Maurice et al., 2008; Vetting et al., 2008; Ramirez and Tolmasky, 2010). The three dimensional structure of AAC(6′)-Ib and AAC(6′)-Ib11 have been experimentally determined in various conditions. AAC(6′)-Ib was crystallized in complex with coenzyme A and also in complex with both coenzyme A and kanamycin. The structures were solved to 1.8 Å and 2.4 Å resolution, respectively (Maurice et al., 2008). The broad spectrum variant AAC(6′)-Ib11 was crystallized in the absence of substrate and the structure was solved to 2.1 Å resolution (Maurice et al., 2008). These studies concluded that AAC(6′)-Ib exists as a monomer while AAC(6′)-Ib11 shows monomer/dimer equilibrium (Maurice et al., 2008). This was a somewhat surprising finding considering that previous studies had shown that two other acetyltransferases, AAC(6′)-Ii and AAC(6′)-Iy, exist as dimers (Wright and Ladak, 1997; Wybenga-Groot et al., 1999; Draker et al., 2003; Vetting et al., 2004; Wright and Berghuis, 2007; Vong et al., 2012). Interestingly, analysis of these crystal structures showed the presence of a flexible flap in AAC(6′)-Ib11 that may be the basis for its ability to utilize amikacin as well as gentamicin as substrates (Maurice et al., 2008). In another study a molecular model of AAC(6′)-Ib-cr has been generated (Maurice et al., 2008; Vetting et al., 2008), which led to postulate that the D181Y substitution is mainly responsible for modification in the strength of binding of the antibiotic substrate and that the substitution W104R stabilizes the positioning of Y181 (Robicsek et al., 2006; Strahilevitz et al., 2009).
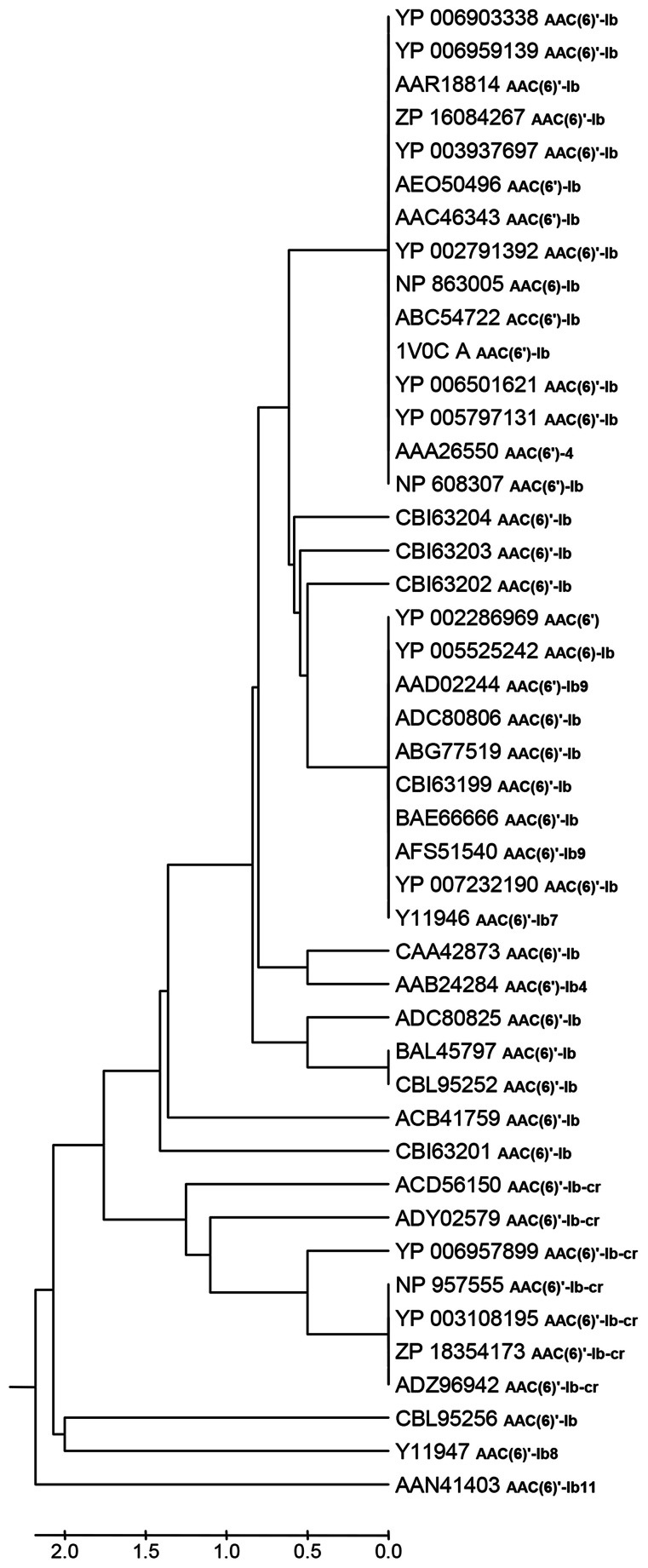
Table 1 shows that there are 45 non-identical AAC(6′)-Ib related entries in the NCBI database, 32 of which have identical name in spite of not having identical amino acid sequence. The N-termini of these proteins show the highest degree of heterogeneity with high variations in length stretching up to 60 amino acids, but these differences were suggested to be irrelevant (Casin et al., 1998; Maurice et al., 2008). Therefore, we defined a highly conserved central region composed of 181 amino acids shared by all proteins, which were compared using the MAFFT alignment algorithm (Katoh and Standley, 2013). Pairwise comparisons show that the sequences have 1 to 8 amino acid differences and a total of 24 positions showed amino acid variations. Moreover, clustering using the UPGMA algorithm (Sneath and Sokal, 1973) defined 18 sequence clusters, 14 of which consist of a singleton, and 4 of which include 2–16 proteins (Figure 1). Different clusters can exhibit similar properties while others show substantial differences in their characteristics such as those cases in which there are significant specificity variations like extended substrate spectrum as described in the above paragraphs.
Figure 1.
UPGMA clustering analyses of 45 AAC(6′)-Ib protein sequences. The optimal tree with the sum of branch length = 20.70628249 is shown. The evolutionary distances were computed using the number of differences method and are in the units of the number of amino acid differences per sequence. All positions containing gaps and missing data were eliminated. There were a total of 181 positions in the final dataset. Evolutionary analyses were conducted in MEGA5.
The aac(6′)-Ib gene
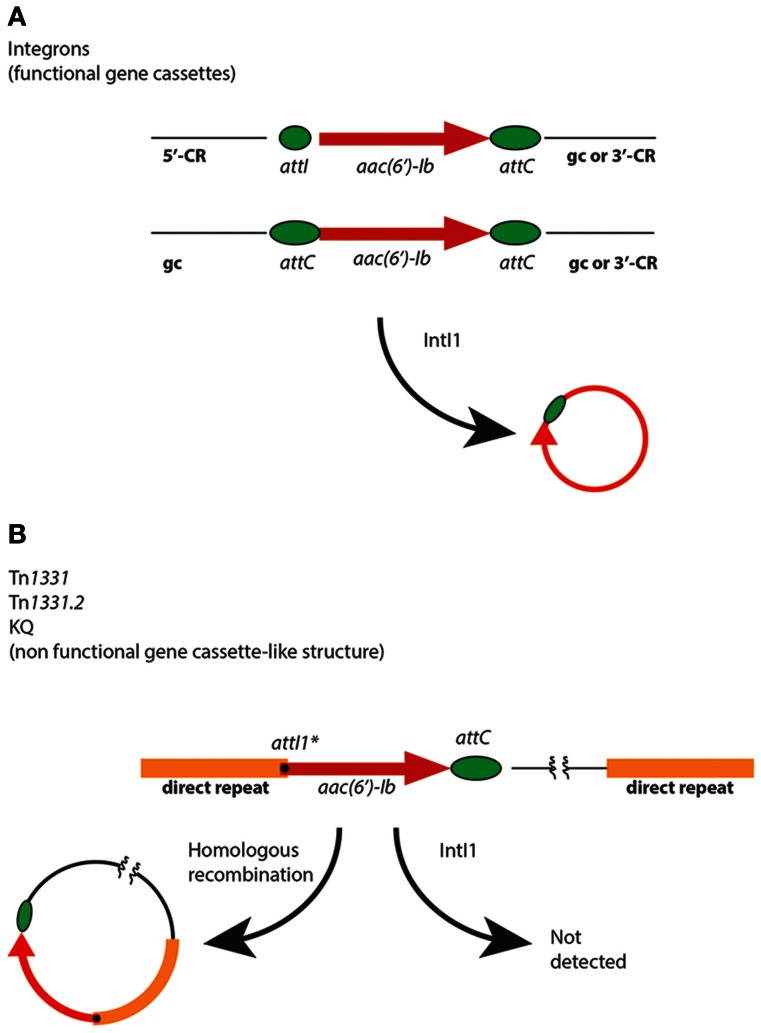
The aac(6′)-Ib genes are usually found as fully functional or deficient gene cassettes associated to class 1 integrons, insertion sequences such as IS26, and truncated or disrupted integrons (Figure 2 and Table 1) (Sarno et al., 2002; Woodford et al., 2009; Ramirez and Tolmasky, 2010). These genetic elements may be part of plasmids, transposons, genomic islands, or other structures such as the KQ element (Rice et al., 2008), which together contribute to the gene's ability to disseminate at the cellular and molecular levels (Tolmasky, 2007b). When present in integrons, aac(6′)-Ib gene cassettes can be found located adjacent to the 5′-conserved region, i.e., flanked by attI and attC, or internal to the variable portion containing attC loci at both ends (Figure 2). In both cases, as expected, the gene cassette can be mobilized by the integrase IntI1 (Figure 2) (Cambray et al., 2010; Hall, 2012). In addition, a gene cassette-like structure containing aac(6′)-Ib, composed of a copy of attI1* at the beginning of the structural gene and a regular attC downstream of it (see Figure 2), was found as part of a region resembling the variable portion of integrons in Tn1331 (Woloj et al., 1986; Tolmasky et al., 1988; Tolmasky, 1990; Tolmasky and Crosa, 1993; Sarno et al., 2002), Tn1331.2 (Tolmasky and Crosa, 1991), Tn1332 (Poirel et al., 2006), the KQ element (Rice et al., 2008), a Tn1331 derivative recently isolated from a clinical Klebsiella pneumoniae strain belonging to the ST512, which derived from the ST258, known to be spread worldwide (Chen et al., 2012; Garcia-Fernandez et al., 2012; Warburg et al., 2012), and a complex mosaic region present in the chromosome of Proteus mirabilis JIE273 (Zong et al., 2009). Assays overexpressing IntI1 in cells containing Tn1331 were unable to detect any excision of the aac(6′)-Ib gene cassette-like structure, suggesting that it is nonfunctional or it excises at an extremely low efficiency (Ramirez et al., 2008). Interestingly, despite the IntI1-mediated lack of mobility of this DNA region, the gene could be mobilized by means of a mechanism recently proposed by Zong et al. that occurs through homologous recombination between 520-bp direct repeats located upstream and downstream of the gene cassette-like structure (Figure 2) (Zong et al., 2009).
Figure 2.
Mobilization of aac(6′)-Ib. (A) Generic genetic maps of integrons in which an aac(6′)-Ib gene cassette is located immediately following the 5′ conserved region (5′-CR) (top map) or following one or more gene cassettes (gc) inside the variable portion, and followed by other gene cassettes or the 3′ conserved region (3′-CR) (bottom map). The small green ellipse represents attI and the big green ellipses represent attC. (B) Relevant portion of the Tn1331, Tn1331.2, and KQ elements (Tolmasky and Crosa, 1987; Tolmasky et al., 1988; Sarno et al., 2002; Rice et al., 2008). For clarity Tn1332, which has a more complicated structure in its direct repeats (Poirel et al., 2006), is not shown, but it could experience mobilization by homologous recombination as shown. The black dot represents attI1*. The homologous recombination pathway for generation of an aac(6′)-Ib-containing circular molecule has been proposed by Zong et al. (2009).
These multiple locations taken together with the ability of the genetic elements to spread at the molecular and cellular level provide aac(6′)-Ib genes with the capability to reach virtually all gram-negatives and other undetermined bacteria such as those that are still unculturable. The gene has also been found in plasmids harboring resistance genes of high importance such as the recently described ndm-1 (Yong et al., 2009; Bonnin et al., 2012; Villa et al., 2012).
Inhibition
The rise in drug resistance affects all known antibiotics and has been identified as one of the greatest threats to human health. Therefore, there is an immediate need for new agents with activity against multiresistant bacteria and at the present moment there is no evidence that this need will be fully met in the near future (Boucher et al., 2009). A group of pathogens that cause the majority of hospital infections, named ESKAPE (Enterococcus faecium, Staphylococcus aureus, K. pneumoniae, Acinetobacter baumannii, P aeruginosa, and Enterobacter), is becoming highly resistant to antibiotics including aminoglycosides (Rice, 2008). They carry aminoglycoside modifying enzymes genes, and one of the most common in the gram-negative members is aac(6′)-Ib (Ramirez and Tolmasky, 2010; Shaul et al., 2011; Herzog et al., 2012). An obvious solution to this problem would be the development of new aminoglycosides, a strategy that is being pursued using numerous approaches (Green et al., 2010, 2011; Houghton et al., 2010). A variety of new aminoglycoside derivatives including chemical modification of existing aminoglycosides, aminoglycoside dimers, or aminoglycoside-small molecule conjugated are being produced and tested (reviewed in Houghton et al., 2010). In particular plazomicin (ACHN-490), a novel neoglycoside derived from sisomicin that carries a hydroxymethyl group at position 6′, has shown enhanced activity against multiresistance gram-negatives and gram-positives including strains carrying aac(6′)-Ib (Endimiani et al., 2009; Landman et al., 2011).
Others are approaching the problem in such a way that the existing aminoglycosides continue to be effective by designing enzymatic inhibitors that can act in combination with the antibiotic, mimicking the strategy successfully used to curb resistance to β-lactams (Williams and Northrop, 1979; Daigle et al., 1997; Haddad et al., 1999; Liu et al., 2000; Burk and Berghuis, 2002; Boehr et al., 2003; Draker et al., 2003; Gao et al., 2005, 2006; Welch et al., 2005; Lombes et al., 2008; Magalhaes et al., 2008; De Pascale and Wright, 2010; Drawz and Bonomo, 2010; Green et al., 2012; Vong et al., 2012). However, these efforts are still scarce when one compares them to those invested to discover and design β-lactamase inhibitors. Furthermore, the attempts to find inhibitors of AAC(6′)-Ib have only yielded a compound, synthesized using non-aminoglycoside-like fragments, with a rather modest level of inhibition of AAC(6′)-Ib (Lombes et al., 2008).
An alternative approach that is being explored is silencing expression of the resistance gene. Early attempts at interfering with expression of aac(6′)-Ib consisted of identifying regions available for interaction with antisense oligonucleotides in a monocistronic in vitro synthesized mRNA by RNase H mapping in combination with computer prediction of its secondary structure (Sarno et al., 2003). The selected sites were used as targets for a collection of oligodeoxynucleotides, of which some had the ability to induce RNase H-mediated in vitro degradation of the mRNA, inhibited in vitro synthesis of the enzyme in coupled transcription/translation assays, and upon delivery by electroporation significantly reduced the number of cells surviving after exposure to amikacin (Sarno et al., 2003). The mechanism of this in vivo inhibition is most probably through RNase H digestion of the mRNA, but other possibilities such as steric hindrance cannot be discarded at this time. Alternatively, modest but significant inhibition of expression of aac(6′)-Ib was achieved by applying EGS technology, in which short antisense RNA molecules, known as external guide sequences, are used to elicit RNase P-mediated degradation of a target mRNA (Guerrier-Takada et al., 1997; Lundblad and Altman, 2010). Initially, E. coli harboring aac(6′)-Ib were transformed with recombinant clones specifying the appropriate RNA oligonucleotide sequences under an inducible promoter. The transformed derivatives were then cultured in the presence of amikacin under conditions of expression of the external guide sequences. The results showed that in a few cases the external guide sequences induced a reduction of the minimal inhibitory concentration of amikacin (Soler Bistue et al., 2007). These results were considered proof of concept, but the strategy was not viable because antisense oligonucleotides must be added from the milieu and find their way inside the cells without being degraded. Thus, nuclease resistant oligonucleotide analogs that still induce inhibition of gene expression by RNase P activation had to be found. Out of a variety of oligoribonucleotide analogs including 2′-O-methyl oligoribonucleotides, phosphorodiamidate morpholino oligomers, phosphorothioate oligodeoxynucleotides, or locked nucleic acids (LNA)/DNA co-oligomers that were tested, LNA/DNA co-oligomers with certain configurations were found to be capable of eliciting RNase P-mediated cleavage of mRNA in vitro (Soler Bistue et al., 2009). Following this finding, a selected LNA/DNA co-oligomer was added to the hyperpermeable E. coli AS19 harboring aac(6′)-Ib and it was found that growth was inhibited in the presence of amikacin, indicating that the compound may have induced RNase P-mediated inhibition of expression of the gene (Soler Bistue et al., 2009). These results were encouraging but it must be noted that inhibition of expression of aac(6′)-Ib is still far from being a viable option to overcome aminoglycoside resistance in the clinical setting. Several problems remain to be solved like inducing penetration of the oligonucleotide analogs inside wild type cells in enough quantities to exert the biological activity or achieve enough inhibition levels in spite of the usual presence of multiple copies of the gene due to its inclusion in high copy number plasmids. Toward finding solutions to these problems, recent experiments suggest that LNA/DNA co-oligomers may be able to reach the cytoplasm of untreated cells at low efficiency (Traglia et al., 2012). Strategies will have to be developed to increase the efficiency of delivery inside bacterial cells.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors' work cited in this review article was funded by Public Health Service grant 2R15AI047115 (to Marcelo E. Tolmasky) from the National Institutes of Health. María S. Ramirez is a research career member of C.O.N.I.C.E.T.
References
- Avent M. L., Rogers B. A., Cheng A. C., Paterson D. L. (2011). Current use of aminoglycosides: indications, pharmacokinetics and monitoring for toxicity. Intern. Med. J. 41, 441–449 10.1111/j.1445-5994.2011.02452.x [DOI] [PubMed] [Google Scholar]
- Bakker E. P. (1992). Aminoglycoside and aminocyclitol antibiotics: hygromycin B is an atypical bactericidal compound that exerts effects on cells of Escherichia coli characteristics for bacteriostatic aminocyclitols. J. Gen. Microbiol. 138, 563–569 [DOI] [PubMed] [Google Scholar]
- Belousoff M. J., Graham B., Spiccia L., Tor Y. (2009). Cleavage of RNA oligonucleotides by aminoglycosides. Org. Biomol. Chem. 7, 30–33 10.1039/b813252f [DOI] [PubMed] [Google Scholar]
- Bidou L., Allamand V., Rousset J. P., Namy O. (2012). Sense from nonsense: therapies for premature stop codon diseases. Trends Mol. Med. 18, 679–688 10.1016/j.molmed.2012.09.008 [DOI] [PubMed] [Google Scholar]
- Boehr D. D., Draker K. A., Koteva K., Bains M., Hancock R. E., Wright G. D. (2003). Broad-spectrum peptide inhibitors of aminoglycoside antibiotic resistance enzymes. Chem. Biol. 10, 189–196 10.1016/S1074-5521(03)00026-7 [DOI] [PubMed] [Google Scholar]
- Bonnin R. A., Poirel L., Carattoli A., Nordmann P. (2012). Characterization of an IncFII plasmid encoding NDM-1 from Escherichia coli ST131. PLoS ONE 7:e34752 10.1371/journal.pone.0034752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher H. W., Talbot G. H., Bradley J. S., Edwards J. E., Gilbert D., Rice L. B., et al. (2009). Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48, 1–12 10.1086/595011 [DOI] [PubMed] [Google Scholar]
- Brossier F., Veziris N., Aubry A., Jarlier V., Sougakoff W. (2010). Detection by Geno- type MTBDRsl test of complex resistance mechanisms to second-line drugs and ethambutol in multidrug-resistant Mycobacterium tuberculosis complex isolates. J. Clin. Microbiol. 48, 1683–1689 10.1128/JCM.01947-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryskier A. (2005). Antibiotics and antibacterial agents: classifications and structure- activity relationships, in Antimicrobial Agents, ed Bryskier A. (Washington, DC: ASM Press; ), 13–38 [Google Scholar]
- Burk D. L., Berghuis A. M. (2002). Protein kinase inhibitors and antibiotic resistance. Pharmacol. Ther. 93, 283–292 10.1016/S0163-7258(02)00197-3 [DOI] [PubMed] [Google Scholar]
- Busse H. J., Wostmann C., Bakker E. P. (1992). The bactericidal action of streptomycin: membrane permeabilization caused by the insertion of mistranslated proteins into the cytoplasmic membrane of Escherichia coli and subsequent caging of the antibiotic inside the cells due to degradation of these proteins. J. Gen. Microbiol. 138, 551–561 [DOI] [PubMed] [Google Scholar]
- Cambray G., Guerout A. M., Mazel D. (2010). Integrons. Annu. Rev. Genet. 44, 141–166 10.1146/annurev-genet-102209-163504 [DOI] [PubMed] [Google Scholar]
- Cambray G., Mazel D. (2008). Synonymous genes explore different evolutionary landscapes. PLoS Genet. 4:e1000256 10.1371/journal.pgen.1000256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casin I., Bordon F., Bertin P., Coutrot A., Podglajen I., Brasseur R., et al. (1998). Aminoglycoside 6′-N-acetyltransferase variants of the Ib type with altered substrate profile in clinical isolates of Enterobacter cloacae and Citrobacter freundii. Antimicrob. Agents Chemother. 42, 209–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casin I., Hanau-Bercot B., Podglajen I., Vahaboglu H., Collatz E. (2003). Salmonella enterica serovar Typhimurium bla(PER-1)-carrying plasmid pSTI1 encodes an extended-spectrum aminoglycoside 6′-N-acetyltransferase of type Ib. Antimicrob. Agents Chemother. 47, 697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoir V., Nordmann P. (2009). Plasmid-mediated quinolone resistance in gram-negative bacterial species: an update. Curr. Med. Chem. 16, 1028–1046 10.2174/092986709787581879 [DOI] [PubMed] [Google Scholar]
- Chavideh R., Sholly S., Panaite D., Tolmasky M. E. (1999). Effects of F171 mutations in the 6′-N-acetyltransferase type Ib [AAC(6′)-Ib] enzyme on susceptibility to aminoglycosides. Antimicrob. Agents Chemother. 43, 2811–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Chavda K. D., Mediavilla J. R., Zhao Y., Fraimow H. S., Jenkins S. G., et al. (2012). Multiplex real-time PCR for detection of an epidemic KPC-producing Klebsiella pneumoniae ST258 clone. Antimicrob. Agents Chemother. 56, 3444–3447 10.1128/AAC.00316-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Biswas T., Porter V. R., Tsodikov O. V., Garneau-Tsodikova S. (2011). Unusual regioversatility of acetyltransferase Eis, a cause of drug resistance in XDR-TB. Proc. Natl. Acad. Sci. U.S.A. 108, 9804–9808 10.1073/pnas.1105379108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang W. C., Nilsson M., Jensen P. O., Hoiby N., Nielsen T. E., Givskov M., et al. (2013). Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 57, 2352–2361 10.1128/AAC.00001-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle D. M., McKay G. A., Wright G. D. (1997). Inhibition of aminoglycoside antibiotic resistance enzymes by protein kinase inhibitors. J. Biol. Chem. 272, 24755–24758 10.1074/jbc.272.40.24755 [DOI] [PubMed] [Google Scholar]
- De Pascale G., Wright G. D. (2010). Antibiotic resistance by enzyme inactivation: from mechanisms to solutions. Chembiochem 11, 1325–1334 10.1002/cbic.201000067 [DOI] [PubMed] [Google Scholar]
- Dery K. J., Chavideh R., Waters V., Chamorro R., Tolmasky L. S., Tolmasky M. E. (1997). Characterization of the replication and mobilization regions of the multiresistance Klebsiella pneumoniae plasmid pJHCMW1. Plasmid 38, 97–105 10.1006/plas.1997.1303 [DOI] [PubMed] [Google Scholar]
- Dery K. J., Soballe B., Witherspoon M. S., Bui D., Koch R., Sherratt D. J., et al. (2003). The aminoglycoside 6′-N-acetyltransferase type Ib encoded by Tn1331 is evenly distributed within the cell's cytoplasm. Antimicrob. Agents Chemother. 47, 2897–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Toro M., Rodriguez I., Rojo-Bezares B., Helmuth R., Torres C., Guerra B., et al. (2013). pMdT1, a small ColE1-like plasmid mobilizing a new variant of the aac(6′)-Ib-cr gene in Salmonella enterica serovar Typhimurium. J. Antimicrob. Chemother. [Epub ahead of print]. 10.1093/jac/dkt001 [DOI] [PubMed] [Google Scholar]
- Draker K. A., Northrop D. B., Wright G. D. (2003). Kinetic mechanism of the GCN5-related chromosomal aminoglycoside acetyltransferase AAC(6′)-Ii from Enterococcus faecium: evidence of dimer subunit cooperativity. Biochemistry 42, 6565–6574 10.1021/bi034148h [DOI] [PubMed] [Google Scholar]
- Drawz S. M., Bonomo R. A. (2010). Three decades of beta-lactamase inhibitors. Clin. Microbiol. Rev. 23, 160–201 10.1128/CMR.00037-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois V., Poirel L., Marie C., Arpin C., Nordmann P., Quentin C. (2002). Molecular characterization of a novel class 1 integron containing bla(GES-1) and a fused product of aac3-Ib/aac6′-Ib′ gene cassettes in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 46, 638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyda F., Klein D. C., Hickman A. B. (2000). GCN5-related N-acetyltransferases: a structural overview. Annu. Rev. Biophys. Biomol. Struct. 29, 81–103 10.1146/annurev.biophys.29.1.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endimiani A., Hujer K. M., Hujer A. M., Armstrong E. S., Choudhary Y., Aggen J. B., et al. (2009). ACHN-490, a neoglycoside with potent in vitro activity against multidrug-resistant Klebsiella pneumoniae isolates. Antimicrob. Agents Chemother. 53, 4504–4507 10.1128/AAC.00556-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galani I., Xirouchaki E., Kanellakopoulou K., Petrikkos G., Giamarellou H. (2002). Transferable plasmid mediating resistance to multiple antimicrobial agents in Klebsiella pneumoniae isolates in Greece. Clin. Microbiol. Infect. 8, 579–588 10.1046/j.1469-0691.2002.00391.x [DOI] [PubMed] [Google Scholar]
- Gao F., Yan X., Baettig O. M., Berghuis A. M., Auclair K. (2005). Regio- and chemoselective 6′-N-derivatization of aminoglycosides: bisubstrate inhibitors as probes to study aminoglycoside 6′-N-acetyltransferases. Angew. Chem. Int. Ed. Engl. 44, 6859–6862 10.1002/anie.200501399 [DOI] [PubMed] [Google Scholar]
- Gao F., Yan X., Shakya T., Baettig O. M., Ait-Mohand-Brunet S., Berghuis A. M., et al. (2006). Synthesis and structure-activity relationships of truncated bisubstrate inhibitors of aminoglycoside 6′-N-acetyltransferases. J. Med. Chem. 49, 5273–5281 10.1021/jm060732n [DOI] [PubMed] [Google Scholar]
- Garcia-Fernandez A., Villa L., Carta C., Venditti C., Giordano A., Venditti M., et al. (2012). Klebsiella pneumoniae ST258 producing KPC-3 identified in italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob. Agents Chemother. 56, 2143–2145 10.1128/AAC.05308-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green K. D., Chen W., Garneau-Tsodikova S. (2011). Effects of altering aminoglycoside structures on bacterial resistance enzyme activities. Antimicrob. Agents Chemother. 55, 3207–3213 10.1128/AAC.00312-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green K. D., Chen W., Garneau-Tsodikova S. (2012). Identification and characterization of inhibitors of the aminoglycoside resistance acetyltransferase Eis from Mycobacterium tuberculosis. ChemMedChem 7, 73–77 10.1002/cmdc.201100332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green K. D., Chen W., Houghton J. L., Fridman M., Garneau-Tsodikova S. (2010). Exploring the substrate promiscuity of drug-modifying enzymes for the chemoenzymatic generation of N-acylated aminoglycosides. Chembiochem 11, 119–126 10.1002/cbic.200900584 [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Salavati R., Altman S. (1997). Phenotypic conversion of drug-resistant bacteria to drug sensitivity. Proc. Natl. Acad. Sci. U.S.A. 94, 8468–8472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad J., Vakulenko S. B., Mobashery S. (1999). An antibiotic cloaked by its own resistance enzyme. J. Am. Chem. Soc. 121, 11922–11923 [Google Scholar]
- Hall R. M. (2012). Integrons and gene cassettes: hotspots of diversity in bacterial genomes. Ann. N.Y. Acad. Sci. 1267, 71–78 10.1111/j.1749-6632.2012.06588.x [DOI] [PubMed] [Google Scholar]
- Hancock R. E. (1981). Aminoglycoside uptake and mode of action–with special reference to streptomycin and gentamicin. I. Antagonists and mutants. J. Antimicrob. Chemother. 8, 249–276 10.1093/jac/8.4.249 [DOI] [PubMed] [Google Scholar]
- Hermann T. (2007). Aminoglycoside antibiotics: old drugs and new therapeutic approaches. Cell. Mol. Life Sci. 64, 1841–1852 10.1007/s00018-007-7034-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog I. M., Green K. D., Berkov-Zrihen Y., Feldman M., Vidavski R. R., Eldar-Boock A., et al. (2012). 6′-Thioether tobramycin analogues: towards selective targeting of bacterial membranes. Angew. Chem. Int. Ed. Engl. 51, 5652–5656 10.1002/anie.201200761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocquet D., Vogne C., El Garch F., Vejux A., Gotoh N., Lee A., et al. (2003). MexXY-OprM efflux pump is necessary for a adaptive resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 47, 1371–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghton J. L., Green K. D., Chen W., Garneau-Tsodikova S. (2010). The future of aminoglycosides: the end or renaissance? Chembiochem 11, 880–902 10.1002/cbic.200900779 [DOI] [PubMed] [Google Scholar]
- Huon L. K., Fang T. Y., Wang P. C. (2012). Outcomes of intratympanic gentamicin injection to treat Meniere's disease. Otol. Neurotol. 33, 706–714 10.1097/MAO.0b013e318259b3b1 [DOI] [PubMed] [Google Scholar]
- Jana S., Deb J. K. (2006). Molecular understanding of aminoglycoside action and resistance. Appl. Microbiol. Biotechnol. 70, 140–150 10.1007/s00253-005-0279-0 [DOI] [PubMed] [Google Scholar]
- Kandasamy J., Atia-Glikin D., Shulman E., Shapira K., Shavit M., Belakhov V., et al. (2012). Increased selectivity toward cytoplasmic versus mitochondrial ribosome confers improved efficiency of synthetic aminoglycosides in fixing damaged genes: a strategy for treatment of genetic diseases caused by nonsense mutations. J. Med. Chem. 55, 10630–10643 10.1021/jm3012992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermayer R. (2006). Translational readthrough induction of pathogenic nonsense mutations. Eur. J. Med. Genet. 49, 445–450 10.1016/j.ejmg.2006.04.003 [DOI] [PubMed] [Google Scholar]
- Kim C., Villegas-Estrada A., Hesek D., Mobashery S. (2007). Mechanistic characterization of the bifunctional aminoglycoside-modifying enzyme AAC(3)-Ib/AAC(6′)-Ib' from Pseudomonas aeruginosa. Biochemistry 46, 5270–5282 10.1021/bi700111z [DOI] [PubMed] [Google Scholar]
- Kim Y. T., Jang J. H., Kim H. C., Kim H., Lee K. R., Park K. S., et al. (2011). Identification of strain harboring both aac(6′)-Ib and aac(6′)-Ib-cr variant simultaneously in Escherichia coli and Klebsiella pneumoniae. BMB Rep. 44, 262–266 10.5483/BMBRep.2011.44.4.262 [DOI] [PubMed] [Google Scholar]
- Kohanski M. A., Dwyer D. J., Hayete B., Lawrence C. A., Collins J. J. (2007). A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130, 797–810 10.1016/j.cell.2007.06.049 [DOI] [PubMed] [Google Scholar]
- Kondo J., Hainrichson M., Nudelman I., Shallom-Shezifi D., Barbieri C. M., Pilch D. S., et al. (2007). Differential selectivity of natural and synthetic aminoglycosides towards the eukaryotic and prokaryotic decoding A sites. Chembiochem 8, 1700–1709 10.1002/cbic.200700271 [DOI] [PubMed] [Google Scholar]
- Labaune J. M., Bleyzac N., Maire P., Jelliffe R. W., Boutroy M. J., Aulagner G., et al. (2001). Once-a-day individualized amikacin dosing for suspected infection at birth based on population pharmacokinetic models. Biol. Neonate 80, 142–147 10.1159/000047133 [DOI] [PubMed] [Google Scholar]
- Lambert T., Ploy M. C., Courvalin P. (1994). A spontaneous point mutation in the aac(6′)-Ib' gene results in altered substrate specificity of aminoglycoside 6′-N-acetyltransferase of a Pseudomonas fluorescens strain. FEMS Microbiol. Lett. 115, 297–304 [DOI] [PubMed] [Google Scholar]
- Landman D., Kelly P., Backer M., Babu E., Shah N., Bratu S., et al. (2011). Antimicrobial activity of a novel aminoglycoside, ACHN-490, against Acinetobacter baumannii and Pseudomonas aeruginosa from New York City. J. Antimicrob. Chemother. 66, 332–334 10.1093/jac/dkq459 [DOI] [PubMed] [Google Scholar]
- Liu M., Haddad J., Azucena E., Kotra L. P., Kirzhner M., Mobashery S. (2000). Tethered bisubstrate derivatives as probes for mechanism and as inhibitors of aminoglycoside 3′-phosphotransferases. J. Org. Chem. 65, 7422–7431 10.1021/jo000589k [DOI] [PubMed] [Google Scholar]
- Lombes T., Begis G., Maurice F., Turcaud S., Lecourt T., Dardel F., et al. (2008). NMR-guided fragment-based approach for the design of AAC(6′)-Ib ligands. Chembiochem 9, 1368–1371 10.1002/cbic.200700677 [DOI] [PubMed] [Google Scholar]
- Lundblad E. W., Altman S. (2010). Inhibition of gene expression by RNase P. N. Biotechnol. 27, 212–221 10.1016/j.nbt.2010.03.003 [DOI] [PubMed] [Google Scholar]
- Macleod D. L., Nelson L. E., Shawar R. M., Lin B. B., Lockwood L. G., Dirk J. E., et al. (2000). Aminoglycoside-resistance mechanisms for cystic fibrosis Pseudomonas aeruginosa isolates are unchanged by long-term, intermittent, inhaled tobramycin treatment. J. Infect. Dis. 181, 1180–1184 10.1086/315312 [DOI] [PubMed] [Google Scholar]
- Magalhaes M. L., Vetting M. W., Gao F., Freiburger L., Auclair K., Blanchard J. S. (2008). Kinetic and structural analysis of bisubstrate inhibition of the Salmonella enterica aminoglycoside 6′-N-acetyltransferase. Biochemistry 47, 579–584 10.1021/bi701957c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnet S., Blanchard J. S. (2005). Molecular insights into aminoglycoside action and resistance. Chem. Rev. 105, 477–498 10.1021/cr0301088 [DOI] [PubMed] [Google Scholar]
- Magnet S., Smith T. A., Zheng R., Nordmann P., Blanchard J. S. (2003). Aminoglycoside resistance resulting from tight drug binding to an altered aminoglycoside acetyltransferase. Antimicrob. Agents Chemother. 47, 1577–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder K., Wei L., Annedi S., Kotra L. P. (2007). Aminoglycoside antibiotics, in Enzyme-Mediated Resistance to Antibiotics: Mechanisms, Dissemination, and Prospects for Inhibition, eds Bonomo R. A., Tolmasky M. E. (Washington, DC: ASM Press; ), 7–20 [Google Scholar]
- Maurice F., Broutin I., Podglajen I., Benas P., Collatz E., Dardel F. (2008). Enzyme structural plasticity and the emergence of broad-spectrum antibiotic resistance. EMBO Rep. 9, 344–349 10.1038/embor.2008.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta R., Champney W. S. (2003). Neomycin and paromomycin inhibit 30S ribosomal subunit assembly in Staphylococcus aureus. Curr. Microbiol. 47, 237–243 [DOI] [PubMed] [Google Scholar]
- Mendes R., Toleman M., Ribeiro J., Sader H., Jones R., Walsh T. (2004). Integron carrying a novel metallo-b-lactamase gene, blaIMP-16, and a fused form of aminoglycoside-resistance gene aac(6′)-30/aac(6′)-Ib: report from the SENTRY antimicrobial surveillance program. Antimicrob. Agents Chemother. 48, 4693–4702 10.1128/AAC.48.12.4693-4702.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies D., Benedetti A., Paydar A., Martin I., Royce S., Pai M., et al. (2009). Effect of duration and intermittency of rifampin on tuberculosis treatment outcomes: a systematic review and meta-analysis. PLoS Med. 6:e1000146 10.1371/journal.pmed.1000146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen N. E., Brannvall M., Virtanen A., Kirsebom L. A. (1999). Inhibition of RNase P RNA cleavage by aminoglycosides. Proc. Natl. Acad. Sci. U.S.A. 96, 6155–6160 10.1073/pnas.96.11.6155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G. H., Sabatelli F. J., Hare R. S., Glupczynski Y., Mackey P., Shlaes D., et al. (1997). The most frequent aminoglycoside resistance mechanisms–changes with time and geographic area: a reflection of aminoglycoside usage patterns? Aminoglycoside Resistance Study Groups. Clin. Infect. Dis. 24Suppl. 1, S46–S62 10.1093/clinids/24.Supplement_1.S46 [DOI] [PubMed] [Google Scholar]
- Morita Y., Tomida J., Kawamura Y. (2012). MexXY multidrug efflux system of Pseudomonas aeruginosa. Front. Microbiol. 3:408 10.3389/fmicb.2012.00408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir M. E., Van Heeswyck R. S., Wallace B. J. (1984). Effect of growth rate on streptomycin accumulation by Escherichia coli and Bacillus megaterium. J. Gen. Microbiol. 130, 2015–2022 [DOI] [PubMed] [Google Scholar]
- Neuwald A. F., Landsman D. (1997). GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem. Sci. 22, 154–155 10.1016/S0968-0004(97)01034-7 [DOI] [PubMed] [Google Scholar]
- O'Connor M., De Stasio E. A., Dahlberg A. E. (1991). Interaction between 16S ribosomal RNA and ribosomal protein S12: differential effects of paromomycin and streptomycin. Biochimie 73, 1493–1500 [DOI] [PubMed] [Google Scholar]
- Over U., Gur D., Unal S., Miller G. H., Aminoglycoside Resistance Study Group (2001). The changing nature of aminoglycoside resistance mechanisms and prevalence of newly recognized resistance mechanisms in Turkey. Clin. Microbiol. Infect. 7, 470–478 10.1046/j.1198-743x.2001.00284.x [DOI] [PubMed] [Google Scholar]
- Pacheu-Grau D., Perez-Delgado L., Gomez-Diaz C., Fraile-Rodrigo J., Montoya J., Ruiz-Pesini E. (2012). Mitochondrial ribosome and Meniere's disease: a pilot study. Eur. Arch. Otorhinolaryngol. 269, 2003–2008 10.1007/s00405-012-2066-8 [DOI] [PubMed] [Google Scholar]
- Panaite D. M., Tolmasky M. E. (1998). Characterization of mutants of the 6′-N-acetyltransferase encoded by the multiresistance transposon Tn1331: effect of Phen171-to-Leu171 and Tyr80-to-Cys80 substitutions. Plasmid 39, 123–133 10.1006/plas.1997.1330 [DOI] [PubMed] [Google Scholar]
- Poirel L., Cabanne L., Collet L., Nordmann P. (2006). Class II transposon-borne structure harboring metallo-beta-lactamase gene blaVIM-2 in Pseudomonas putida. Antimicrob. Agents Chemother. 50, 2889–2891 10.1128/AAC.00398-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourreza A., Witherspoon M., Fox J., Newmark J., Bui D., Tolmasky M. E. (2005). Mutagenesis analysis of a conserved region involved in acetyl coenzyme A binding in the aminoglycoside 6′-N-acetyltransferase type Ib encoded by plasmid pJHCMW1. Antimicrob. Agents Chemother. 49, 2979–2982 10.1128/AAC.49.7.2979-2982.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiroga M. P., Andres P., Petroni A., Soler Bistue A. J., Guerriero L., Vargas L. J., et al. (2007). Complex class 1 integrons with diverse variable regions, including aac(6′)-Ib-cr, and a novel allele, qnrB10, associated with ISCR1 in clinical enterobacterial isolates from Argentina. Antimicrob. Agents Chemother. 51, 4466–4470 10.1128/AAC.00726-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez M. S., Parenteau T. R., Centron D., Tolmasky M. E. (2008). Functional characterization of Tn1331 gene cassettes. J. Antimicrob. Chemother. 62, 669–673 10.1093/jac/dkn279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez M. S., Tolmasky M. E. (2010). Aminoglycoside modifying enzymes. Drug Resist. Updat. 13, 151–171 10.1016/j.drup.2010.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rather P. N., Munayyer H., Mann P. A., Hare R. S., Miller G. H., Shaw K. J. (1992). Genetic analysis of bacterial acetyltransferases: identification of amino acids determining the specificities of the aminoglycoside 6′-N-acetyltransferase Ib and IIa proteins. J. Bacteriol. 174, 3196–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice L. B. (2008). Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J. Infect. Dis. 197, 1079–1081 10.1086/533452 [DOI] [PubMed] [Google Scholar]
- Rice L. B., Carias L. L., Hutton R. A., Rudin S. D., Endimiani A., Bonomo R. A. (2008). The KQ element, a complex genetic region conferring transferable resistance to carbapenems, aminoglycosides, and fluoroquinolones in Klebsiella pneumoniae. Anti-microb. Agents Chemother. 52, 3427–3429 10.1128/AAC.00493-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich D. P., Anderson M. P., Gregory R. J., Cheng S. H., Paul S., Jefferson D. M., et al. (1990). Expression of cystic fibrosis transmembrane conductance regulator corrects defective chloride channel regulation in cystic fibrosis airway epithelial cells. Nature 347, 358–363 10.1038/347358a0 [DOI] [PubMed] [Google Scholar]
- Robicsek A., Strahilevitz J., Jacoby G. A., Macielag M., Abbanat D., Park C. H., et al. (2006). Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12, 83–88 10.1038/nm1347 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Martinez J. M., Cano M. E., Velasco C., Martinez-Martinez L., Pascual A. (2011). Plasmid-mediated quinolone resistance: an update. J. Infect. Chemother. 17, 149–182 10.1007/s10156-010-0120-2 [DOI] [PubMed] [Google Scholar]
- Romanowska J., Reuter N., Trylska J. (2013). Comparing aminoglycoside binding sites in bacterial ribosomal RNA and aminoglycoside modifying enzymes. Proteins 81, 63–80 10.1002/prot.24163 [DOI] [PubMed] [Google Scholar]
- Ruiz J., Pons M. J., Gomes C. (2012). Transferable mechanisms of quinolone resistance. Int. J. Antimicrob. Agents 40, 196–203 10.1016/j.ijantimicag.2012.02.011 [DOI] [PubMed] [Google Scholar]
- Salipante S. J., Hall B. G. (2003). Determining the limits of the evolutionary potential of an antibiotic resistance gene. Mol. Biol. Evol. 20, 653–659 10.1093/molbev/msg074 [DOI] [PubMed] [Google Scholar]
- Sarno R., Ha H., Weinsetel N., Tolmasky M. E. (2003). Inhibition of aminoglycoside 6′-N-acetyltransferase type Ib-mediated amikacin resistance by antisense oligodeoxynucleotides. Antimicrob. Agents Chemother. 47, 3296–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarno R., McGillivary G., Sherratt D. J., Actis L. A., Tolmasky M. E. (2002). Complete nucleotide sequence of Klebsiella pneumoniae multiresistance plasmid pJHCMW1. Antimicrob. Agents Chemother. 46, 3422–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt E., Galimand M., Panvert M., Courvalin P., Mechulam Y. (2009). Structural bases for 16 s rRNA methylation catalyzed by ArmA and RmtB methyltransferases. J. Mol. Biol. 388, 570–582 10.1016/j.jmb.2009.03.034 [DOI] [PubMed] [Google Scholar]
- Shaul P., Green K. D., Rutenberg R., Kramer M., Berkov-Zrihen Y., Breiner-Goldstein E., et al. (2011). Assessment of 6′- and 6′”-N-acylation of aminoglycosides as a strategy to overcome bacterial resistance. Org. Biomol. Chem. 9, 4057–4063 10.1039/c0ob01133a [DOI] [PubMed] [Google Scholar]
- Shaw K. J., Rather P. N., Hare R. S., Miller G. H. (1993). Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57, 138–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmara A., Weinsetel N., Dery K. J., Chavideh R., Tolmasky M. E. (2001). Systematic analysis of a conserved region of the aminoglycoside 6′-N-acetyltransferase type Ib. Antimicrob. Agents Chemother. 45, 3287–3292 10.1128/AAC.45.12.3287-3292.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneath P. H., Sokal R. R. (1973). Numerical Taxonomy. The Principles and Practice of Numerical Classification. San Francisco, CA: W. H. Freeman [Google Scholar]
- Soler Bistue A. J., Birshan D., Tomaras A. P., Dandekar M., Tran T., Newmark J., et al. (2008). Klebsiella pneumoniae multiresistance plasmid pMET1: similarity with the Yersinia pestis plasmid pCRY and integrative conjugative elements. PLoS ONE 3:e1800 10.1371/journal.pone.0001800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler Bistue A. J., Ha H., Sarno R., Don M., Zorreguieta A., Tolmasky M. E. (2007). External guide sequences targeting the aac(6′)-Ib mRNA induce inhibition of amikacin resistance. Antimicrob. Agents Chemother. 51, 1918–1925 10.1128/AAC.01500-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler Bistue A. J., Martin F. A., Petroni A., Faccone D., Galas M., Tolmasky M. E., et al. (2006). Vibrio cholerae InV117, a class 1 integron harboring aac(6′)-Ib and blaCTX-M-2, is linked to transposition genes. Antimicrob. Agents Chemother. 50, 1903–1907 10.1128/AAC.50.5.1903-1907.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler Bistue A. J., Martin F. A., Vozza N., Ha H., Joaquin J. C., Zorreguieta A., et al. (2009). Inhibition of aac(6′)-Ib-mediated amikacin resistance by nuclease-resistant external guide sequences in bacteria. Proc. Natl. Acad. Sci. U.S.A. 106, 13230–13235 10.1073/pnas.0906529106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahilevitz J., Jacoby G. A., Hooper D. C., Robicsek A. (2009). Plasmid-mediated quinolone resistance: a multifaceted threat. Clin. Microbiol. Rev. 22, 664–689 10.1128/CMR.00016-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taber H. W., Mueller J. P., Miller P. F., Arrow A. S. (1987). Bacterial uptake of aminoglycoside antibiotics. Microbiol. Rev. 51, 439–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolmasky M. (2007a). Aminogylcoside-modifying enzymes: characteristics, localization, and dissemination, in Enzyme-Mediated Resistance to Antibiotics: Mechanisms, Dissemination, and Prospects for Inhibition, eds Bonomo R., Tolmasky M. (Washington, DC: ASM Press; ), 35–52 [Google Scholar]
- Tolmasky M. E. (2007b). Overview of dissemination mechanisms of genes coding for resistance to antibiotics, in Enzyme-Mediated Resistance to Antibiotics: Mechanisms, Dissemination, and Prospects for Inhibition, eds Bonomo R., Tolmasky M. E. (Washington, DC: ASM Press; ), 267–270 [Google Scholar]
- Tolmasky M. E. (1990). Sequencing and expression of aadA, bla, and tnpR from the multiresistance transposon Tn1331. Plasmid 24, 218–226 [DOI] [PubMed] [Google Scholar]
- Tolmasky M. E., Chamorro R. M., Crosa J. H., Marini P. M. (1988). Transposon-mediated amikacin resistance in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 32, 1416–1420 10.1128/AAC.32.9.1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolmasky M. E., Crosa J. H. (1987). Tn1331, a novel multiresistance transposon encoding resistance to amikacin and ampicillin in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 31, 1955–1960 10.1128/AAC.31.12.1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolmasky M. E., Crosa J. H. (1991). Regulation of plasmid-mediated iron transport and virulence in Vibrio anguillarum. Biol. Met. 4, 33–35 [DOI] [PubMed] [Google Scholar]
- Tolmasky M. E., Crosa J. H. (1993). Genetic organization of antibiotic resistance genes (aac(6′)-Ib, aadA, and oxa9) in the multiresistance transposon Tn1331. Plasmid 29, 31–40 10.1006/plas.1993.1004 [DOI] [PubMed] [Google Scholar]
- Traglia G., Davies Sala C., Fuxman Bass J., Soler Bistué A., Zorreguieta A., Ramirez M. S., et al. (2012). Internalization of locked nucleic acids/DNA hybrid oligomers into Escherichia coli. Biores. Open Access 1, 260–263 10.1089/biores.2012.0257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakulenko S. B., Mobashery S. (2003). Versatility of aminoglycosides and prospects for their future. Clin. Microbiol. Rev. 16, 430–450 10.1128/CMR.16.3.430-450.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van ‘t Veen A., Van Der Zee A., Nelson J., Speelberg B., Kluytmans J. A., Buiting A. G. (2005). Outbreak of infection with a multiresistant Klebsiella pneumoniae strain associated with contaminated roll boards in operating rooms. J. Clin. Microbiol. 43, 4961–4967 10.1128/JCM.43.10.4961-4967.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetting M. W., Lp S. D. C., Yu M., Hegde S. S., Magnet S., Roderick S. L., et al. (2005). Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 433, 212–226 10.1016/j.abb.2004.09.003 [DOI] [PubMed] [Google Scholar]
- Vetting M. W., Magnet S., Nieves E., Roderick S. L., Blanchard J. S. (2004). A bacterial acetyltransferase capable of regioselective N-acetylation of antibiotics and histones. Chem. Biol. 11, 565–573 10.1016/j.chembiol.2004.03.017 [DOI] [PubMed] [Google Scholar]
- Vetting M. W., Park C. H., Hegde S. S., Jacoby G. A., Hooper D. C., Blanchard J. S. (2008). Mechanistic and structural analysis of aminoglycoside N-acetyltransferase AAC(6′)-Ib and its bifunctional, fluoroquinolone-active AAC(6′)-Ib-cr variant. Biochemistry 47, 9825–9835 10.1021/bi800664x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicens Q., Westhof E. (2003). Molecular recognition of aminoglycoside antibiotics by ribosomal RNA and resistance enzymes: an analysis of x-ray crystal structures. Biopolymers 70, 42–57 10.1002/bip.10414 [DOI] [PubMed] [Google Scholar]
- Villa L., Poirel L., Nordmann P., Carta C., Carattoli A. (2012). Complete sequencing of an IncH plasmid carrying the blaNDM-1, blaCTX-M-15 and qnrB1 genes. J. Antimicrob. Chemother. 67, 1645–1650 10.1093/jac/dks114 [DOI] [PubMed] [Google Scholar]
- Vong K., Tam I. S., Yan X., Auclair K. (2012). Inhibitors of aminoglycoside resistance activated in cells. ACS Chem. Biol. 7, 470–475 10.1021/cb200366u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachino J., Arakawa Y. (2012). Exogenously acquired 16S rRNA methyltransferases found in aminoglycoside-resistant pathogenic Gram-negative bacteria: an update. Drug Resist. Updat. 15, 133–148 10.1016/j.drup.2012.05.001 [DOI] [PubMed] [Google Scholar]
- Warburg G., Hidalgo-Grass C., Partridge S. R., Tolmasky M. E., Temper V., Moses A. E., et al. (2012). A carbapenem-resistant Klebsiella pneumoniae epidemic clone in Jerusalem: sequence type 512 carrying a plasmid encoding aac(6′)-Ib. J. Antimicrob. Chemother. 67, 898–901 10.1093/jac/dkr552 [DOI] [PubMed] [Google Scholar]
- Welch K. T., Virga K. G., Whittemore N. A., Ozen C., Wright E., Brown C. L., et al. (2005). Discovery of non-carbohydrate inhibitors of aminoglycoside-modifying enzymes. Bioorg. Med. Chem. 13, 6252–6263 10.1016/j.bmc.2005.06.059 [DOI] [PubMed] [Google Scholar]
- Williams J. W., Northrop D. B. (1979). Synthesis of a tight-binding, multisubstrate analog inhibitor of gentamicin acetyltransferase I. J. Antibiot. 32, 1147–1154 [DOI] [PubMed] [Google Scholar]
- Woloj M., Tolmasky M. E., Roberts M. C., Crosa J. H. (1986). Plasmid-encoded amikacin resistance in multiresistant strains of Klebsiella pneumoniae isolated from neonates with meningitis. Antimicrob. Agents Chemother. 29, 315–319 10.1128/AAC.29.2.315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodford N., Carattoli A., Karisik E., Underwood A., Ellington M. J., Livermore D. M. (2009). Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob. Agents Chemother. 53, 4472–4482 10.1128/AAC.00688-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright G. D. (1999). Aminoglycoside-modifying enzymes. Curr. Opin. Microbiol. 2, 499–503 10.1016/S1369-5274(99)00007-7 [DOI] [PubMed] [Google Scholar]
- Wright G. D., Berghuis A. M. (2007). Structural aspects of aminoglycoside modifying enzymes, in Enzyme-Mediated Resistance to Antibiotics: Mechanisms, Dissemination, and Prospects for Inhibition, eds Bonomo R., Tolmasky M. (Washington, DC: ASM Press; ), 21–33 [Google Scholar]
- Wright G. D., Ladak P. (1997). Overexpression and characterization of the chromosomal aminoglycoside 6′-N-acetyltransferase from Enterococcus faecium. Antimicrob. Agents Chemother. 41, 956–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wybenga-Groot L. E., Draker K., Wright G. D., Berghuis A. M. (1999). Crystal structure of an aminoglycoside 6′-N-acetyltransferase: defining the GCN5-related N-acetyltransferase superfamily fold. Structure 7, 497–507 [DOI] [PubMed] [Google Scholar]
- Yao J., Moellering R. (2007). Antibacterial agents, in Manual of Clinical Microbiology, eds Murray P., Baron E., Jorgensen J., Landry M., Pfaller M. (Washington, DC: American Society for Microbiology Press; ), 1077–1113 [Google Scholar]
- Yong D., Toleman M. A., Giske C. G., Cho H. S., Sundman K., Lee K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher H. S., Green R. (2009). Fidelity at the molecular level: lessons from protein synthesis. Cell 136, 746–762 10.1016/j.cell.2009.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingman L. V., Park S., Olson T. M., Alekseev A. E., Terzic A. (2007). Aminoglycoside-induced translational read-through in disease: overcoming nonsense mutations by pharmacogenetic therapy. Clin. Pharmacol. Ther. 81, 99–103 10.1038/sj.clpt.6100012 [DOI] [PubMed] [Google Scholar]
- Zong Z., Partridge S. R., Iredell J. R. (2009). A blaVEB-1 variant, blaVEB-6, associated with repeated elements in a complex genetic structure. Antimicrob. Agents Chemother. 53, 1693–1697 10.1128/AAC.01313-08 [DOI] [PMC free article] [PubMed] [Google Scholar]