Abstract
Brefeldin A-inhibited guanine nucleotide-exchange protein 1 (BIG1) is an ≈200-kDa brefeldin A-inhibited guanine nucleotide-exchange protein that preferentially activates ADP-ribosylation factor 1 (ARF1) and ARF3. BIG1 was found in cytosol in a multiprotein complex with a similar ARF-activating protein, BIG2, which is also an A kinase-anchoring protein. In HepG2 cells growing with serum, BIG1 was primarily cytosolic and Golgi-associated. After incubation overnight without serum, a large fraction of endogenous BIG1 was in the nuclei. By confocal immunofluorescence microscopy, BIG1 was localized with nucleoporin p62 at the nuclear envelope (probably during nucleocytoplasmic transport) and also in nucleoli, clearly visible against the less concentrated overall matrix staining. BIG1 was also identified by Western blot analyses in purified subnuclear fractions (e.g., nucleoli and nuclear matrix). Antibodies against BIG1, nucleoporin, or nucleolin coimmunoprecipitated the other two proteins from purified nuclei. In contrast, BIG2 was not associated with nuclear BIG1. Also of note, ARF was never detected among proteins precipitated from purified nuclei by anti-BIG1 antibodies, although microscopically the two proteins do appear sometimes to be colocalized in the nucleus. These data are consistent with independent intracellular movements and actions of BIG1 and BIG2, and they are also evidence of the participation of BIG1 in both Golgi and nuclear functions.
ADP-ribosylation factors (ARF) are 20-kDa GTPases of the ras superfamily that regulate vesicular transport in eukaryotic cells. There are three classes of ARFs: class I (ARF1–3), which function in endoplasmic reticulum-Golgi trafficking; the much less studied class II (ARF4–5); and class III (ARF6), with significant roles in endocytotic pathways and cytoskeletal dynamics near the cell surface (1–4). ARFs are activated by guanine nucleotide-exchange proteins (GEPs), which accelerate the replacement of bound GDP with GTP (5), and inactivated by GTPase-activating proteins, which are required to stimulate hydrolysis of bound GTP to generate ARF–GDP (6).
GEPs differ in molecular weight, domain structure, and sensitivity to inhibition by brefeldin A, a fungal fatty acid metabolite that can block protein secretion (5). All GEPs contain an ≈200-aa Sec7 domain (7) that is responsible for ARF activation (8). Among the main GEP families, the Sec7/brefeldin A-inhibited GEP (BIG) proteins have long been of interest to us because of their potential to serve scaffolding and catalytic functions in multimolecular complexes. BIG1 and BIG2, which preferentially activate ARF1 and ARF3, were initially isolated together from bovine brain cytosol as components of an ≈670-kDa macromolecular complex on gel filtration (9–12). On immunofluorescence microscopy of HepG2 cells, the two molecules seemed to be concentrated in the perinuclear region and distributed in a punctate pattern in the cytoplasm (13). Until recently, the Sec7 domain had been the focus of most structure function studies, although Mansour et al. (14) had reported that the N-terminal one-third of BIG1 was required for its Golgi localization. More recently, using a yeast two-hybrid system, the BIG2 molecule was shown to have three protein kinase A-anchoring domains (15). The N terminus of BIG1 was found to interact with FK506-binding protein 13 (FKBP13), and the immunosuppressant drug FK506 increased BIG1 binding to Jurkat cell membranes (16).
Here, we describe the localization of BIG1, but not BIG2, at several nuclear sites in serum-starved HepG2 cells. Immunoreactive BIG1 was identified in the purified nuclear matrix; it was partially colocalized with nucleoporin p62 on confocal microscopy, and antibodies against either protein precipitated the other. BIG1 was also concentrated in nucleolar areas and was coimmunoprecipitated with nucleolin. All data are consistent with a previously unsuspected role(s) for BIG1 in nuclear processes.
Materials and Methods
Materials. HepG2 human liver carcinoma cells were from the American Type Culture Collection, FK506 was from Calbiochem, and penicillin, streptomycin, and goat and horse serum were from GIBCO.
Cell Fractionation and Western Blotting. Confluent HepG2 cells were grown (37°C, 5% CO2/95% air) in DMEM with 10% FBS (GIBCO), 100 units/ml penicillin G, and 100 μg/ml streptomycin on collagen I (Sigma)-coated dishes (Becton Dickinson). Unless otherwise indicated, cells were incubated overnight (16–18 h) in the same medium without FBS for all experiments. Cells harvested by scraping in ice-cold PBS (0.14 M NaCl/8.1 mM Na2HPO4/1.5 mM KH2PO4) (BioSource International, Camarillo, CA) were washed twice with the same solution before isolation, purification (17), and fractionation (18) of nuclei. Briefly, confluent cells from ten 15-cm plates (≈3 × 108 cells total) were sedimented by centrifugation at 1,000 × g for 5 min and homogenized with 10 strokes in a 7-ml Dounce tissue grinder (Wheaton Scientific) in 4 ml of TKMS buffer (50 mM Tris, pH 7.5/25 mM KCl/5 mM MgCl2/250 mM sucrose) containing 0.5 mM 4-(2)-aminomethylbenzenesulfonyl fluoride, 10 μg/ml each leupeptin and aprotinin, and 1 μg/ml pepstatin A. The homogenate was centrifuged at 4,000 × g for 10 min to sediment nuclei, unbroken cells, and cell debris (crude nuclear fraction). The supernatant was centrifuged at 100,000 × g for 1.5 h at 4°C to separate cytosol (supernatant) from membranes (precipitate).
To obtain pure nuclei, the crude nuclear fraction was washed once with TKMS buffer, incubated at 37°C for 45 min in 2 ml of TKMS buffer, washed twice with TKMS buffer, and applied to the top of a sucrose gradient (2-ml layers of TKMS buffer containing 2.5, 2.25, 2, 1.75, and 1.5 M sucrose), which was then centrifuged at 100,000 × g for 1.5 h at 4°C. Pure nuclei were collected at the 1.75–2 M interface and washed twice with TKMS buffer.
To obtain different subnuclear fractions (18), pure nuclei were successively washed and centrifuged (400 × g at 4°C for 10 min) three times with 1.5 ml of 10 mM Tris, pH 7.5/200 μM MgCl2 (low-salt washes), once with 10 mM Tris, pH 7.5/200 μM MgCl2/2 M NaCl (high-salt wash), 1% Triton X-100 in 10 mM Tris, pH 7.5/5 mM MgCl2 (Triton wash), and with 10 mM Tris, pH 7.5/5 mM MgCl2 (matrix wash). The pellet (nuclear matrix) was dispersed in 2 ml of 10 mM Tris, pH 7.5/5 mM MgCl2. Samples (100 μg) of proteins from each fraction and wash [except for nuclear matrix (30 μg)] were separated by SDS/PAGE in 15-well 8–16% gels (Invitrogen) and transferred to nitrocellulose membranes, which were divided for reaction with antibodies against BIG1 (0.5 μg/ml), BIG2 (1 μg/ml), nucleolin (Research Diagnostics, Flanders, NJ; 1:5000), nucleoporin p62 (Transduction Laboratories, Lexington, KY; 1:5,000), polyADP-ribose polymerase (PARP; Biomol, Plymouth Meeting, PA; 1:5,000), nuclear mitotic apparatus protein (Transduction Laboratories; 1:500), exportin or CRM1 (Transduction Laboratories; 1:5,000), lamin B (Transduction Laboratories; 1:5,000), GM130 (Transduction Laboratories; 1:1,000), FKBP13 (0.5 μg/ml), and/or ARF (ABR, Golden, CO; 1:500). Secondary antibodies [goat anti-rabbit and goat anti-mouse IgG conjugated to horseradish peroxidase (Promega)] were detected with Super-Signal chemiluminescent substrate (Pierce), and densitometry was done by using ChemiImager 5500 (Alpha Innotech, San Leandro, CA). Preparation and purification of antibodies against BIG1, BIG2, and FKBP13 have been reported (13, 16).
Purification of Nucleoli. Nucleoli were isolated from HepG2 cells according to Muramatsu et al. (19), with slight modifications. HepG2 cells (≈1 × 108) were washed three times with 50 ml of ice-cold PBS, suspended in 20 ml of RSB buffer (10 mM Tris·HCl, pH 7.5/10 mM NaCl/1.5 mM MgCl2), incubated on ice for 10 min, and centrifuged at 600 × g for 10 min at 4°C. Cells were suspended in 4 ml of RSB buffer containing 0.3% (wt/vol) (octylphenoxy)polyethoxyethanol (IGEPAL CA-630, Sigma), which is chemically identical to Nonidet P-40, and 0.2% sodium deoxycholate and homogenized (Dounce; 10 strokes with a tight pestle). The homogenate was centrifuged at 1,200 × g for 5 min at 4°C, and the nuclear pellet (dispersed in 2 ml of 0.25 M sucrose containing 3.3 mM CaCl2) was layered over an equal volume of 0.88 M sucrose. After centrifugation at 1,200 × g for 10 min at 4°C), the pelleted nuclei were dispersed in 2 ml of 0.34 M sucrose and sonified (three pulses, 15 times at 5-s intervals; power setting, 50) by using a microtip probe and a Branson Sonifier 450 (VWR Scientific, West Chester, PA). Sonified nuclei were layered over 1 ml of 0.88 M sucrose and centrifuged at 2,200 × g for 15 min at 4°C. The nucleolar pellet was washed twice with 0.34 M sucrose and stored at -80°C in 1 ml of the same solution. Proteins (100 μg) from each fraction were separated by SDS/PAGE in 15-well 8–16% gels, transferred to nitrocellulose, reacted with the indicated antibodies, and detected by using chemiluminescence.
Immunoprecipitation. HepG2 cells, after incubation as indicated, were fractionated as described above. Samples of nuclear pellets were homogenized (Dounce; 10 strokes) in TKMS buffer containing 0.5% (wt/vol) IGEPAL. For immunoprecipitation (all procedures at 4°C), samples (200 μg of protein in 1.5 ml of TKMS buffer) were cleared by incubation overnight (16–18 h) with 50 μl of 50% (vol/vol) slurry of protein A Sepharose CL-4B (Amersham Pharmacia) in TKMS buffer, and then with the indicated antibodies for 8 h. Protein A Sepharose (60 μl, 50% slurry) was added, and, after incubation for 16 h, beads were washed three times with 0.5-ml volumes of each of three buffers: 50 mM Tris-HCl, pH 7.5/150 mM NaCl/1% IGEPAL/0.5% sodium deoxycholate; 50 mM Tris-HCl, pH 7.5/500 mM NaCl/0.1% IGEPAL/0.05% sodium deoxycholate; and 10 mM Tris-HCl, pH 7.5/0.1% IGEPAL/0.5% sodium deoxycholate. All buffers contained the same concentrations of protease inhibitors as the homogenizing buffer. Bound proteins were eluted by boiling beads in 100 μl of gel loading buffer, 15-μl samples were subjected to SDS/PAGE in 15-well 8–16% gels, and proteins were transferred to nitrocellulose for reaction with the indicated primary and secondary antibodies followed by detection of chemiluminescent horseradish peroxidase product.
Immunofluorescence Microscopy. HepG2 cells were transferred from collagen I-coated dishes to four-well collagen-coated culture slides (<5 × 104 cells per well) (Becton Dickinson), grown for 24 h in the same medium without or with FBS, treated as described in figure legends, and washed three times with PBSCM (PBS containing 1 mM CaCl2 and 1 mM MgCl2) before fixation with 3% paraformaldehyde (Electron Microscopy Services, Washington, PA) in PBSCM for 20 min at room temperature (as were subsequent procedures, unless otherwise specified). Cells were washed three times with PBSCM, permeabilized (3 min, 0.05% Triton X-100 in PBSCM), washed three times with PBSCM, and incubated for 1–2 h in 0.5 ml of PBSCM containing 6% BSA, 5% goat serum, and 5% horse serum (blocking buffer). After three washes with PBSCM, samples were incubated for 16 h in blocking buffer at 4°C with primary antibody (0.5 μg/ml anti-BIG1, 1 μg/ml anti-FKBP13, anti-nucleolin at 1:1,500, anti-nucleoporin at 1:5,000, and anti-ARF at 1:500) and washed three times with PBSCM before incubation for 2 h with secondary antibodies, fluorescein isothiocyanate-labeled anti-rabbit IgG or Texas red-labeled anti-mouse IgG antibodies (Vector Laboratories), which had been diluted in blocking buffer. After washing with PBSCM, mounting medium without or with 4′,6-diamidino-2-phenylindole, dihydrochloride (Vectashield, Vector Laboratories) was added, and coverslips were sealed with clear nail polish (Electron Microscopy Services). Images were collected by using a Zeiss LSM 510 laser scanning confocal microscope.
Results
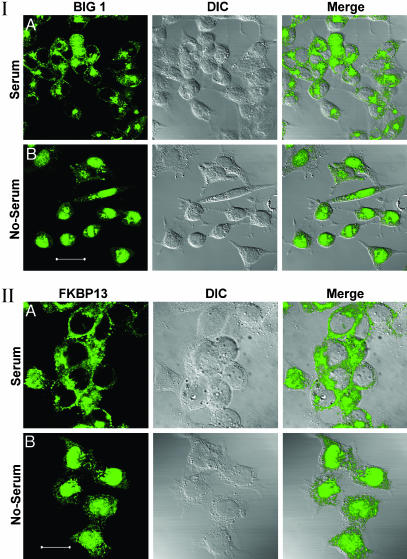
Effects of FBS Starvation on Intracellular Distribution of BIG1 and FKBP13. In HepG2 cells growing in medium with 10% FBS, endogenous BIG1 (Fig. 1I) was most often seen by confocal immunofluorescence microscopy in the perinuclear region and in a punctate distribution through the cytoplasm, but usually not in nuclei. After incubation of cells for 16–18 h in medium without FBS, essentially all of the BIG1 was present in the nucleus of ≈80% of cells, but large differences in BIG1 distribution were observed among serum-starved cells (see Fig. 4). FKBP13 was mostly confined to the cytoplasm of cells in medium with FBS, where its appearance differed from that of BIG1 (Fig. 1II), which was of interest because BIG1 coimmunoprecipitated with FKBP13 from membranes of Jurkat cells growing with FBS, and the proteins interacted in a yeast two-hybrid screen (16). Incubation of those cells with FK506 had increased the recovery of both BIG1 and ARF in membrane fractions, but no proteins were precipitated by the FKBP13 antibodies after FK506 treatment, presumably because FK506 binding alters the FKBP13 epitope (16). In HepG2 cells incubated without FBS, virtually all nuclei contained some FKBP13, but the fraction in the cytoplasm was greater than that of BIG1 (Fig. 1II).
Fig. 1.
Effect of FBS on localization of endogenous BIG1 (I) and FKBP13 (II) in HepG2 cells. Cells were incubated for 18 h without or with 10% FBS before preparation for confocal immunofluorescence microscopy. All observations were replicated three or more times. (Scale bars: 40 μm in I and 20 μm in II.)
Fig. 4.
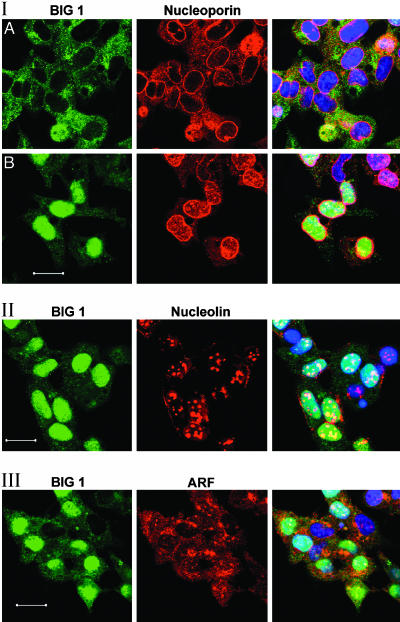
Localization of BIG1 with nucleoporin (I), nucleolin (II), and ARF (III) in FBS-starved cells by confocal immunofluorescence microscopy. All observations have been replicated three or more times. (Scale bars: 25 μm.)
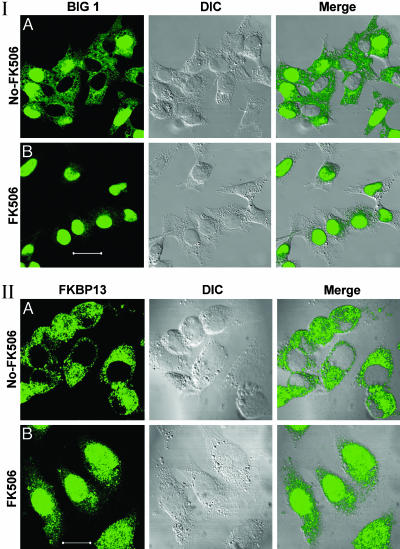
Effects of FK506 on Distribution of BIG1 and FKBP13. After incubation of FBS-starved HepG2 cells with 100 nM FK506, every nucleus contained BIG1 and there was very little to be seen in the cytoplasm (Fig. 2I). FK506, similarly, caused the appearance of FKBP13 in every nucleus, but its distribution appeared quite different from that of BIG1, and a larger fraction of FKBP13 than of BIG1 remained in the cytoplasm (Fig. 2II). Cyclosporin and rapamycin had no apparent effect on the distribution of BIG1 or FKBP13 (data not shown).
Fig. 2.
Effect of FK506 on localization of endogenous BIG1 (I) and FKBP13 (II). FBS-starved HepG2 cells were incubated for 30 min without or with 100 nM FK506 before preparation for microscopy. All observations were replicated three or more times. (Scale bars: 40 μm in I and 20 μm in II.)
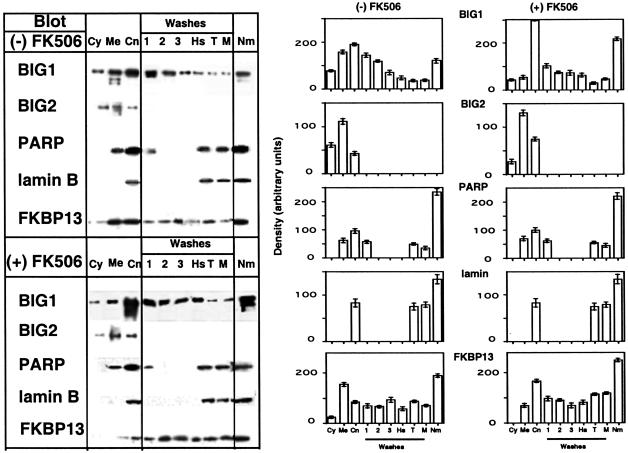
Subcellular fractionation confirmed a shift of BIG1 and FKBP13 from membrane to nuclear fractions after incubation with FK506 (Fig. 3 Left), which densitometry made more clear (Fig. 3 Right). There was no similar change in distribution of BIG2, PARP, or lamin B after incubation of FBS-starved cells with FK506. HepG2 cell lysates were separated into cytosol, membrane, and crude nuclear fractions; the last was fractionated to yield subfractions and purified nuclear matrix. Microscopically, BIG1 and FKBP13 appeared associated with different structural elements in the cytoplasm. In cell homogenates, some BIG1, but not FKBP13, was always seen in cytosol (Fig. 3). After incubation of cells with FK506, the percentages of BIG1 and FKBP13 in the crude nuclei and nuclear subfractions were clearly increased (Fig. 3). Of note, there appeared to be preferential translocation of two faster-migrating forms of immunoreactive BIG1 from membrane to nuclear fractions, and specifically to the purified nuclear matrix.
Fig. 3.
Effect of FK506 on localization of endogenous BIG1 and other proteins by subcellular fractionation. FBS-starved HepG2 cells were incubated without or with 100 nM FK506 for 30 min before preparation and fractionation of homogenates. Samples [100 μg of protein, except for nuclear matrix (30 μg)] of each fraction were subjected to SDS/PAGE, and blots were reacted with antibodies against BIG1, BIG2, PARP, lamin B, or FKBP13. (Left) Representative Western blots. (Right) Data are means (± one-half of the range) of densitometric values from duplicate samples in one experiment representative of three. Fractions are cytosol (Cy), membranes (Me), and crude nuclei (CN). Washes: 1, first low-salt; 2, second low-salt; 3, third low-salt; Hs, high-salt; T, Triton X-100; M, matrix wash; Nm, nuclear matrix. Data were similar in two other experiments.
BIG2 was present in both cytosol and membrane fractions, with a small amount in the crude nuclei, but it was not detected in nuclear subfractions (Fig. 3), nor was it seen in nuclei by immunofluorescence microscopy (results not shown). PARP (also present in membranes) and lamin B were similarly distributed among different fractions (Fig. 3). In other experiments (data not shown), CRM1 (exportin) was identified in several nuclear subfractions, as was nuclear mitotic apparatus protein, which was also abundant in the membrane fraction, whereas GM130, a Golgi protein, was detected only in membranes and, to a small extent, in the crude nuclear fraction but not in any of its subfractions.
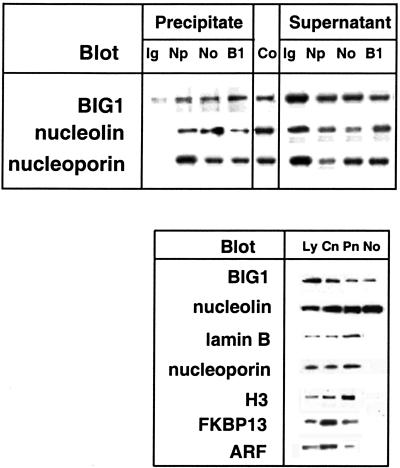
Intranuclear Localization of BIG1. On confocal immunofluorescence microscopy, nuclear BIG1 staining varied in intensity, with small areas of higher concentration, from which we suspected a nucleolar localization. As shown in Fig. 4II, BIG1 was partially colocalized with nucleolin in nuclei but not in cytoplasm, where, especially near the plasma membrane, collections of nucleolin were clearly not associated with BIG1. In Fig. 4I, nucleolin is seen without BIG1 in nuclei of two cells that lack visible intranuclear BIG1. Differences between nucleoporin p62 distribution in cells with and without BIG1 in the nucleus are shown in fields of FBS-starved cells (Fig. 4I). In cells with little or no nuclear BIG1 (Fig. 4IA), nucleoporin p62 is clearly concentrated at the periphery of most nuclei (presumably the nuclear envelope), with some scattered through the cytoplasm and little to be seen within the nucleus. In contrast, cells with essentially all BIG1 in the nucleus (Fig. 4IB) contain seemingly more nucleoporin near the nuclear envelope, and the inner portion appears to coincide with BIG1; the outer nucleoporin (red) does not. There is also less nucleoporin p62 in the cytoplasm and more scattered through the nuclei in these cells (Fig. 4IB). Sites of apparent coincidence of BIG1 and nucleoporin staining within the nucleus clearly differ in morphology from double-staining nucleoli in Fig. 4II. Nucleoporin p62 was not detected in purified nucleoli, which contained BIG1 and nucleolin but lacked also FKBP13, ARF, lamin B, and histone H3 (Fig. 5).
Fig. 5.
Interactions of BIG1 with nuclear proteins. (Upper) Immunoprecipitation of BIG1, nucleolin, and nucleoporin from HepG2 nuclei. Samples (200μg of protein) of purified nuclei from FBS-starved cells were incubated with rabbit IgG (Ig), anti-nucleoporin (Np), anti-nucleolin (No), or anti-BIG1 (B1) antibodies. Samples (20 μl) of supernatant or of immunoprecipitated proteins eluted from beads in 100 μl of gel-loading buffer, and 1.5 μg of total nuclear protein (Co) were separated by SDS/PAGE and transferred to nitrocellulose, which was divided for reaction with antibodies against BIG1, nucleolin, or nucleoporin. (Lower) BIG1 and nucleolin in purified nucleoli. Samples (100 μg of protein) of lysate (Ly), crude nuclear (Cn), pure nuclear (Pn), and pure nucleolar (No) fractions were separated by SDS/PAGE and reacted on blots with antibodies against BIG1, nucleolin, nucleoporin, lamin B, histone H3, FKBP13, or ARF. Data were similar in two other experiments.
Coimmunoprecipitation of BIG1 and Nucleolin. From HepG2 cell nuclei purified by density gradient centrifugation, antibodies against BIG1, nucleoporin, or nucleolin each precipitated also the other two proteins (Fig. 5). Antibodies against BIG1 and nucleolin failed to precipitate ARF, FKBP13, lamin B, CRM1 (exportin), PARP, Ran, or RanBP (data not shown).
Discussion
After observing the interaction of BIG1 with FKBP13, the effects of FK506 on distribution of BIG1 in Jurkat cells, and cAMP effects on BIG1 (and BIG2) distribution in HepG2 cells, we looked for effects of serum starvation on the BIG proteins. HepG2 cells were selected for these experiments because they seemed more suitable than Jurkat cells for immunofluorescence microscopy, although in earlier studies we had not detected endogenous BIG1 in HepG2 (or HeLa) cell nuclei (13). Cells growing in medium containing FBS had been used for previous experiments (13, 16), and we found, as reported here, that incubation of HepG2 cells without FBS resulted in a striking accumulation of BIG1 in most nuclei. Cells that had been incubated 16–18 h without FBS were then used for further experiments to characterize BIG1 in the nucleus microscopically and by subcellular fractionation.
BIG1 was essentially completely concentrated in nuclei of FBS-starved cells incubated for 30 min with FK506. Some FKBP13 was also present in nuclei in those cells, but the fraction still in the cytoplasm was much greater than the fraction of BIG1 that remained. It seemed clear also that BIG1 and FKBP13 were quite differently localized in the nuclei, just as they were in the cytoplasm. Differences were evident microscopically as well as by immunoblotting of electrophoretically separated proteins in subcellular and nuclear fractions. In addition, FKBP13 was not detected among proteins immunoprecipitated by antibodies against BIG1 from nuclei purified from FBS-starved HepG2 cells. FKBP13 had been precipitated by anti-BIG1 antibodies from membranes of Jurkat cells growing in medium with FBS (16). Whether these differences in molecular interactions are due to differences in growth conditions or differences in types of cells or both, we have not yet determined. We need also to learn to what extent the effects of FBS may be due to alterations in morphology during the cell cycle and/or to effects of specific FBS components via other signaling pathways on structure and function.
Microscopically, BIG1 was distributed unevenly throughout the nuclei of FBS-starved cells. BIG1's association with nuclear pore structures was demonstrated by confocal immunofluorescence and by coimmunoprecipitation with antibodies against nucleoporin p62 (20). BIG1 was also identified in purified nuclear matrix, where it could be involved in processes of DNA repair, replication, and transcription (21, 22), as well as in nucleoli (23). This ubiquitous distribution seems quite consistent with the increasingly recognized dynamic nature of nuclear architecture, the magnitude (rate and volume) of protein exchange between nucleus and cytoplasm, and the energy-independent unobstructed movement of most proteins within the nucleus (24, 25). Localization of BIG1 in the nucleolus is of interest because of the structure and multiple functions of this organelle, both of which are maintained while the component proteins are transient. Residence times of many nucleolar proteins are <1 min (24). Such dynamic characteristics are crucial for regulation of the complex molecular machinery through which nucleolar operations, including transcription, RNA processing, translation, and ribonucleoprotein assembly, are accomplished. This environment enables the alterations necessary during the cell cycle as well as in response to other signals, but it also causes problems in defining nuclear regulatory mechanisms and difficulties for us in establishing procedures for consistent replication of the observations described here.
BIG1 was, in part, colocalized with nucleoporin p62 at the nuclear envelope, probably in transit between nuclear and cytoplasmic compartments. Their colocalization (or coimmunoprecipitation) does not, of course, mean a direct molecular interaction of BIG1 and nucleoporin, which seems, in fact, unlikely. Similarly, the demonstrated coimmunoprecipitation of BIG1 and nucleolin from nuclei and their colocalization in nucleoli (microscopically and in purified preparations) probably do not reflect a direct interaction. Piñol-Roma (26) reported the disruption of association of nucleolin with several proteins in ribonucleoprotein complexes from HeLa cells by RNase, but not by DNase.
The dynamic nature of nuclear morphology is remarkable; it is also critical for precision of timing, specificity, and continuity in effector responses (e.g., transcription, translation) as well in signal transduction (27, 28). Misteli (29) considered several dynamic intracellular systems in a valuable review of “self-organization,” defined as “the capacity of a macromolecular complex or organelle to determine its own structure based on the functional interactions of its components.” The nucleus and the Golgi complex were discussed as examples of this paradigm. It now appears that BIG1 is a participant in both nuclear and Golgi systems. We believe that BIG1 may be a signaling, as well as a scaffolding, molecule for the necessary integration of complex alterations that both of these organelles must undergo throughout the cell cycle, most dramatically during mitosis.
BIG1 is a protein of 1,849 aa with no function(s) assigned to most of its structure. Amino acids 711–715 (KKPKR) contain a putative nuclear localization signal. The central ≈200-aa Sec7 domain is present in all ARF-activating GEPs; it is responsible for the acceleration of guanine nucleotide exchange on ARF (8), the action that is required for initiation of vesicle formation from Golgi membranes (2), as well as for the inhibition of BIG1 by brefeldin A (30), and was the basis for its purification (9, 10). The N-terminal 331 aa of BIG1 contain sequence that interacts with FKBP13 (16); however, we do not know how or whether BIG1 and FKBP13 are associated when they enter the nucleus in response to FK506. We do know that they are seen at different intranuclear sites. Also in the N-terminal region of BIG1 is a protein kinase A-anchoring sequence identical to that in BIG2, which was shown to bind each of the four kinase regulatory subunits (15); its functionality in BIG1 has not been investigated. Overall, more than one-half of the BIG1 molecule, including >900 C-terminal amino acids, is without known interactions and may perform many other functions.
The role of ARF, if any, in the translocation of BIG1 and/or its function in the nucleus remains uncertain. There is no reason to expect that all BIG1 actions involve ARF. It has been difficult to be convinced that ARF is, in fact, in the nucleus or is colocalized with BIG1, although by immunofluorescence microscopy it often appears to be (Fig. 4III). We have never detected ARF among proteins precipitated from purified nuclei by anti-BIG1 antibodies. This may reflect, among other things, a relative low affinity of ARF interactions or the rapid inactivation of ARF and dissociation of complexes perhaps induced by experimental manipulations, including enzyme activities released during cell breakage. Questions regarding ARF and, of course, the nuclear action of BIG1 in relation to its other functions are among many that we are trying to answer.
Acknowledgments
We thank Dr. Christian Combs (National Heart, Lung, and Blood Institute Confocal Microscopy Core Facility) and Michael W. Spencer for invaluable help and Dr. Vincent C. Manganiello for helpful discussions and manuscript review.
Abbreviations: ARF, ADP-ribosylation factor; GEP, guanine nucleotide-exchange protein; BIG1, brefeldin A-inhibited GEP; FKBP, FK506-binding protein; PARP, polyADP-ribose polymerase; IGEPAL, (octylphenoxy)polyethoxyethanol.
References
- 1.Tsuchiya, M., Price, S. R., Tsai, S.-C., Moss, J. & Vaughan, M. (1991) J. Biol. Chem. 206, 2772-2777. [PubMed] [Google Scholar]
- 2.Rothman, J. E. & Wieland, F. T. (1996) Science 272, 227-234. [DOI] [PubMed] [Google Scholar]
- 3.Moss, J. & Vaughan, M. (1998) J. Biol. Chem. 273, 21431-21434. [DOI] [PubMed] [Google Scholar]
- 4.Donaldson, J. G. (2003) J. Biol. Chem. 278, 41573-41576. [DOI] [PubMed] [Google Scholar]
- 5.Jackson, C. L. & Casanova, J. E. (2000) Trends Cell Biol. 10, 60-67. [DOI] [PubMed] [Google Scholar]
- 6.Donaldson, J. G. (2000) Proc. Natl. Acad. Sci. USA 97, 3792-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franzusoff, A. & Schekman, R. (1989) EMBO J. 8, 2695-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chardin, P., Paris, S., Antonny, B., Robineau, S., Beraud-Dufour, S., Jackson, C. L., and Chabre, M. (1996) Nature 384, 484-484. [DOI] [PubMed] [Google Scholar]
- 9.Morinaga, N., Tsai, S.-C., Moss, J. & Vaughan, M. (1996) Proc. Natl. Acad. Sci. USA 93, 12856-12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morinaga, N., Moss, J. & Vaughan, M. (1997) Proc. Natl. Acad. Sci. USA 94, 12926-12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morinaga, A, Adamik, R., Moss, J. & Vaughan, M. (1999) J. Biol. Chem. 274, 17417-17423. [DOI] [PubMed] [Google Scholar]
- 12.Togawa, A., Morinaga, N., Ogasawara, M., Moss, J. & Vaughan, M. (1999) J. Biol. Chem. 274, 12308-12315. [DOI] [PubMed] [Google Scholar]
- 13.Yamaji, R., Adamik, R., Takeda, K., Togawa, A., Pacheco-Rodriguez, G., Ferrans, V., Moss, J. & Vaughan, M. (1999) Proc. Natl. Acad. Sci. USA 97, 2567-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mansour, S. J., Skaug, J., Zhao, X. H., Giordano, J., Sherere, S. W. & Melancon, P. (1999) Proc. Natl. Acad. Sci. USA 96, 7968-7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, H., Adamik, R., Pacheco-Rodriguez, G., Moss, J. & Vaughan, M. (2003) Proc. Natl. Acad. Sci. USA 88, 6677-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Padilla, P. I., Chang, M.-J., Pacheco-Rodriguez, G., Adamik, R., Moss, J. & Vaughan, M. (2003) Proc. Natl. Acad. Sci. USA 100, 2322-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Resendez-Perez, D., Barrera-Saldana, H. A., Morales-Vallarta, M. R., Ramirez-Bon, E., Leal-Garza, C. H., Feria-Velazco, A. & Sanchez-Anzaldo, F. J. (1984) Placenta 5, 523-532. [DOI] [PubMed] [Google Scholar]
- 18.Alvarez-Gonzalez, R. & Ringer, D. (1988) FEBS Lett. 236, 362-366. [DOI] [PubMed] [Google Scholar]
- 19.Muramatsu, M., Hayashi, Y., Onishi, T., Sakai, M., Takai, K. & Kashiyama, T. (1974) Exp. Res. Commun. 88, 345-351. [DOI] [PubMed] [Google Scholar]
- 20.Suntharalingam, M. & Wente, S. (2003) Dev. Cell 4, 775-789. [DOI] [PubMed] [Google Scholar]
- 21.Nickerson, J. (2001) J. Cell Sci. 114, 463-474. [DOI] [PubMed] [Google Scholar]
- 22.Berezney, R. (2002) Adv. Enzyme Regul. 42, 39-52. [DOI] [PubMed] [Google Scholar]
- 23.Ginistry, H., Sicard, H., Roger, B. & Bouvet, P. (1999) J. Cell Sci. 112, 761-772. [DOI] [PubMed] [Google Scholar]
- 24.Roix, J. & Misteli, T. (2002) Histochem. Cell Biol. 118, 105-116. [DOI] [PubMed] [Google Scholar]
- 25.Misteli, T. (2003) Nat. Cell Biol. 5, 183-184. [DOI] [PubMed] [Google Scholar]
- 26.Piñol-Roma, S. (1999) Mol. Biol. Cell 10, 77-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Misteli, T. (2001) Science 291, 843-847. [DOI] [PubMed] [Google Scholar]
- 28.Wolfe, A. P. & Hansen, J. C. (2001) Cell 104, 631-634. [DOI] [PubMed] [Google Scholar]
- 29.Misteli, T. (2001) J. Cell Biol. 155, 181-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sata, M., Moss, J. & Vaughan, M. (1999) Proc. Natl. Acad. Sci. USA 96, 2752-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]