Abstract
Despite many years of genetic and biochemical studies on the λ integrase (Int) recombination system, it is still not known whether the Int protein is competent for DNA cleavage as a monomer. We have addressed this question, as part of a larger study of Int functions critical for the formation of higher-order complexes, by isolating “multimer-specific” mutants. We identify a pair of oppositely charged residues, E153 and R169, that comprise an intermolecular salt bridge within a functional Int multimer. Mutation of either of these residues significantly reduces both the cleavage of full-att sites and the resolution of Holliday junctions without compromising the cleavage of half-att site substrates. Allele-specific suppressor mutations were generated at these residues. Their interaction with wild-type Int on preformed Holliday junctions indicates that the mutated residues comprise an intermolecular salt bridge. We have also shown that the most C-terminal seven residues of Int, which comprise another previously identified subunit interface, inhibit DNA cleavage by monomeric but not multimeric Int. Taken together, our results lead us to conclude that Int can cleave DNA as a monomer. We also identify and discuss unique structural features of Int that act negatively to reduce its activity as a monomer and other features that act positively to enhance its activity as a multimer.
The integrase protein (Int) of Escherichia coli phage λ (1), which mediates the integration and excision of the viral genome in to and out of its host chromosome by recombination at specific loci (att sites) (2), belongs to the large λ Int family of recombinases (for review see refs. 3 and 4). They use transient covalent phospho-tyrosine intermediates to first generate and then resolve Holliday junction (HJ) recombination intermediates by means of two sequential pairs of strand exchanges. The locus of these reactions on each partner DNA duplex is a pair of 9- to 13-bp inverted binding sites for the recombinase (core-type sites) separated by a 6- to 8-bp overlap region (7 bp in the case of λ) whose boundaries are defined by the staggered and precisely positioned DNA cleavage sites. The att sites of the λ pathway, and other “heterobivalent” recombinases have the additional complexity of nearby binding sites for accessory DNA bending proteins and binding sites for a second, N-terminal, recombinase domain that binds distant “arm-type” DNA sites.
Mechanistic studies of the λ Int family have benefited enormously from the availability of crystal structures of several family members (reviewed in ref. 5), the most relevant to this work being the structures for the catalytic domain of λ Int (6), the covalent complex with DNA of the λ Int C75 domain (which is analogous to the monovalent recombinases Cre and Flp) (7), and the Cre recombinase complexed with a HJ and a recombination synapse (8, 9). The common motif that emerges from all of the λ-family structures is a C-terminal catalytic domain (containing the tyrosine nucleophile and active site pocket) appropriately positioned for DNA cleavage on one face of a “core-type” binding site. On the other face of the DNA helix, and connected by a long linker, is a second domain [the N-terminal domain in the case of Cre and Flp and the central, or core binding (CB), domain in the case of λ Int]. For λ Int, most of the affinity for a core-type site resides in the CB domain. The cocrystals of a HJ with Cre (8) or Flp (10) dramatically reveal pseudo fourfold symmetric arrangement of recombinases around the four-way junction, with the four N-terminal domains interacting on one side of the approximately planar HJ and the four catalytic domains interacting on the other face of the HJ. These structures, as well as a considerable amount of genetic and biochemical data, indicate that the C-terminal residues of each recombinase participate in trans (intermolecular) interactions that seem to have a regulatory function, albeit by different mechanisms in each system (8, 10–13). Some of the experiments in this report are designed to reconcile and expand on the existing structural, genetic, and biochemical data pertaining to the role of the C-terminal tail of λ Int (6, 7, 14–16).
Despite many years of genetic and biochemical studies on the λ Int recombination system, it is still not known whether the Int recombinase is competent for DNA cleavage as a monomer. When Kikuchi and Nash first purified Int, they showed that at high salt it is a monomer in solution (17). However, at more physiological salt concentrations Int has a strong tendency to form multimers, both in the absence and presence of DNA (18). Although it is clear that Int functions as a multimeric complex during recombination, it has been difficult to establish the functional competency of a single Int protomer. Even when Int's basic nicking/ligation activity is assayed as a topoisomerase, relaxing a supercoiled DNA substrate lacking specific protein binding sites (19), one cannot rule out the action of transient multimers as the minimal functional unit.
We have addressed this question, as part of a larger study of the multimeric functions of Int, by using the following logic. If a half-att site suicide substrate can be cleaved by a single Int protomer (acting on its own), then it might be possible to isolate mutants that are competent to cleave this substrate but impaired for the cleavage of a full-att site and/or the resolution of HJs. We have identified a pair of oppositely charged residues, E153 and R169, that we propose comprise an intermolecular salt bridge within a functional Int multimer. The effects of allele-specific suppressor mutations at these residues demonstrate that they comprise an ion pair that makes an intermolecular interaction between Int subunits. We have also studied the previously isolated and well characterized truncation mutant, W350ter, in which the seven C-terminal residues (after Glu-349) have been deleted (15, 20, 21). We find that W350ter is much more efficient than wild-type Int in cleaving half-att sites, but it is less efficient than wild-type in cleaving full-att sites. Taken together, the results lead us to conclude that Int can cleave DNA as a monomer. We also identify and discuss structural features of Int that act negatively to reduce its activity as a monomer and other features that act positively to enhance its activity as a multimer.
Materials and Methods
Oligonucleotides and Protein Preparation. The wild-type and mutant Int proteins were prepared from expression plasmids as described (22). The QuikChange site-directed mutagenesis kit (Stratagene) was used to generate mutations in the Int gene on the pRT101 plasmid (23) using HPLC purified oligonucleotides (Operon Technologies, Alameda, CA): (R169A) 5′-GTCGCTGCCACTGCCGCAGCAAAATCAGAGG-3′; (E153A) 5′-GATGCATTCCGAGCGGCAATAGCTGAAGGCC-3′; (R169D) 5′-CCATGTCGCTGCCACTGACGCAGCAAAATCAGAGG-3′; (E153R) 5′-GCGATGCATTCCGACGGGCAATAGCTGAAGGCC-3′; as primers. The double mutants E153R;R169D and E153A;R169D were derived from the R169D mutant by using the E153R and E153A primers, respectively. Full-att and half-att site suicide substrates and the small synthetic HJ (CM7) oligonucleotides were designed as described (24, 25) and prepared as HPLC-purified oligos by Operon Technologies. The sequence of the top strand of the half-att site and the full-att site up to the nick is 5′-TTAAGGTTGAATCATATTT. In the full-att site, the top-strand sequence after the nick is 5′-TTTCGACGAGCT (see Fig. 1). The BOC′ strand from the HJ and the top strand from the half-att site and full-att site were 5′ end-labeled with [γ-32P]ATP (NEN) using T4 polynucleotide kinase and precipitated in 0.3 M Na-acetate and 95% ethanol.
Fig. 1.
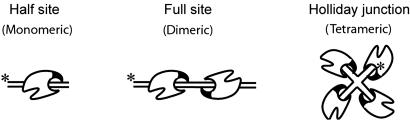
Schematic representation of the three different DNA substrates used in this study. The half-att site, the full-att site, and the HJ have one, two, and four core-type Int binding sites, respectively. The two core-type Int binding sites in the full site are inverted repeats separated by a 7-bp overlap region and containing a nick in the top strand between the third and fourth base pair (COC′ suicide substrate) (see Materials and Methods). The half-att site contains only the first three base pairs of the overlap region, i.e., extends 3 bp beyond the Int cleavage site. In both of these substrates, cleavage by Int releases a three-base oligonucleotide that diffuses away, thus removing the 5′ hydroxyl that would otherwise attack and reverse the 3′ phosphotyrosine bond between Int and the att site. The trapped covalent Int–DNA complex is readily separated from the uncleaved substrate by SDS gel electrophorsesis. The positions of the 32P-labeled 5′ termini of the substrates in this report are indicated by asterisks.
Cleavage Assays of Full-att and Half-att Site Suicide Substrates and HJ Resolution Assay. DNA cleavage and recombination assays of wild-type and mutant Int proteins were carried out at 19°C in 100-μl reactions containing 10 mM Tris·HCl (pH 8.00), 55 mM NaCL, 5 mM DTT, 0.5 mg/ml BSA, and 20 nM of the indicated suicide substrate DNA. Ten-microliter aliquots were removed at the indicated time points and mixed with 4 μl of gel loading solution containing SDS. For the HJ resolution mixing experiment, the E153R;R169D double mutant (400 nM) was initially added to the 10-μl reaction along with 10 nM HJ substrate, and then wild-type Int was added as indicated. The resulting products were separated by electrophoresis on a 7% polyacrylamide/0.4% SDS gel at 200 V for 30 min. The radiolabeled bands were detected by autoradiography and quantitated by using a Fuji BAS 2500 PhosphorImager system.
Results
Multimer-Specific Interactions. To identify residues that might differentiate between the monomeric and multimeric functions of Int, we made two tentative assumptions. The first was to assume that some of the residues important for the multimeric functions are located on the surface of the Int crystal structure. The second was that cleavage of a half-att site suicide substrate involved a single Int protomer, whereas cleavage of a full-att site suicide substrate involved two or more Int protomers. The design of the two suicide substrates and the mechanism by which they trap covalent Int cleavage complexes are described in Fig. 1 and in Materials and Methods.
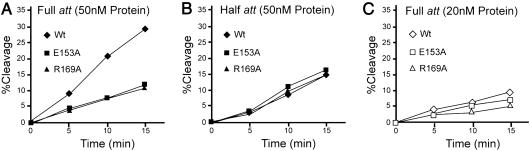
Inspection of the central, core-binding domain of Int (the CB domain) yields a number of charged residues exposed on the surface. The most attractive candidates for residues functioning within higher-order recombination complexes came from modeling the cocrystal structure of C75 Int covalently complexed to an att site (7) onto the Cre-recombination synapse cocrystal structure (9). Based on this rough model, we made single alanine substitution mutants at E153 and R169 and found that they were ≈3-fold less efficient than wild-type in cleaving the COC′ suicide substrate (Fig. 2A). However, the mutants were indistinguishable from wild-type Int in the rate at which they cleaved the half-att suicide substrate (Fig. 2B).
Fig. 2.
Cleavage rates for Int and Ala substitution mutants on full-att and half-att site suicide substrates. The amount of covalent complex formed by Int (diamonds), R169A (triangles), and E153A (squares) on a full-att (A and C) or a half-att suicide substrate (B) is shown as a function of time. The reactions containing 20 nM DNA and 50 nM protein (A and B) or 20 nM protein (C) were carried out for the indicated amounts of time, terminated with SDS, analyzed by SDS/PAGE, and quantitated on a PhosphorImager as described in Materials and Methods. The full-att and half-att site substrates are described in Fig. 1 and Materials and Methods.
At low protein concentrations, the mutants and wild-type Int have the same DNA cleavage activity on COC′, in accordance with their similar activities on the half-att substrate at higher protein concentrations (Fig. 2C). At low protein concentrations, only one of the two Int-binding sites in COC′ is expected to be filled, and the COC′ site behaves like the half-att site. We take this as support of our assumptions that at higher Int concentrations the two substrates do indeed distinguish between monomeric and multimeric Int activities. We conclude that E153A and R169A have normal catalytic proficiency for DNA cleavage as monomers, but cleavage activity requiring the formation of functional multimeric complexes is severely depressed in the mutants. This phenotype is consistent with that of an E153K excision-defective mutant, previously isolated in the Gardner laboratory, by virtue of its inability to form attL complexes in vivo and shown to have topoisomerase and HJ resolution activity in vitro (15).
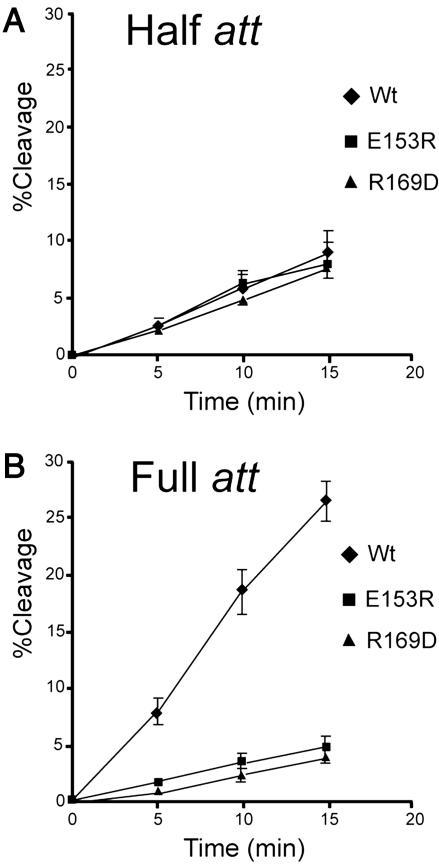
Allele-Specific Intermolecular Intragenic Suppression. Because the Ala substitutions at 153 and 169 have a multimer-specific phenotype and the parent residues have the potential to form an ion pair, we constructed two charge reversal mutants, E153R and R169D, and showed that individually they have the same phenotype as mutants with Ala substitutions at these positions (Fig. 3).
Fig. 3.
Cleavage rates for Int and charge reversal substitution mutants on full-att and half-att site suicide substrates. The amount of covalent complex formed by Int (diamonds), R169D (triangles), and E153R (squares) on a full-att (A) or a half-att (B) suicide substrate is shown as a function of time. The reactions, containing 20 nM DNA and 50 nM protein, were carried out and analyzed as described in Fig. 2.
To determine whether E153 and R169 form an ion pair critical for the multimeric function of Int, we took advantage of the HJ resolution assay (26). The substrate in this assay is a small synthetic four-way DNA junction containing four inverted core-type Int binding sites in a configuration that mimics a single-strand exchange intermediate of Int-mediated recombination (26–28). Int cleavage at one pair of “partner” att sites generates one pair of duplex DNA products, whereas cleavage at the other pair of partner att sites generates the alternative pair of duplex DNA products. Even though resolution of the HJ intermediate involves DNA cleavage at only two sites, at least three Int protomers, and possibly four, must bind to the HJ to catalyze this reaction (29).
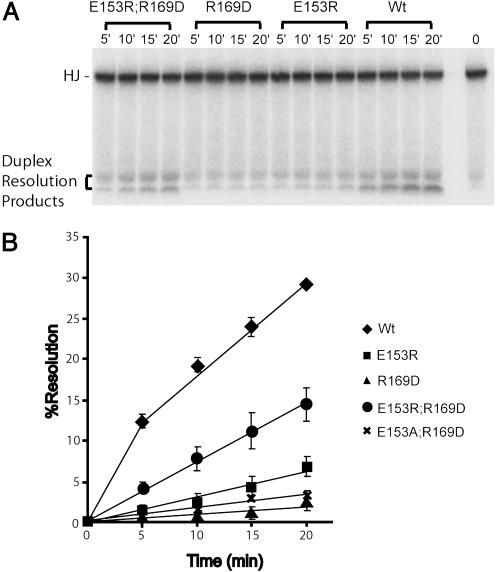
Consistent with the results obtained in the COC′ suicide cleavage assays, the two single mutants E153R and R169D are both defective for resolving the HJ (Fig. 4). However, when an E153R;R169D double mutant was constructed, it was found to be clearly more efficient than either of the single mutants, although it is not as proficient as wild-type Int. The mutual suppression of the two mutant phenotypes is highly specific for this particular ion pair, and it is not observed with either E153A;R169D (Fig. 4B) or E153R;R169E double mutants (data not shown; see also Discussion). It is not surprising to find a narrow window of parameters within which allele-specific second-site suppression is observed because the swapping of charged residues may have secondary unfavorable effects, either within an Int protomer or on the interfaces between Int subunits.
Fig. 4.
HJ resolution by mutant and wild-type Ints. (A) HJ assays were carried out with the indicated proteins as described in Materials and Methods and analyzed by SDS/PAGE and autoradiography. (B) Quantitation of the gel profiles on a PhosphorImager was used to determine the amount of the two resolution products and the percent resolution product is plotted as a function of time for each of the proteins: Int (diamonds), E153R (squares), R169D (triangles), E153R;R169D (circles), and E153A;R169D (crosses).
The observed allele-specific intragenic suppression could result from the formation of an intramolecular ion pair or from an intermolecular ion pair between different Int monomers within higher-order complexes. To distinguish these possibilities, we challenged the double-mutant Int with increasing amounts of wild-type Int in a HJ resolution assay. If the mutant suppression involves an intramolecular ion pair, then we would expect the displacement of mutant subunits by wild-type Int to result in a simple monotonic transition from the mutant resolution efficiency to the wild-type resolution efficiency. However, if the suppression involves an intermolecular ion pair, the transition should be more complex. In this case, the double-mutant should not interact effectively with a wild-type Int monomer because the intermolecular pairings of residues 153 and 169 oppose two like charges that would repel one another instead of forming a salt bridge. The admixture of wild-type subunits into complexes of double mutant Ints should therefore be inactivating (see cartoon in Fig. 5A).
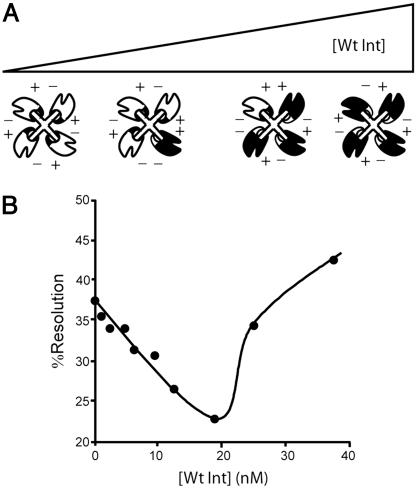
Fig. 5.
HJ resolution with varying ratios of the double mutant E153R/R169D and wild-type Int. (A) Cartoon illustrating the transition of a HJ complex formed entirely with the double mutant (white shapes), through a mixed complex with both kinds of Int, to a complex formed entirely with wild-type Int (black shapes), as the ratio of the two proteins in the reaction is varied. The relative location of the positively (+) and negatively (-) charged residues on the wild-type and double mutant highlights the transition from 100% attractive intermolecular ion pairs for the two homogeneous HJ complexes (all mutant or all wild-type Ints) and one or more repulsive intermolecular ion pairs for each of the heterogeneous HJ complexes (containing a mixture of mutant and wild-type Ints). The cartoon is not intended to depict any particular order of displacement during the titration. (B) The percent of HJ resolution product formed in 1 h in the presence of a constant amount of the double mutant and increasing amounts of wild-type Int. Each reaction contained 10 nM HJ and 400 nM of the E153R;R169D double mutant, to which was added the indicated concentration of wild-type Int. Because the wild-type Int is so much more active than the double mutant, it comprises <15% of the total protein in the reaction.
To test this idea, a series of resolution reactions containing a fixed amount of the double-mutant were titrated with increasing amounts of wild-type Int. Fig. 5B shows that the addition of small amounts of wild-type Int depresses the amount of resolution relative to that of the double-mutant alone. This result is consistent with the proposal that mixed complexes inactivate HJ resolution because of unfavorable interactions between mutant and wild-type subunits involving the modified residues at positions 153 and 169. Resolution efficiency is restored by the addition of more wild-type Int (Fig. 5), which presumably displaces the mutant subunits completely. We take this to reflect the displacement of all of the mutant Ints and formation of purely wild-type complexes. The small amount of wild-type Int required to compete out all of the double-mutant in the resolution reaction agrees with the observation (noted above) that the double-mutant is not as proficient as wild-type Int.
A Multimer-Specific Function of the C-Terminal Residues. In addition to the interactions involving the CB domain, we also investigated the C-terminal region of Int's catalytic domain, which had previously been implicated in intermolecular interactions. This suggestion was initiated by the interesting finding by the Gardner and Gumport laboratories that an Int mutant (W350ter) lacking the C-terminal seven residues (C-terminal tail) retains topoisomerase activity but is unable to carry out recombination (15). Additional genetic and biochemical experiments led to the suggestion that the C-terminal tail is somehow involved in coordinating the multiple DNA cleavage/ligation reactions of recombination (7, 14, 16).
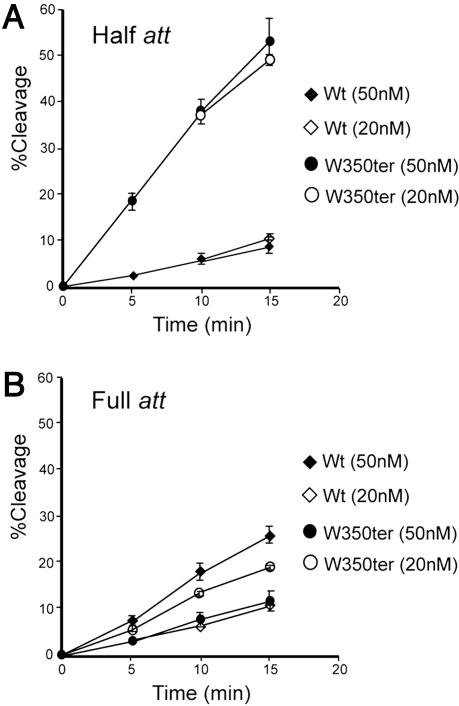
Having demonstrated the usefulness of the half-att/full-att site comparisons, we applied this assay to understanding the role of the C-terminal tail. When a W350ter mutant and wild-type Int are compared for their respective abilities to cleave a half-att site, it is seen that the mutant makes covalent cleavage complex ≈6-fold more efficiently than the wild-type (Fig. 6A). The simplest interpretation of this result is that the C-terminal tail inhibits Int function because its removal activates DNA cleavage activity (16). However, when the two proteins are compared on a COC′ full-att site suicide substrate, the relative activities are reversed and the wild-type Int now makes covalent cleavage complex ≈2 times more efficiently than the W350ter mutant. In this assay for single-site DNA cleavage by a multimeric Int, the presence of the C-terminal tail is clearly important for maximal activity. Is the difference between the two assays due to some (possibly artifactual) difference between the half-att and full-att site DNA substrates? To test this possibility, the full-att site cleavage assay was carried out at 2.5-fold lower Int concentrations, where there is less tendency to form multimeric Int complexes. Under these conditions, the full-att site results are similar to those observed with the half-att site, i.e., W350ter is more efficient than wild-type Int because the C-terminal tail only affords an advantage to multimeric Int.
Fig. 6.
Cleavage rates for Int and W350ter on half-att and full-att site suicide substrates. The rate of covalent complex formation on a half-att (A) or a full-att (B) site was determined at two different protein concentrations as described in Fig. 1 and Materials and Methods. The DNA concentration was 20 nM and the protein concentration of wild-type Int (diamonds) or W350ter (circles) was either 50 nM (filled symbols) or 20 nM (open symbols).
Discussion
As predicted from our rough structural modeling (see also below), mutation of either E153 or R169 generates a phenotype that is observed only in multimeric complexes: DNA cleavage activity on a half-att site is unaffected and activity on a full-att site and/or a HJ is significantly reduced (Figs. 2, 3, 4). Restoration of HJ resolution in the double mutant E153R;R169D (Fig. 4) strongly suggests that these two residues interact, and the inhibition of the double mutant by wild-type Int (Fig. 5) further indicates an intermolecular ion pair that is critical for the multimeric functions of Int.
If this suppression of one mutant by the other were due to the formation of an essential salt bridge, it should depend on having oppositely charged partner residues and might be very sensitive to their precise position and orientation. Accordingly, neither the E153A;R169D double mutant (Fig. 4) nor an E153R;R169E double mutant (data not shown) is any better at resolving HJs than the single mutants by themselves. The failure of the glutamate substitution at the position of R169 to suppress the phenotype of the E153R could be due to an unfavorable interaction with D149 from an opposing residue. The E153R;R169D allele-specific suppression was not observed in the suicide cleavage assay or in a full recombination reaction (data not shown), presumably because the less-than-perfect salt bridge is stabilized more effectively in complexes with a preformed HJ in comparison to complexes with either the COC′ DNA or a synaptic pairing of recombining attP and attB substrates. It is also likely that the recombination reaction is more demanding of a perfect salt bridge than the HJ resolution reaction. For a discussion of the constraints on allele-specific suppression involving swapping of complementary charges, see ref. 30.
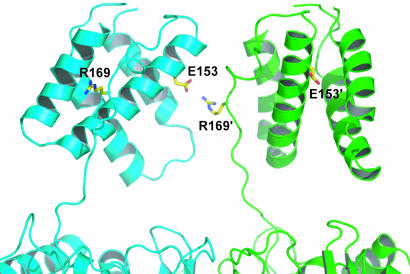
When the structure of Int C75 (7) is superimposed onto the Cre subunits of a recombination synapse cocrystal structure (9), it is evident that R169 of one Int subunit is pointed toward and within 4 Å of E153 of a neighboring Int with a geometry that is consistent with the formation of a salt bridge (31) (Fig. 7) and is consistent with a recent model of the CB–CB domain interface (B. Swalla, personal communication). It can also be seen from the simple superposition in Fig. 7 that the four predicted salt bridges of an Int tetramer are arranged in a cyclical fashion, such that on each Int protomer E153 bridges toward a neighbor in one direction and R169 bridges in the opposite direction to a different Int neighbor.
Fig. 7.
A model of the λ Int dimer showing a possible intermolecular interaction between R169 and E153. The λ Int C75 structure (1P7D) (7) was fitted manually on the crystal structure of a Cre–DNA complex (4CRX) (9), such that the two λ Int molecules pack reasonably well. The side-chain torsion angles of R169 were adjusted manually. The CB domains of λ Int are shown at the top with part of the catalytic domains shown at the bottom. The bound DNA, clamped by the two domains, was omitted for clarity. In this model, the intermolecular distance between R169 and E153 on adjacent protomers is 4 Å, consistent with the ion pair identified by the biochemical experiments reported here. Also illustrated here is the large distance, which precludes intramolecular interactions, between R169 and E153 on the same protomer (labeled with or without a prime). The cyclic pattern of interactions between R169 and E153 is not immediately obvious because only two of the protomers in the tetrameric complex are shown.
The C-terminal seven residues of Int (referred to as the C-terminal tail) have been seen in x-ray crystal structures to assume two very different orientations. In a structure of the catalytic domain (residues 170–356), the C-terminal tail packs against the β2 and β3 sheets of the same protomer (i.e., in cis), such that the tyrosine nucleophile is on a disordered loop and pulled out of the catalytic pocket. A more recent crystal structure of an Int monomer complexed to DNA shows the CB and catalytic domains (residues 75–356) trapped as a recombination intermediate, covalently linked to an att site substrate via the normally transient phospho-tyrosine bond (7). Here the C-terminal tail is observed to pack (in trans) against the β2–β3 sheets of the other protomer in the unit cell. As a result of the rearrangement of the C-terminal tail, the tyrosine nucleophile is now well positioned in the catalytic pocket. Although we believe this observed trans tail interaction is a mechanism for regulating DNA cleavage in monomeric versus multimeric complexes, the fortuitous packing of two independent Int protomers in the crystallographic unit cell is unlikely to mimic the arrangement of Int protomers in the higher-order complexes on DNA. Biochemical verification of the proposed mechanism is therefore especially important.
One of the most useful mutants for studying the role of the C-terminal tail is a deletion of the seven most distal residues (W350ter), which results in a hyperactive topoisomerase (14, 15, 20, 21). Previous results indicating that W350ter is similar to wild-type Int in its ability to cleave both half- and full-att suicide substrates (14) contrasts with the results from our laboratory. We had reported previously (16), and confirm in this report, that W350ter is considerably more active than wild-type Int on a half-att suicide substrate (Fig. 6A). We also show here that W350ter is significantly less active than wild-type Int on a full-att site. We suggest two reasons that may account for the different results from the two laboratories. The experiments reported here used highly purified preparations of Int, as opposed to partially purified extracts, and were carried out with Int concentrations just below the concentration where protein aggregation becomes a complicating factor. We believe that additional considerations, such as the correspondence with the x-ray crystal structures and relative topoisomerase activities (discussed below), lend further weight to the data and conclusions presented here.
The results with W350ter reported here are in excellent agreement with the models proposed on the basis of the x-ray crystal structures (6, 7). On a half-att site, where the monomeric Int is “encouraged” to keep its C-terminal tail in cis because it lacks the opportunity for appropriate trans interactions, the tail is inhibitory. Deletion of the tail in W350ter removes the cis inhibition and facilitates an enhanced rate of DNA cleavage (Fig. 6A and ref. 16). The multimeric complexes that form on a full-att site provide the opportunity for trans tail interactions that are no longer inhibitory, as suggested by the x-ray crystal structure. However, the data presented here, like those from earlier work (14, 16), suggest that the trans tail interactions do more than just neutralize the inhibited cis configuration. When the DNA cleavage rates from Figs. 3 and 6 are normalized to the number of Int binding sites in each substrate (two for the full-att and one for the half-att site), wild-type Int cleaves the full-att site approximately six times faster than it cleaves the half-att site. For R169D and E153R the normalized cleavage rates are the same on both a full-att and a half-att site. In contrast, W350ter cleaves the full-att site approximately half as well as it cleaves the half-att site, suggesting that in the absence of a C-terminal tail aberrant multimers are formed on a full-att site.
All of the results reported here are most reasonably interpreted within the framework of the proposal that the half-att site is cleaved by a monomeric Int, whereas the full-att site is cleaved by a multimer. Indeed, it is very difficult to understand the properties and behavior of the mutants characterized here by any other plausible stoichiometry. Additional support for the proposed stoichiometry comes from the observation that with respect to DNA cleavage rates, a full-att site behaves like a half-att site at low Int concentrations (when it is more likely that only one of the two Int binding sites on the full-att will be occupied) (Fig. 2C). A secondary implication of these results concerns the stoichiometry of Int when it is acting as a nonspecific topoisomerase, something that has been very difficult to determine for λ Int. The fact that W350ter shows the same enhanced activity (relative to wild-type Int) in cleaving a half-att site (Fig. 6 and ref. 16) and in relaxing a non-att-containing supercoiled molecule (15) is consistent with monomeric Int effectively catalyzing the topoisomerase activity.
In conclusion, we note that the observed differences between the mutant and wild-type Ints on the half- and full-att site substrates is primarily due to the fact that the mutants, unlike wild-type Int, do not enjoy the benefit of sisterhood that is afforded by the full-att site. The E153R and R169D single mutants lack the benefit of intermolecular salt bridges between the CB domains, and the W350ter mutant lacks the benefit of trans tail interactions between the catalytic domains. The latter mutant additionally highlights a feature of Int whose primary role is to suppress the activity of monomers while enabling full activity within multimeric complexes. This primary modulation of cleavage activity is distinct from the previously proposed role of the tail in coordinating the activities of individual protomers within the higher-order recombination complex (14, 16).
Acknowledgments
We thank Christine Lank for technical assistance, Joan Boyles for manuscript preparation, members of the Landy and Ellenberger laboratories, especially Dane Hazelbaker, Marta Radman-Livaja, Jeffrey Mumm, David Warren, and Tapan Biswas, for their comments and suggestions, and Reid Johnson, Christie Papagiannis, Bob Weisberg, Peter Droge, and Brian Swalla for critical reading of the manuscript and helpful comments. This work was supported by National Institutes of Health Grants GM62723 and GM33928 (to A.L.) and GM59902 (to T.E.).
Abbreviations: Int, integrase; HJ, Holliday junction; CB, core binding.
References
- 1.Nash, H. A. (1974) Nature 247, 543-545. [DOI] [PubMed] [Google Scholar]
- 2.Campbell, A. M. (1962) in Advances in Genetics, eds. Caspari, E. W. & Thoday, J. M. (Academic, New York), pp. 101-145.
- 3.Azaro, M. A. & Landy, A. (2002) in Mobile DNA II, eds. Craig, N. L., Craigie, R., Gellert, M. & Lambowitz, A. (Am. Soc. Microbiol. Press, Washington, DC), pp. 118-148.
- 4.Grainge, I. & Jayaram, M. (1999) Mol. Microbiol. 33, 449-456. [DOI] [PubMed] [Google Scholar]
- 5.Van Duyne, G. D. (2002) in Mobile DNA II, eds. Craig, N. L., Craigie, R., Gellert, M. & Lambowitz, A. M. (Am. Soc. Microbiol. Press, Washington, DC), pp. 93-117.
- 6.Kwon, H. J., Tirumalai, R. S., Landy, A. & Ellenberger, T. (1997) Science 276, 126-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aihara, H., Kwon, H. J., Nunes-Düby, S. E., Landy, A. & Ellenberger, T. (2003) Mol. Cell 12, 187-198. [DOI] [PubMed] [Google Scholar]
- 8.Gopaul, D. N., Guo, F. & Van Duyne, G. D. (1998) EMBO J. 17, 4175-4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo, F., Gopaul, D. N. & Van Duyne, G. D. (1999) Proc. Natl. Acad. Sci. USA 96, 7143-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, Y., Narendra, U., Iype, L. E., Cox, M. M. & Rice, P. A. (2000) Mol. Cell 6, 885-897. [PubMed] [Google Scholar]
- 11.Spiers, A. J. & Sherratt, D. J. (1999) Mol. Microbiol. 32, 1031-1042. [DOI] [PubMed] [Google Scholar]
- 12.Strizhov, N., Soukovatitsin, V., Ksenzenko, V., Tikhomirova, L. & Bayev, A. (1980) Gene 12, 201-214. [DOI] [PubMed] [Google Scholar]
- 13.Lee, J., Jayaram, M. & Grainge, I. (1999) EMBO J. 18, 784-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kazmierczak, R. A., Swalla, B. M., Burgin, A. B., Gumport, R. I. & Gardner, J. F. (2002) Nucleic Acids Res. 30, 5193-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han, Y. W., Gumport, R. I. & Gardner, J. F. (1994) J. Mol. Biol. 235, 908-925. [DOI] [PubMed] [Google Scholar]
- 16.Tekle, M., Warren, D. J., Biswas, T., Ellenberger, T., Landy, A. & Nunes-Düby, S. E. (2002) J. Mol. Biol. 324, 649-665. [DOI] [PubMed] [Google Scholar]
- 17.Kikuchi, Y. & Nash, H. A. (1978) J. Biol. Chem. 253, 7149-7157. [PubMed] [Google Scholar]
- 18.Jessop, L., Bankhead, T., Wong, D. & Segall, A. M. (2000) J. Bacteriol. 182, 1024-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kikuchi, Y. & Nash, H. A. (1979) Proc. Natl. Acad. Sci. USA 76, 3760-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enquist, L. W. & Weisberg, R. A. (1977) J. Mol. Biol. 111, 97-120. [DOI] [PubMed] [Google Scholar]
- 21.Bear, S. E., Clemens, J. B., Enquist, L. W. & Zagursky, R. J. (1987) J. Bacteriol. 169, 5880-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarkar, D., Radman-Livaja, M. & Landy, A. (2001) EMBO J. 20, 1203-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tirumalai, R. S., Kwon, H., Cardente, E., Ellenberger, T. & Landy, A. (1998) J. Mol. Biol. 279, 513-527. [DOI] [PubMed] [Google Scholar]
- 24.Nunes-Düby, S. E., Radman-Livaja, M., Kuimelis, R. G., Pearline, R. V., McLaughlin, L. W. & Landy, A. (2002) J. Bacteriol. 184, 1385-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radman-Livaja, M., Shaw, C., Azaro, M., Biswas, T., Ellenberger, T. & Landy, A. (2003) Mol. Cell 11, 783-794. [DOI] [PubMed] [Google Scholar]
- 26.Hsu, P.-L. & Landy, A. (1984) Nature 311, 721-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holliday, R. (1964) Genet. Res. 5, 282-304. [Google Scholar]
- 28.Kitts, P. A. & Nash, H. A. (1988) Nucleic Acids Res. 16, 6839-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kho, S. H. & Landy, A. (1994) EMBO J. 13, 2714-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaywitz, A. J., Dove, S. L., Kornhauser, J. M., Hochschild, A. & Greenberg, M. E. (2000) Mol. Cell. Biol. 20, 9409-9422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar, S. & Nussinov, R. (2002) ChemBioChem 3, 604-617. [DOI] [PubMed] [Google Scholar]