Abstract
High-throughput genetic interaction screens have enabled functional genomics on a network scale. Groups of cofunctional genes commonly exhibit similar interaction patterns across a large network, leading to novel functional inferences for a minority of previously uncharacterized genes within a group. However, such analyses are often unsuited to cases with a few relevant gene variants or sparse annotation. Here we describe an alternative analysis of cell growth signaling using a computational strategy that integrates patterns of pleiotropy and epistasis to infer how gene knockdowns enhance or suppress the effects of other knockdowns. We analyzed the interaction network for RNAi knockdowns of a set of 93 incompletely annotated genes in a Drosophila melanogaster model of cellular signaling. We inferred novel functional relationships between genes by modeling genetic interactions in terms of knockdown-to-knockdown influences. The method simultaneously analyzes the effects of partially pleiotropic genes on multiple quantitative phenotypes to infer a consistent model of each genetic interaction. From these models we proposed novel candidate Ras inhibitors and their Ras signaling interaction partners, and each of these hypotheses can be inferred independent of network-wide patterns. At the same time, the network-scale interaction patterns consistently mapped pathway organization. The analysis therefore assigns functional relevance to individual genetic interactions while also revealing global genetic architecture.
Keywords: genetic interaction, pleiotropy, epistasis, genetic network, signaling network
The systematic study of genetic interactions has proven a powerful means to map genetic networks (Boone et al. 2007). Genome-scale interaction analysis has provided a global view of gene function in yeast (Costanzo et al. 2010), and studies focused on specific processes have mapped large-scale networks in yeast (Collins et al. 2007; Drees et al. 2005; Segre et al. 2005; St Onge et al. 2007), worm (Byrne et al. 2007; Lehner et al. 2006), and fly (Horn et al. 2011; Yamamoto et al. 2009). Analyses of statistical epistasis, the population-level manifestation of genetic interaction, have identified important effects in mouse (Li and Churchill 2010; Reifsnyder et al. 2000; Shao et al. 2008) and human (McKinney and Pajewski 2011; Ritchie 2011) genetics. These studies indicate that genetic interactions reveal underlying structure in biological networks and map complex genetic architecture. Advances in study design and the characterization of genetic populations have been accompanied by parallel progress in quantitative phenotyping. Multidimensional phenotypic characterization is becoming increasingly common, often including multiple physiological traits coupled with thousands of molecular measures such as protein and transcript abundances (Andreux et al. 2012; Chen et al. 2012). Such research ultimately aims to provide a genetically precise and phenotypically predictive approach to medicine. Success of this approach is contingent on the development of analytical methods to extract quantitative models from genetic interactions across multiple phenotypes. These methods will increase the power to formulate precise biological hypotheses to potentially address the complex genetics that underlie human health and disease.
To date, studies have primarily used statistical concordance of interaction patterns across multiple genes to infer the role of previously uncharacterized genes. This strategy is often referred to as guilt-by-association (GBA). Advanced GBA approaches, such as clustering genes based on correlated interaction spectra across multiple interaction partners (Carter et al. 2009; Collins et al. 2007; Costanzo et al. 2010; Drees et al. 2005; Segre et al. 2005), have successfully mapped genetic architecture on a large scale. In these networks genes often form highly connected communities, or gene modules, that are enriched in one or more functional annotations. The principle of GBA dictates that a minority of uncharacterized genes within a module can be assigned the dominant function of the module.
While successful on a large scale, GBA-based methods have multiple limitations. First, they require large data sets to generate adequate statistical power to resolve modules, and can therefore be limited in populations with a small number of relevant mutations such as studies of specific developmental or signaling processes, drivers of cancer evolution, or interacting candidates in genome-wide associations. Second, GBA relies on the availability of functional annotations for the vast majority of interacting genes. Third, GBA approaches often generate implicit predictions of gene function without providing explicit predictions of the effects of a mutation or combination of mutations, thereby limiting the power to generate directly testable hypotheses. Fourth, large-scale GBA approaches rarely take advantage of the complementary information in multiple phenotypes. In cases when multiple phenotypes are considered, the analysis is usually based on coincidence of interactions derived independently for each phenotype (Horn et al. 2011; Michaut and Bader 2012). Finally, it has been proposed that GBA results may be driven by a small number of critical interactions and therefore network associations are not generally reliable (Gillis and Pavlidis 2012).
Here we use an approach based on the combined analysis of pleiotropy and epistasis to infer the genetic architecture of growth-related signaling in Drosophila melanogaster. This strategy infers and interprets genetic interaction data in terms of quantitative variant-to-variant and variant-to-phenotype influences, rather than non-directional epistasis. This results in a network model that maps how each specific variant affects each other and, in turn, multiple related phenotypes. This method is applicable to a range of genetic diversity from a few genes to genome-scale screens. It integrates information from multiple phenotype measures to generate specific hypotheses for genetic interactions. We recently demonstrated the utility of this method in a population of yeast strains by mapping relationships of the yeast mating pathway (Carter et al. 2012). Here, we extend the method to a large set of double knockdowns of genes involved in signal transduction on a common genetic background (Horn et al. 2011). This represents a much larger network of potential interacting genes and involves multiple signaling pathways. The analysis exploits the subtle differences between the regulation of cell proliferation and nuclear size across 93 mutations. We obtain an interaction network of knockdown-to-knockdown influences that identifies novel suppressors and enhancers of Ras signaling, demonstrating the method’s applicability to complex networks and large-scale genetic screens of multiple phenotypes. At a local level, the network provides specific hypotheses of how each gene knockdown modifies the activity of other gene knockdowns in the same pathway and across antagonistic pathways, narrowing the possible molecular mechanisms that underlie the observed genetic interactions.
Methods
Data source
Data were obtained from a study of double-stranded RNA (dsRNA) gene knockdowns (Horn et al. 2011) that is briefly summarized as follows. Schneider S2 cells were exposed to two independent dsRNA molecules to knock down 93 genes in all 4278 pair-wise combinations. Genes were chosen based on prior annotation for mitogen-activated protein kinase (MAPK) signaling or prior annotation as a protein or lipid phosphatase expressed in S2 cells. High-throughput fluorescent imaging and automated image analysis were used to quantify three non-redundant features: total cell number, mean nuclear area per cell, and nuclear fluorescence intensity. Data for each knockdown pair were averaged and log-transformed, and each phenotype distribution was mean-centered and normalized to a standard deviation of 1. At a population level, each phenotype resembled the combination of a normal distribution and a long tail below the mean that was populated by lines with knockdowns of a few especially strong perturbations such as Pvr, pnt, and drk (Table 1 and see Supporting Information, Figure S1).
Table 1. Genes with significant main effects used as covariates for all pair-wise scans, with prior pathway annotation (FlyBase 2004).
| Knockdown | βET1 | βET2 | Pathway |
|---|---|---|---|
| alph | 0.0093 | Ras | |
| CG3573 | 0.0141 | Ras | |
| Cka | −0.0126 | Ras | |
| dome | −0.0205 | Ras | |
| drk | −0.0373 | 0.0092 | Ras |
| Dsor1 | −0.0084 | 0.0209 | Ras |
| Gap1 | 0.0152 | −0.0099 | Ras inhibitor |
| lic | −0.0105 | JNK | |
| mop | −0.0089 | Ras | |
| msk | −0.0095 | ||
| mtm | 0.0086 | Ras inhibitor | |
| mts | −0.0255 | 0.0215 | Ras |
| pnt | −0.0382 | −0.0108 | Ras |
| PpV | −0.0101 | Ras | |
| Pten | 0.0110 | ||
| Ptp69D | 0.0109 | Ras inhibitor | |
| PTP-ER | 0.0087 | Ras inhibitor | |
| puc | −0.0153 | 0.0133 | Ras |
| Pvr | −0.0600 | −0.0189 | Ras |
| pyd | 0.0091 | JNK | |
| Ras85D | −0.0101 | 0.0108 | Ras |
| Rho1 | 0.0139 | 0.0741 | |
| rl | −0.0106 | 0.0106 | Ras |
| Sos | −0.0104 | 0.0108 | Ras |
| Src42A | −0.0163 | JNK | |
| stg | −0.0123 | 0.0220 | Ras |
βET1, main effect on eigentrait 1; βET2, main effect on eigentrait 2.
Interaction model
Our data analysis technique is described at length in a previous publication (Carter et al. 2012) and is summarized here. We first perform singular value decomposition (SVD) on the phenotype matrix to maximize orthogonality. In this case, we analyzed the first two left singular vectors, referred to hereafter as eigentraits, since the third singular value represented less than 2% of the global variance and was therefore unlikely to encode significant biology (Results). With these eigentraits denoted and for sample i, we performed linear regression for each of the 93 perturbed genes in isolation to identify strong-effect knockdowns to be treated as additive covariates in subsequent pair-wise regressions. For each locus we considered the model:
| (1) |
The index i is from 1 to and j is 1 or 2. The variable is the probability of the gene variant at the locus in the strain i, is the effect on the eigentrait i, and is the residual error. For our data all were binary, corresponding to the presence (1) or absence (0) of dsRNA that target the locus. We define strong-effect knockdowns as those with a significant effect (see below) and condition each subsequent knockdown-pair scan by including strong-effect knockdowns as covariates for the associated phenotype. We next model every possible knockdown pair with main effects and an interaction term. For two knockdowns labeled 1 and 2 we have:
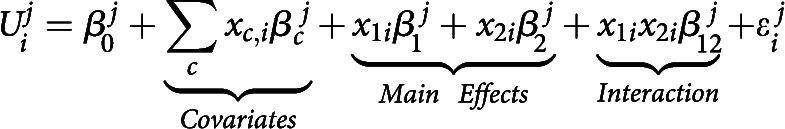 |
(2) |
The variables are defined as in Equation 1 with the additional interaction coefficient and sum over strong-effect knockdowns as covariates (excluding knockdowns 1 and 2). This step is conceptually similar to the original analysis of the data (Horn et al. 2011), which identified pair-wise interactions for each phenotype independently.
To derive a model in terms of knockdown-to-knockdown influences we consider how each knockdown affects the activity of each other knockdown, first in terms of modified knockdown activity and then in terms of the knockdown-to-knockdown influences that account for the modified activity level. We first recast the interaction coefficients in terms of modified knockdown activity parameters and that are independent of the eigentrait j. The activity variables are computed by matrix inversion:
| (3) |
The variables and are an exact reparametrization of the interaction coefficients. We next compute the directional influence coefficients that generate the modified activities. These knockdown-to-knockdown influences, and , are:
| (4) |
By substituting the solution of Equation 3 into Equation 4 we obtain the influence coefficients as a function of the regression parameters. This defines a model in terms of knockdown-to-knockdown influence coefficients () and main-effect, knockdown-to-eigentrait coefficients (). Without loss of information, we multiply the eigentrait coefficient matrix by the other center and left singular value matrices to recompose knockdown-to-phenotype coefficients for the original phenotypes.
Calculation of significance
We assess the significance of the influence coefficients, , and knockdown-to-phenotype coefficients using standard error analysis methods on the regression parameters (Bevington 1969). For example, the variance of is estimated by differentiating with respect to all model parameters:
| (5) |
The indices i and k run over main effect and interaction coefficients, and j and l run from 1 to . The first sum is over individual parameters and the second double sum is the cross terms. Variances and covariances are estimated from the least-squares regression using standard methods (Bevington 1969).
After computing models for all knockdown pairs, we obtained two knockdown-to-knockdown influence coefficients between each knockdown pair (one in each direction) and knockdown-to-phenotype coefficients. To determine significance we used effect size divided by estimated standard error as our test statistic because it could be computed for both regression coefficients and variables computed from them (e.g., ). We selected the coefficient of median standard effect size to represent overall knockdown-to-phenotype coefficients. We first determined significance for single-locus scans. We performed 2000 permutations of the genotype data and fit an extreme value distribution (EVD) for the maximum test statistic from each permutation. This accounted for multiple tests and empirically estimated the likelihood of chance association. We determined that a test statistic of 5.18 or greater corresponds to P < 0.001. We repeated the procedure for pair-wise scans, permuting the genotypes of the two knockdowns being tested in tandem. This procedure retained the structure between all other knockdowns (including covariates) and all three phenotypes, and thereby randomized only the marginal effects after conditioning on covariates. We collected test statistics for 700 permutations to obtain null distributions for knockdown-to-phenotype and knockdown-to-knockdown influence coefficients. We computed an empirical P value for each coefficient. The family wise error rate was controlled by adjusting P values using a step-down procedure (Holm 1979). Step-down EVDs were not used because the empirical distributions had slightly greater support at higher values than fitted EVDs and thus the EVDs would artificially inflate significance. A significance cutoff of adjusted P < 0.01 was used in our network (Figure 3) and all estimated P values are reported in Table S1.
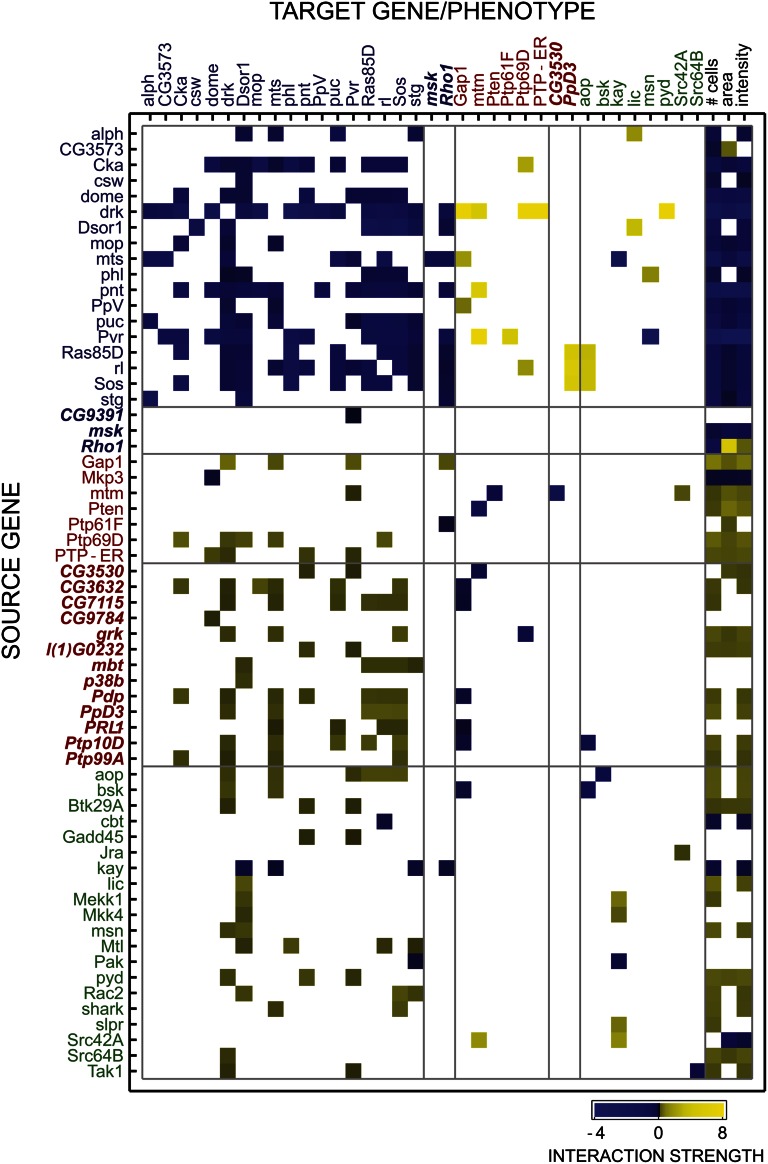
Figure 3.
Adjacency matrix of significant interactions for knockdowns of Ras (blue labels), Ras inhibitors (red labels), and JNK (green labels) genes. Previously uncharacterized gene names are in italics. Color denotes interaction sign and intensity of directional interactions. Overall, within-pathway interactions are overwhelmingly suppressive (blue squares), whereas interactions across antagonistic pathways are enhancing (yellow squares). Ras knockdowns generally reduce cellular phenotypes, while Ras inhibitors and JNK knockdowns increase phenotypes. Empty squares did not meet the significance threshold of P < 0.01 (see text).
Results
The assayed phenotypes shared many genetic components and therefore exhibited substantial pleiotropy. All three phenotypes were significantly correlated, with Pearson coefficients as follows: cell number and nuclear area, 0.65; cell number and nuclear intensity, 0.90; and nuclear area and nuclear intensity, 0.85. However, there was variation across the tested knockdowns. For example, the Rho1 defect in cytokinesis caused decreased cell numbers and increased nuclear area, whereas Ras signaling knockdowns such as drk decreased both numbers and area (Horn et al. 2011). To maximize complementarity and dimensionally reduce the phenotype data, we performed singular value decomposition (SVD) to define two composite eigentraits, each an orthogonal combination of the phenotypes (Methods). The eigentraits were orthogonal, normalized combinations of the phenotypes (Figure 1) and represented 87%, 11.5%, and 1.5% of the variance in the data. Since the bulk of the variance was present in the first two eigentraits we discarded the third. We refer to these composite phenotypes as eigentrait 1 (ET1) and eigentrait 2 (ET2). ET1 is a common signal to all three phenotypes, whereas ET2 primarily differentiates nuclear area from cell number and nuclear intensity. Thus, the inferences made for genetic interactions will be mostly based on combining an overall common signal with a signal that separates nuclear area from the other two, highly correlated phenotypes.
Figure 1.
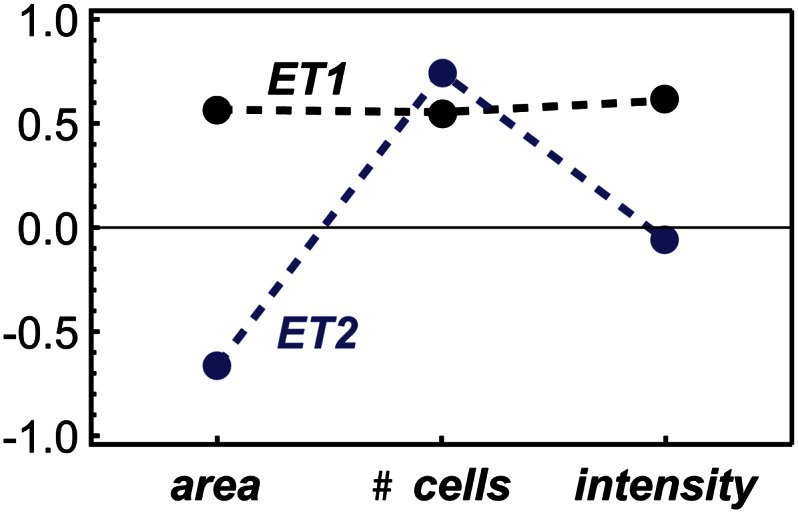
Eigentrait compositions in terms of measured phenotypes. Eigentrait 1 (ET1) is the signal common to all three phenotypes, whereas eigentrait 2 (ET2) encodes the differences.
For each eigentrait we performed linear regression for each knockdown individually, similar to the method in the original study (Horn et al. 2011). This identified significant knockdowns to be used as covariates in pair-wise scans (Methods). We identified 21 knockdowns as covariates for ET1 and 17 knockdowns for ET2 (Table 1, Figure S1 and Figure S2). We next computed a pair-wise model for each of the 4278 knockdown pairs to obtain quantitative interactions between knockdowns (Table S1). We reparametrized the two interaction coefficients from pair-wise linear regression on the eigentraits (Methods, Equation 2) in terms of two knockdown-to-knockdown influences between perturbations (Methods, Equation 4) (Carter et al. 2012). To determine the allelic effects on the original cellular phenotypes, we recomposed the phenotype SVD for each pair-wise model and averaged all models to obtain knockdown-to-phenotype influence coefficients (Methods). Errors were estimated for all coefficients using standard least-squares regression and error propagation formulas for quantities computed from regression coefficients (Methods, Equation 5).
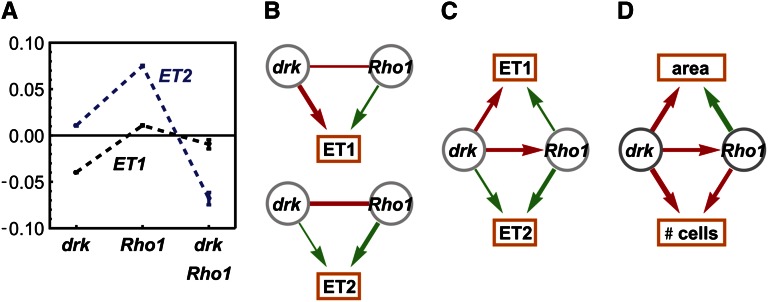
A representative interaction is illustrated in Figure 2. The drk and Rho1 knockdowns have opposite effects on the first eigentrait (Figure 2A) and a negative regression interaction coefficient (Figure 2B). For the second eigentrait, both knockdowns have positive effects and the regression interaction coefficient is negative and larger than that for ET1. Reparametrizing these results in terms of our genetic influence coefficients inferred a significant negative influence from drk to Rho1 (Figure 2C); that is, the drk knockdown suppressed the effects of the Rho1 knockdown. The complementary information in the two eigentraits ruled out alternative hypotheses such as a Rho1 enhancement of the drk effect that would have been consistent with ET1 but not ET2. Finally, we recomposed the eigentraits into the original phenotypes (Figure 2D), an operation that does not modify the inferred genetic interaction. This analysis recapitulates the interpretation in Horn et al. (2011) with the advantage of a mathematically defined procedure that has been systematically applied to all pairs of knockdowns in the data set. In total, we obtained a network of 290 significant knockdown-to-knockdown edges and 146 significant knockdown-to-phenotype edges. Interactions for 61 genes with existing or novel (see below) functional annotations are shown in Figure 3.
Figure 2.
Genetic interactions between drk and Rho1 knockdowns. (A) Inferred main and interaction effects of drk and Rho1 mutations on eigentraits ET1 and ET2. (B) Significant main and interaction effects shown in terms of positive (green) and negative (red) influences. Edge width represents interaction strength. (C) Interaction model consistent with both ET1 and ET2, in which drk knockdown suppresses Rho1 knockdown. (D) The same model expressed in terms of the original phenotypes of total cell number and nuclear area (total fluorescent intensity is similar to cell number and not shown).
Sixty-three knockdowns had a significant main effect (P < 0.01; Methods) on at least one of the three phenotypes, and 29 had significant influences on all three phenotypes (Figure 3). Knockdowns of the 18 genes previously annotated for Ras signaling had 48 negative effects, compared to only one positive effect [CG3573 (Ocrl) knockdown on nuclear area]. In contrast, knockdowns of the 29 genes previously annotated for JNK cascade and Ras inhibition had 48 positive main effects, compared to 13 negative effects. These findings are consistent with Ras signaling activation of cell proliferation and growth and the generally antagonistic relationship between Ras and JNK signaling. For each knockdown, main effects were generally consistent across all partners in each pair-wise scan; no specific combination of knockdowns revealed a uniquely significant effect for one of the genes.
Our total of 290 significant knockdown-to-knockdown interactions out of 8556 possible (two for each pair) was numerically comparable to the 531 significant interactions out of 12,834 possible (three phenotypes for each pair) obtained with the same threshold in the previous study (Horn et al. 2011). Gene pairs with at least one significant interaction in both analyses strongly overlapped, as 55% (126 of 229 pairs) of our significant pairs were also significant in at least one phenotype in Horn, et al. (8 × 10−88, Fisher’s exact test). Pairs that did not overlap nevertheless tended to have low q values in Horn, et al. For each of our pairs we identified the lowest q value across all three phenotypes, and found a median of 0.006 as compared to a global median lowest q value of 0.38. This demonstrates that our analysis generally agreed with more standard measures of epistasis and, in some cases, improved significance by reformulation in terms of directional model parameters.
Global genetic interaction patterns revealed large-scale gene function and module/pathway organization (Figure 3 and Figure S3). Knockdown-to-knockdown influences between knockdowns of documented Ras signaling genes were fairly common (63% of possible interactions were significant) and always negative. We interpret this as genes with redundant signaling functions. Although it is suggestive that downstream knockdowns should suppress upstream knockdowns (Avery and Wasserman 1992; Carter et al. 2012), the majority of pair-wise knockdowns of known Ras genes exhibited significant negative interactions in both directions. In cases of unidirectional suppression, some known downstream knockdowns (e.g., pnt) tended to suppress upstream proteins (e.g., Ras85D and Sos) but others (e.g., rl) were suppression targets more often than not. Overall, there is a clear pattern of negative interactions between within-pathway genes. In contrast, Ras inhibitor knockdowns tended to enhance the effect of knockdowns of Ras signaling genes. This is consistent with standard interpretations of genetic interactions in signaling. For example, PTP-ER is named for its mutant alleles that enhance Ras signaling in eye development (Karim and Rubin 1999). Furthermore, knockdowns of genes documented or predicted to be involved in JNK signaling generally enhanced Ras mutations, consistent with the antagonistic relationship between JNK and Ras signaling.
Specific edges in the interaction network allowed us to infer the function of many uncharacterized genes, based on the rule that knockdowns functioning in the same signaling process will exhibit suppressive interactions whereas knockdowns functioning in antagonistic processes will exhibit enhancing interactions (Avery and Wasserman 1992). We assessed the interactions for 45 genes not previously associated with Ras or JNK signaling and one gene (Rho1) which had been annotated for both (Flybase 2004; Noselli and Agnes 1999). Novel pathway assignments were made when an unannotated knockdown exhibited interactions with Ras knockdowns annotated in Horn, et al. (Horn et al. 2011) or curated data (Flybase 2004). Negative interaction partners were candidate Ras signaling genes while positive interaction partners were designated as Ras inhibitors. We assigned Ras signaling function to three knockdowns that suppressed the effect of one or more Ras signaling knockdowns and/or had its effect enhanced by one or more Ras inhibitor knockdowns (Table 2). These novel Ras signaling knockdowns had generally negative influences on cell number and nuclear area (Figure 3), consistent with other Ras signaling knockdowns. The multiannotated gene Rho1 was in this group, suggesting that for the measured phenotypes it operates as a Ras signaling component. We assigned Ras inhibitor function to 13 knockdowns that enhanced the effect of one or more Ras signaling knockdowns and/or had its effect enhanced by one or more Ras signaling knockdowns (Table 2 and Figure 3). In most cases these novel Ras inhibitor knockdowns suppressed the knockdown of a fellow Ras inhibitor knockdown and the vast majority had a positive influence on cell number and nuclear area (Figure 3), consistent with other Ras inhibitors. Ten of these genes are annotated as phosphatases and, in many cases, we identified specific Ras signaling kinases for which knockdown effects are unidirectionally enriched by one or more phosphatase knockdowns (Figure 4). These provide specific hypotheses for the prioritization of phosphatase-kinase targeting experiments. The inferred Ras-inhibitor phosphatase PpD3 exhibited mutual enhancing interactions with Ras knockdowns Sos, Ras85D, and rl (Figure 3), which suggests a more complex role in Ras inhibition. Taken in sum, these 16 new functional hypotheses can be added to the set of approximately 20 novel pathway assignments inferred in the original, classifier-based analysis of the interaction data (Horn et al. 2011).
Table 2. New candidate Ras signaling genes and Ras inhibitors, with relevant GO annotations for the knockdown (Ashburner et al. 2000; FlyBase 2004).
| Pathway | Knockdown | Interaction Partner(s) | Annotated Function |
|---|---|---|---|
| Ras signaling | CG9391 | Pvr | Inositol monophosphate 1-phosphatase |
| msk | mts | Ran GTPase binding, protein transport | |
| Rho1 | drk, Dsor1, mts, pnt, Pvr, Ras85D, rl, Sos, stg | Kinase binding (JNK cascade and GTPase signal transduction both predicted) | |
| Ras inhibitor | CG3530 | pnt, Pvr | Protein tyrosine/serine/threonine phosphatase |
| CG3632 | Cka, drk, mop, mts, puc, Sos | Protein tyrosine/serine/threonine phosphatase | |
| CG7115 | drk, mts, puc, Ras85D, rl, Sos | Protein serine/threonine phosphatase, cell adhesion | |
| CG9784 | dome | Inositol trisphosphate phosphatase | |
| grk | drk, mts, Sos | Epidermal growth factor receptor binding | |
| l(1)G0232 | pnt, Pvr | Non-membrane spanning protein tyrosine phosphatase | |
| mbt | Dsor1, Ras85D, rl, Sos, stg | Protein serine/threonine kinase, negative regulation of cell size | |
| p38b | Dsor1 | Protein serine/threonine MAP kinase, stress response, positive regulation of cell size | |
| Pdp | Cka, drk, mts, pnt, Ras85D, rl, Sos | Protein serine/threonine phosphatase | |
| PpD3 | drk, mts, Ras85D, rl, Sos | Protein serine/threonine phosphatase, mitotic cell cycle | |
| PRL-1 | mts, puc, rl, Sos | Prenylated protein tyrosine phosphatase | |
| Ptp10D | drk, mts, puc, Ras85D, Sos | Protein tyrosine phosphatase, central nervous system development | |
| Ptp99A | Cka, drk, mts, Sos | Transmembrane receptor protein tyrosine phosphatase, motor axon guidance |
GO, gene ontology.
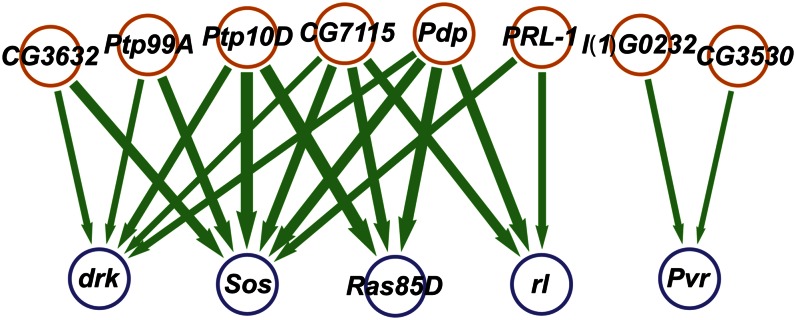
Figure 4.
Network of novel candidate Ras inhibitor phosphatases and Ras kinases inferred from interaction patterns. Knockdowns of phosphatase genes (orange nodes) enhance the effects of Ras kinase knockdowns (blue nodes), similar to known Ras inhibitors and JNK signaling genes (Figure 3). Edge width denotes relative strength of enhancement.
Discussion
We have used a strategy that combines patterns of epistasis and pleiotropy to derive a large-scale network mapping how mutations in signaling genes interact to affect distinct phenotypes. The approach integrates information in multiple phenotypes to resolve ambiguities in model interpretation that arise when individual phenotypes are analyzed in isolation. We have demonstrated the applicability of this strategy to large-scale data, in this case involving 4278 cell lines that mix 93 genetic perturbations. Significant knockdown-to-knockdown interactions were obtained among 68 knockdowns even though each was fairly rare in the population (present in 2.2% of lines). While the original analysis of these data (Horn et al. 2011) revealed patterns of genetic interactions separately for each phenotype, this analysis combines the multiphenotype patterns of epistasis to derive directional models of genetic interactions that are consistent across all three phenotypes. This additional mathematical analysis recovered previous findings and was used to propose additional functional relationships between gene pairs.
Many of the interactions in the network (Figure 3) recapitulated known relationships. Genes that function in the same pathways show consistent, suppressive genetic interactions across a spectrum of interaction partners. This is consistent with the idea that mutations with redundant effects suppress each other, particularly in signaling pathways. The spectral consistency for interactions, which has been previously referred to as “genetic monochromaticity” (Segre et al. 2005), is maintained after their reparametrization in terms of directional influences. A similar spectral consistency is demonstrated for enhancing interactions from pairs spanning multiple pathways. We interpret this enhancement as a signature of an antagonistic relationship. For instance, a knockdown of a Ras inhibitor will increase Ras signaling activity, and therefore a subsequent Ras knockdown will have an enhanced effect since the signaling baseline is increased (cf. PTP-ER, Results). Following this interpretation, in addition to spectral consistency, these interactions can map activation and suppression across the Ras and JNK pathways. The specificity represents de novo derivation of key functional relationships that are more difficult to detect when analyzing each phenotype independently.
We note that classical synthetic effects, in which two perturbations with insignificant main effects generate a strong effect in combination, would also produce enhancing interactions. We did not observe any such cases in our data. When we broaden the definition to include knockdowns with main effects and greater than additive combinatorial effects, all with the same sign (positive or negative), we find only two candidate interactions. Gap1 knockdown enhances Rho1 knockdown activity and both positively affect nuclear area and nuclear intensity; however they show opposite effects on cell number and therefore the effect is not consistently synthetic (Figure 3). A less ambiguous candidate is the strong enhancement of kay knockdown effects by Src42A knockdown. Both perturbations significantly decrease nuclear intensity, and this effect becomes significantly greater when the knockdowns are combined. These genes, both annotated as JNK pathway elements, provide the only unambiguous instance of synthesis in the data.
Our analysis provides new and refined information on gene-gene relationships and places this information in a network context. At a single-interaction level, we identified novel candidate Ras inhibitors and coactivators (Table 2 and Figure 4). At a network-wide level, these genes were spectrally consistent with other genes of similar known function (Figure 3 and Figure S3). Thus, although prior information is usually helpful in placing new candidate mutations in a functional context, the interactions we inferred represent a de novo model of enhancement or suppression with functional implications. Furthermore, some of the network inferences propose refinements of current knowledge, such as the hypothesis that Rho1 is primarily a Ras signaling gene in the context of the measured phenotypes. Likewise, the interaction patterns and phenotype influences of the kayak (kay) knockdown suggest that its functional role in cell proliferation is more similar to Ras signaling genes than JNK cascade genes, in contrast to its similarity to JNK in dorsal closure (Harden 2002). Specifically, the kay knockdown resembled Ras signaling genes by suppressing the knockdown effect of multiple Ras signaling genes, being enhanced by the knockdown of multiple JNK cascade genes, and having a negative main effect on cell number (Figure 3).
In particular, our analysis suggested novel Ras inhibitors not detected the previous study using the same data (Horn et al. 2011). In most cases, inhibitor knockdowns redundantly enhanced multiple kinases (Figure 4). This could be due to non-specific phosphatase activity, but is potentially a result of functional redundancy among the kinases that leads to similar genetic interactions between a phosphatase and both its direct target and genes that cofunction with that target in a signaling cascade. One possible avenue to resolve this ambiguous redundancy would be to extend the interaction analysis beyond independent pair-wise analysis to models that find the most parsimonious model for three or more genes (Carter et al. 2007). We note that the majority of these phosphatase knockdowns significantly suppressed the Gap1 knockdown, providing evidence of redundant function with a known Ras85D inhibitor. We also note that a number of additional enhancing interactions from phosphatases to kinases that did not meet our significance threshold were also detected, providing additional evidence of functional coherence but target non-specificity. However, there does seem to be limited specificity in that CG3632 and Ptp99A enhance drk and Sos, genes that fall relatively upstream in the KEGG-annotated Ras pathway, while CG7115 and Pdp knockdowns most strongly enhance the relatively downstream pathway knockdown Ras85D. In contrast, phosphatase knockdowns l(1)G0232 and CG3530 enhance the Pvr knockdown, which is not placed in the KEGG-annotated Ras cascade. The additional information that the Pvr knockdown tends to unidirectionally suppress the KEGG pathway genes suggests that Pvr potentially acts downstream of the KEGG-annotated pathway and is deactivated by the phosphatases l(1)G0232 and CG3530. Thus, even with the ambiguities arising from pair-wise models, we obtained a degree of specificity in hypotheses for Ras inhibitor activity.
The network of directional influences suggests hypotheses that are often amenable to experimental testing (Carter et al. 2007). In contrast with non-directional associations such as coexpression, statistical epistasis, or synthetic lethality, a directional influence identifies a driver mutation that affects the activity of its target, a second gene that, in turn, affects the phenotype. With this in mind, we can predict that removing the influence with an experimental perturbation, such as an exogenous inhibitor molecule, would nullify the effect of mutations in the target gene. Furthermore, the effect type (enhancement or suppression) specifies the nature of the effect on the target gene. We can therefore predict that overexpression of a novel Ras inhibitor will not only affect, but specifically reduce the relevant activity of its enhanced targets, whereas perturbations of the targets are not expected to affect the Ras inhibitor. For example, overexpression of Ptp99A will specifically inhibit the activity of Ras signaling proteins Cka, drk, mts, and Sos. In this way, a network of directional genetic interactions could prove useful in the selection of candidate genes for validation and, in disease-related networks, the prioritization of therapeutic targets.
Our analysis was limited by the genotypes and specific phenotypes selected in the original study. Although the three phenotypes used exhibit differential interaction patterns (Horn et al. 2011), their high correlation limited the complementary information that could better resolve directional interactions. By probing additional phenotypes, new interactions might be detected and existing interactions refined. For instance, we speculate that mutual cosuppression among Ras signal knockdowns is potentially due to measuring insufficiently distinct outputs. Combining detailed assays of eye development and terminal patterns might have revealed more specific roles and pathway order for canonical Ras signaling proteins. Furthermore, although the genetic perturbations are limited to MAPK signaling genes, the methods could be applied to a wider screen or population-based analysis to identify additional pathway relationships. As data collection in experimental systems and clinical studies becomes more diverse and quantitative, our analytical method has the potential to derive more refined and precise network models.
The approach we have applied derives quantitative, directional models of how each gene knockdown modifies the effects of each other knockdown. It represents an alternative to guilt-by-association methods that infer function by scoring correlations across large-scale data. Taken together as a network, the interactions form consistent patterns between and within gene modules. Large-scale data and prior information is helpful but not necessary because guilt precedes association in our analysis. This allowed us, for instance, to propose new candidate Ras inhibitors that are consistent with known Ras inhibitors, but do not necessarily depend on this prior knowledge for functional inference.
Supplementary Material
Acknowledgments
I thank Chad Meyers and Gary Churchill for fruitful discussions and Tonnya Norwood for a critical reading of the manuscript. This work was supported by National Institute of General Medical Sciences (NIGMS) grants K25 GM079404 and P50 GM076468. The content is the sole responsibility of the author and does not necessarily represent the official views of NIGMS or the National Institutes of Health.
Footnotes
Communicating editor: F. P. Roth
Literature Cited
- Andreux P. A., Williams E. G., Koutnikova H., Houtkooper R. H., Champy M. F., et al. , 2012. Systems genetics of metabolism: the use of the BXD murine reference panel for multiscalar integration of traits. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., et al. , 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L., Wasserman S., 1992. Ordering gene function: the interpretation of epistasis in regulatory hierarchies. Trends Genet. 8: 312–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevington P. R., 1969. Data Reduction and Error Analysis for the Physical Sciences. McGraw-Hill, New York [Google Scholar]
- Boone C., Bussey H., Andrews B. J., 2007. Exploring genetic interactions and networks with yeast. Nat. Rev. Genet. 8: 437–449 [DOI] [PubMed] [Google Scholar]
- Byrne A. B., Weirauch M. T., Wong V., Koeva M., Dixon S. J., et al. , 2007. A global analysis of genetic interactions in Caenorhabditis elegans. J. Biol. 6: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter G. W., Prinz S., Neou C., Shelby J. P., Marzolf B., et al. , 2007. Prediction of phenotype and gene expression for combinations of mutations. Mol. Syst. Biol. 3: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter G. W., Galas D. J., Galitski T., 2009. Maximal extraction of biological information from genetic interaction data. PLoS Comput. Biol. 5: e1000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter G. W., Hays M., Sherman A., Galitski T., 2012. Use of pleiotropy to model genetic interactions in a population. PLoS Genet. 8: e1003010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Mias G. I., Li-Pook-Than J., Jiang L., Lam H. Y., et al. , 2012. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 148: 1293–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S. R., Miller K. M., Maas N. L., Roguev A., Fillingham J., et al. , 2007. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446: 806–810 [DOI] [PubMed] [Google Scholar]
- Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., et al. , 2010. The genetic landscape of a cell. Science 327: 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees B. L., Thorsson V., Carter G. W., Rives A. W., Raymond M. Z., et al. , 2005. Derivation of genetic interaction networks from quantitative phenotype data. Genome Biol. 6: R38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FlyBase , 2004. Gene ontology annotation in FlyBase through association of InterPro records with GO terms, http://www.flybase.org
- Gillis J., Pavlidis P., 2012. “Guilt by association” is the exception rather than the rule in gene networks. PLoS Comput. Biol. 8: e1002444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harden N., 2002. Signaling pathways directing the movement and fusion of epithelial sheets: lessons from dorsal closure in Drosophila. Differentiation 70: 181–203 [DOI] [PubMed] [Google Scholar]
- Holm S., 1979. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 6: 65–70 [Google Scholar]
- Horn T., Sandmann T., Fischer B., Axelsson E., Huber W., et al. , 2011. Mapping of signaling networks through synthetic genetic interaction analysis by RNAi. Nat. Methods 8: 341–346 [DOI] [PubMed] [Google Scholar]
- Karim F. D., Rubin G. M., 1999. PTP-ER, a novel tyrosine phosphatase, functions downstream of Ras1 to downregulate MAP kinase during Drosophila eye development. Mol. Cell 3: 741–750 [DOI] [PubMed] [Google Scholar]
- Lehner B., Crombie C., Tischler J., Fortunato A., Fraser A. G., 2006. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat. Genet. 38: 896–903 [DOI] [PubMed] [Google Scholar]
- Li R. H., Churchill G. A., 2010. Epistasis contributes to the genetic buffering of plasma HDL cholesterol in mice. Physiol. Genomics 42A: 228–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney B. A., Pajewski N. M., 2011. Six degrees of epistasis: statistical network models for GWAS. Front. Genet. 2: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaut M., Bader G. D., 2012. Multiple genetic interaction experiments provide complementary information useful for gene function prediction. PLOS Comput. Biol. 8: e1002559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noselli S., Agnes F., 1999. Roles of the JNK signaling pathway in Drosophila morphogenesis. Curr. Opin. Genet. Dev. 9: 466–472 [DOI] [PubMed] [Google Scholar]
- Reifsnyder P. C., Churchill G., Leiter E. H., 2000. Maternal environment and genotype interact to establish diabesity in mice. Genome Res. 10: 1568–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M. D., 2011. Using biological knowledge to uncover the mystery in the search for epistasis in genome-wide association studies. Ann. Hum. Genet. 75: 172–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre D., Deluna A., Church G. M., Kishony R., 2005. Modular epistasis in yeast metabolism. Nat. Genet. 37: 77–83 [DOI] [PubMed] [Google Scholar]
- Shao H., Burrage L. C., Sinasac D. S., Hill A. E., Ernest S. R., et al. , 2008. Genetic architecture of complex traits: large phenotypic effects and pervasive epistasis. Proc. Natl. Acad. Sci. USA 105: 19910–19914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Onge R. P., Mani R., Oh J., Proctor M., Fung E., et al. , 2007. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat. Genet. 39: 199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Anholt R. R., MacKay T. F., 2009. Epistatic interactions attenuate mutations affecting startle behaviour in Drosophila melanogaster. Genet. Res. 91: 373–382 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.