Abstract
The calcium, calmodulin-dependent phosphatase calcineurin, regulates growth and gene expression of striated muscles. The activity of calcineurin is modulated by a family of cofactors, referred to as modulatory calcineurin-interacting proteins (MCIPs). In the heart, the MCIP1 gene is activated by calcineurin and has been proposed to fulfill a negative feedback loop that restrains potentially pathological calcineurin signaling, which would otherwise lead to abnormal cardiac growth. In a high-throughput screen for small molecules capable of regulating MCIP1 expression in muscle cells, we identified a unique 4-aminopyridine derivative exhibiting an embedded partial structural motif of serotonin (5-hydroxytryptamine, 5-HT). This molecule, referred to as pyridine activator of myocyte hypertrophy, acts as a selective agonist for 5-HT2A/2B receptors and induces hypertrophy of cardiac muscle cells through a signaling pathway involving calcineurin and a kinase-dependent mechanism that inactivates class II histone deacetylases, which act as repressors of cardiac growth. These findings identify MCIP1 as a downstream target of 5-HT2A/2B receptor signaling in cardiac muscle cells and suggest possible uses for 5-HT2A/2B agonists and antagonists as modulators of cardiac growth and gene expression.
Numerous agonists that act through G protein-coupled receptors trigger calcium-dependent signal transduction pathways that stimulate cardiac growth and gene expression (reviewed in ref. 1). Postnatal cardiac myocytes respond to such signals by hypertrophic growth, characterized by an increase in myocyte size and protein synthesis, assembly of sarcomeres, and activation of a fetal gene program (reviewed in ref. 2). Activation of the calcium, calmodulin-dependent phosphatase calcineurin, is sufficient and, in many cases, necessary for pathological cardiac hypertrophy (3), a major predictor of human morbidity and mortality (4). Thus, there has been intense interest in identifying novel small molecules capable of therapeutically modulating cardiac calcineurin signaling.
Many calcineurin-sensitive genes are controlled by members of the nuclear factor of activated T-cell (NFAT) family of transcription factors, which translocate to the nucleus when dephosphorylated by calcineurin (reviewed in ref. 5). The calcineurin pathway also stimulates the myocyte enhancer factor-2 (MEF2) transcription factor by multiple mechanisms (6). We have shown that calcineurin activates a kinase that phosphorylates class II histone deacetylases (HDACs), which act as MEF2 corepressors (7). Signal-dependent phosphorylation of class II HDACs triggers their export from the nucleus and activation of MEF2 target genes (8, 9). HDAC mutants lacking the signal-responsive phosphorylation sites are refractory to calcium signaling and prevent cardiomyocyte hypertrophy. Conversely, mice lacking class II HDACs are hypersensitive to the growth-promoting activity of calcineurin (7).
The activity of calcineurin is influenced by cofactors known as modulatory calcineurin-interacting proteins (MCIPs) or calcipressins (reviewed in ref. 10). Recent studies in yeast (11) and mammalian cells (12–14) have revealed both positive and negative roles for these proteins in the control of calcineurin activity. Overexpression of MCIP1 (also called Down syndrome critical region 1), for example, suppresses calcineurin signaling (12). In contrast, MCIP1 also seems to potentiate calcineurin signaling, as demonstrated by the diminution of calcineurin activity in the hearts of MCIP1 knockout mice (13). Intriguingly, the MCIP1 gene is a target of NFAT and is up-regulated in response to calcineurin signaling (15), which has been proposed to create a negative feedback loop that dampens calcineurin activity, which would otherwise lead to abnormal cardiac growth.
In an effort to identify novel small molecules that might prevent pathological cardiac hypertrophy by stimulating MCIP1 expression, we performed a high-throughput screen (HTS) of a chemical library for compounds capable of activating the calcineurin/NFAT-responsive promoter of the MCIP1 gene in muscle cells. We describe a previously uncharacterized 4-aminopyridine that we refer to as pyridine activator of myocyte hypertrophy (PAMH), which induces MCIP1 expression and, unexpectedly, drives cardiomyocyte hypertrophy. PAMH acts as a 5-hydroxytryptamine (5-HT)2A/2B receptor agonist and induces hypertrophy, at least in part, by stimulating nuclear import of NFAT and nuclear export of class II HDACs. These findings shed light on a powerful hypertrophic signaling pathway downstream of 5-HT2A/2B receptor signaling and suggest that chemical modulators of this pathway may be efficacious in the control of cardiac growth and gene expression.
Materials and Methods
Cardiomyocyte Cultures. Neonatal rat ventricular myocytes (NRVMs) were cultured as described (16). For detailed procedures, see Supporting Text, which is published as supporting information on the PNAS web site.
Primary HTS. H9c2 cells (American Type Culture Collection no. CRL-1446; ref. 17) were cultured in DMEM with 10% (vol/vol) FBS/4 mM l-glutamine/1% penicillin/streptomycin. Cells at a concentration of 50,000 cells per ml were transiently transfected in batch with a reporter construct (20 pg per cell) encoding firefly luciferase under control of the exon 4 promoter from the human MCIP1 gene (base pairs -874 to + 30 relative to the beginning of exon 4) and FuGENE transfection reagent (6 × 10-5 μl per cell). Transfected cells were plated on 96-well plates (Packard) at a density of 5,000 cells per well. A 20,000-member library of small molecule test compounds from the Myogen Library (selected for molecular diversity and purchased from ChemBridge, San Diego) were then added by using a BioMEK FX robotic liquid handling system (Beckman Coulter) (one compound per well, 10 μM concentration). Sixteen control wells per plate received vehicle alone (0.1% DMSO, final). Plates were incubated for 48 h and processed for quantification of luciferase activity on a multiwell luminometer (Packard Fusion). Primary HTS hits were defined as compounds that produced an increase in luciferase activity >3 SD from vehicle-only controls and were verified in secondary screens.
Western Blot Analysis and Immunostaining. A peptide corresponding to the C terminus of the murine MCIP1 protein (GenBank accession no. AAF63486; CRPEYTPIHLS) was synthesized (Sigma Genosys), incorporating an N-terminal cysteine residue to facilitate conjugation to keyhole limpet hemocyanin (KLH) carrier. Rabbits were immunized with KLH-conjugated peptide according to standard procedures (Lampire Biological Laboratories, Pipersville, PA). Western blots and methods for immunodetection of β-myosin heavy chain and atrial natriuretic factor (ANF) were performed as described in Supporting Text.
RNA Isolation and Analysis. Analysis of RNA expression was performed by microarray and real-time RT-PCR as described in Supporting Text.
Receptor-Binding Assays. Binding of PAMH to the 5-HT2B receptor was measured as described in Supporting Text.
Results
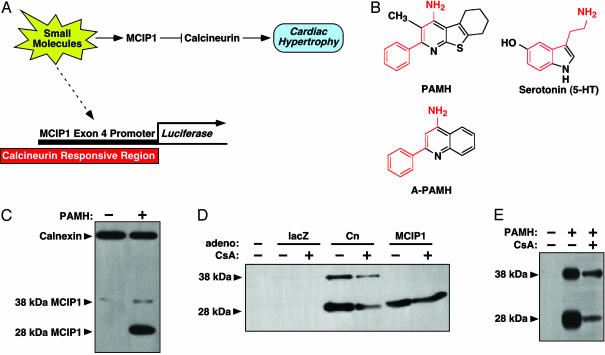
HTS for Molecules That Enhance MCIP1 Expression in Muscle Cells. Based on the resistance to hypertrophy of transgenic mice that overexpress MCIP1 in the heart (18, 19), we sought to identify small molecules capable of increasing MCIP1 expression in muscle cells as a potential means of suppressing pathological cardiac growth. Toward that end, we performed an HTS for compounds able to stimulate expression of a luciferase reporter gene controlled by the alternative promoter upstream of exon 4 of the human MCIP1 gene in the H9c2 muscle cell line (Fig. 1A). This genomic region contains 15 NFAT-binding sites and confers calcineurin responsiveness to MCIP1 (15). Transcripts initiated from the exon 4 promoter encode a 197-aa MCIP1 protein with a Mr ≈ 28 kDa compared with the ≈38-kDa protein encoded by transcripts containing exon 1 (20).
Fig. 1.
Identification of PAMH from a screen for small molecules that enhance MCIP1 expression. (A) Schematic diagram of the screen. A luciferase reporter controlled by the calcineurin-responsive exon 4 promoter of the human MCIP1 gene was transiently transfected into the H9c2 muscle cell line, which was screened in 96-well plates for luciferase expression in the presence of 20,000 individual small molecules. Our hypothesis was that activators of MCIP1 would inhibit cardiac hypertrophy by suppressing calcineurin activity. (B) Structure of PAMH and A-PAMH and their similarity to serotonin. An embedded possible structural motif shared by the three molecules is indicated in red. (C) MCIP1 protein was detected by Western blot after exposure of NRVMs to PAMH (1 μM) for 24 h. Calnexin was detected as a loading control. (D) MCIP1 protein was detected by Western blot in extracts derived from NRVMs infected with adenoviruses encoding lacZ (as a control), activated calcineurin (Cn), or the 28-kDa form of MCIP1 initiated from exon 4 in the presence or absence CsA for 24 h. (E) MCIP1 protein was detected by Western blot in extracts derived from NRVMs after exposure to PAMH (1 μM) in the presence or absence of CsA.
From a screen of 20,000 compounds, we identified 21 that stimulated MCIP1-luciferase expression by 2-fold or greater. The strongest hit, named PAMH, is a previously uncharacterized 4-aminopyridine (5,6,7,8-tetrahydro-3-methyl-2-phenyl[1]benzothieno[2,3-b]pyridin-4-amine), which exhibits an embedded partial structure motif of serotonin (5-HT) (Fig. 1B). Consistent with its ability to stimulate the MCIP1 exon 4 promoter, PAMH selectively and reproducibly induced a dramatic increase in expression of the 28-kDa form of MCIP1 in NRVMs (Fig. 1C).
The sensitivity of the 28-kDa form of MCIP1 to calcineurin signaling is shown in Fig. 1D. Infection of NRVMs with an adenovirus encoding activated calcineurin preferentially upregulates this form of the protein, which comigrates with the MCIP1 protein encoded by a cDNA that initiates from the ATG in exon 4. The 38-kDa form of MCIP that initiates in exon 1 was less responsive but often showed an increase in expression in response to PAMH when cells were maintained at high densities. The variability in responsiveness of the 38-kDa MCIP1 protein may reflect a posttranslational effect of PAMH independent of its effect on the calcineurin-responsive exon 4 promoter. Inhibition of calcineurin activity with cyclosporine A (CsA) diminished the increase in endogenous MCIP1 expression evoked by calcineurin and PAMH (Fig. 1 D and E), further indicating that PAMH activates the calcineurin-responsive MCIP1 promoter.
Stimulation of Cardiomyocyte Hypertrophy by PAMH. Because MCIP1 expression is induced in cardiac myocytes by diverse prohypertrophic agonists (21), we tested positives from the primary screen in a secondary screen for their potential to induce hypertrophy of NRVMs. PAMH potently induced hypertrophy of NRVMs, as measured by assembly of sarcomeres, cell size, and protein synthesis (Fig. 2 A–C). PAMH also activated the expression of ANF and β-myosin heavy chain, markers of the fetal gene program, and promoted hypertrophy as effectively as phenylephrine (PE), a potent hypertrophic agonist that acts through the α-adrenergic signaling pathway (Fig. 2 B–E).
Fig. 2.
Stimulation of cardiac hypertrophy by PAMH. (A) α-Actinin and perinuclear ANF staining of NRVMs in the absence or presence of PAMH (1 μM) for 24 h. PAMH induces hypertrophy, ANF, and sarcomere assembly. (B–E) Cell size, protein content, ANF secretion, and β-myosin heavy chain expression were detected in NRVMs in the absence or presence of PAMH (1 μM) or PE (10 μM) for 24 h. (F) Venn diagram representing array elements regulated by PAMH and PE. Array elements that were regulated at least 2-fold by either PAMH or PE were scored for statistically significant regulation by either compound by using the Affymetrix statistical parameters. The areas of the Venn diagram represent the number of array elements that were significantly regulated by only one drug or by both drugs.
The gene expression patterns in the presence of PAMH and PE, as determined by microarray analysis, were also remarkably similar. Of 15,866 cDNAs analyzed on microarray chips, 175 were upregulated and 226 were down-regulated by at least 2-fold by both compounds (Fig. 2F). The magnitude of changes in gene expression and the rank order of gene responsiveness to the two agonists were also similar with both agonists (Table 1, which is published as supporting information on the PNAS web site, and Supporting Text). MCIP1 mRNA was up-regulated ≈3-fold by PAMH and PE.
Signaling by PAMH Through 5-HT2A/2B Receptors. Following the observation of a partial serotonin pharmacophore embedded in the structure of PAMH (Fig. 1B), and because 5-HT receptor signaling has been implicated in cardiac growth (22–24), we compared the effects of PAMH and serotonin on NRVMs. Serotonin (1 μM) had no observable effects on myocyte growth or MCIP1 expression.
Vigorous contractions were observed after addition of PAMH to NRVMs, suggesting that it acted at the cell surface. To identify cell surface receptors that might mediate its actions, we tested the effects of a series of receptor agonists and antagonists on the ability of PAMH to induce ANF mRNA in NRVMs (Fig. 3A). Despite the similarity between the biological activities of PE and PAMH, the activity of PAMH was unaffected by the α-adrenergic receptor antagonist prazosin or the β-adrenergic receptor antagonist propranolol. In contrast, the general 5-HT receptor antagonist cyproheptadine and the 5-HT2A/2C-selective antagonist, ketanserin, blocked induction of ANF and the 28-kDa form of MCIP1 in response to PAMH (Fig. 3 A and C). AMI-193 and SB204741, which act as 5-HT2A and 5-HT2B selective antagonists, respectively, each had partial inhibitory effects on PAMH action, whereas the 5-HT2B/2C selective antagonist, SB206553, had no effect on PAMH activity (Fig. 3A).
Fig. 3.
Effects of receptor antagonists on the activity of PAMH. (A) NRVMs were pretreated for 1 h with the indicated compound followed by PAMH (1 μM) for 24 h. The ratio of ANF/GAPDH mRNA expression was determined. Values are expressed relative to the ANF/GAPDH ratio in cells treated with PAMH alone, which was assigned a value of 100%. Compounds were used at the concentrations described in Supporting Text. (B) Competition curve showing the percent inhibition of binding of radiolabeled lysergic acid diethylamide to the 5-HT2B receptor by the indicated concentrations of unlabeled PAMH. (C) NRVMs were treated with PAMH (1 μM) and the indicated concentrations of cyproheptadine and ketanserin for 24 h and MCIP1 was detected by Western blot. (D) NRVMs were treated with PAMH (1 μM) in the presence or absence of A-PAMH at the indicated concentrations and secreted ANF was detected. Values are expressed relative to the maximal level of ANF expression in the presence of PAMH. (E) NRVMs were treated with PAMH (1 μM) and A-PAMH at the indicated concentrations, and MCIP1 was detected by Western blot.
We further tested the receptor-binding properties of PAMH by measuring its ability to compete with radiolabeled lysergic acid diethylamide, a standard ligand for the 5-HT2B receptor, in a receptor-binding assay. PAMH bound to the 5-HT2B receptor with high affinity, showing a Ki for the receptor of 64 nM (Fig. 3B). In contrast, PAMH showed no appreciable binding to the α-adrenergic receptor (data not shown). Although PAMH binds the 5-HT2B receptor, the failure of SB206553 to inhibit PAMH activity suggests that the 2A and 2B receptors may play redundant roles in transmitting PAMH signals.
The effects of each of the above antagonists on expression of ANF and MCIP1 and on myocyte hypertrophy and sarcomere assembly in the presence of PAMH were comparable (data not shown). These findings suggest that PAMH acts through 5-HT2A and/or 5-HT2B receptors, and that MCIP1 and hypertrophy are regulated in parallel by PAMH.
In an effort to identify novel PAMH mimetics or antagonists, we compared the activities of a series of structurally related molecules. One such molecule, 2-phenyl-4-quinolinamine, which we refer to as antagonist of PAMH (A-PAMH, Fig. 1B), blocked hypertrophy and induction of MCIP1 in response to PAMH (Fig. 3 D and E).
Analysis of the PAMH Signaling Pathway. To explore the PAMH signaling pathway, we tested the effects of protein kinase and phosphatase inhibitors on PAMH activity in NRVMs. The hypertrophic activity of PAMH was blocked by staurosporin, a general inhibitor of serine, threonine kinases, and by the tyrosine kinase inhibitors AG556 and AG490 (Fig. 4), whereas inhibitors of PKC (Gö6983, Bisindolylmaleimide), PKA (H89, HA1004), PKG (HA1004 and PKG inhibitor), phosphatidylinositol-3 kinase (wortmannin, LY294002), Raf (ZM336372), or MEK1/2 (PD98059) had modest or no effects on PAMH activity. Inhibition of calcineurin by CsA had only a partial effect on the activity of PAMH.
Fig. 4.
Effects of inhibitors on the activity of PAMH. NRVMs were pretreated with the indicated inhibitors for 1 h followed by PAMH (1 μM) for 24 h, and ANF transcripts were detected, as in Fig. 3A. Compounds were used at the concentrations described in Supporting Text.
Transcriptional Responses to PAMH. In light of the sensitivity of the exon 4 promoter of MCIP1 to calcineurin/NFAT signaling (15), we tested whether PAMH promoted the nuclear import of NFAT in NRVMs. PAMH and PE both induced the translocation of a GFP-NFATc fusion protein from the cytoplasm to the nucleus, which suggests that they activate the calcineurin pathway (Fig. 5A).
Fig. 5.
Signaling to NFAT and class II HDACs by PAMH. (A) Stimulation of NFAT nuclear import by PAMH. NRVMs transfected with a GFP-NFATc expression plasmid were exposed to PAMH (1 μM) or PE (10 μM) for 24 h, as indicated, and GFP was detected. In unstimulated cells, GFP-NFATc is localized to the cytoplasm. Both agonists drive GFP-NFATc to the nucleus. (B) Stimulation of GFP-HDAC5 nuclear export by PAMH. NRVMs infected with an adenovirus encoding GFP-HDAC5 were exposed to PAMH (1 μM) or PE (10 μM), as indicated, for 24 h, and GFP was detected. In unstimulated cells, GFP-HDAC5 is localized to the nucleus. Both agonists promote translocation to the cytoplasm, and A-PAMH blocks this effect. (C) Blockade to PAMH activity by signal-resistant HDAC. NRVMs infected with an adenovirus encoding a signal-resistant HDAC5 mutant were exposed to PAMH, as indicated, and stained for α-actinin. The HDAC5 mutant prevents hypertrophy and sarcomere assembly in response to PAMH.
We also examined the effect of PAMH on nuclear localization of HDAC5, which, like other class II HDACs, acts as a suppressor of cardiac hypertrophy (7). Hypertrophic signals lead to the phosphorylation of two critical serine residues in the N-terminal regulatory regions of class II HDACs, which results in their export from the nucleus and derepression of the hypertrophic gene program (8). As shown in Fig. 5B, a GFP-HDAC5 fusion protein was localized to the nucleus in unstimulated NRVMs and became distributed diffusely throughout the cells in the presence of PAMH, suggesting that PAMH activates the kinase-dependent nuclear export of HDAC5. PE also drives nuclear export of GFP-HDAC5. Export was blocked by A-PAMH (data not shown).
To independently test whether HDAC phosphorylation was required for PAMH-mediated hypertrophy, we examined whether a signal-resistant HDAC5 mutant containing alanines in place of serines 259 and 498, which are required for nuclear export, could block the prohypertrophic activity of PAMH. Indeed, NRVMs infected with an adenovirus encoding this HDAC5 mutant showed an increase neither in cell size nor in sarcomere assembly in response to PAMH (Fig. 5C). These findings suggest that hypertrophy in response to PAMH requires transcriptional activation of genes that are repressed by HDAC5, and that PAMH activates a kinase that phosphorylates the regulatory serines that inactivate class II HDACs.
Discussion
In a chemical screen for regulators of MCIP1 expression, we identified a 5-HT2A/2B receptor agonist with potent hypertrophic activity for cardiac muscle cells. This agonist, called PAMH, stimulates the calcineurin-responsive promoter of the MCIP1 gene and promotes myocyte hypertrophy, at least in part, through the calcineurin-dependent nuclear import of NFAT and the kinase-dependent nuclear export of class II HDACs.
Signaling by PAMH Through 5-HT2A/2B Receptors. We speculate that PAMH and serotonin share a partial pharmacophore (Fig. 1B), which directs the activity of PAMH through 5-HT2 receptors. By comparison of a series of PAMH analogs, it appears that the fused cycloalkyl ring is critical for biological activity, and that substitution on the pyridine ring contributes to the prohypertrophic activity. The PAMH antagonist, A-PAMH, which lacks a fused cyclohexyl ring and methyl side chain modification of PAMH, cannot induce hypertrophy, although it can bind 5-HT receptors (data not shown). PAMH is structurally related to a family of molecules that possess anxiolytic activity (25); however, their potential effects on muscle cells have not been reported.
The antihypertrophic activity of A-PAMH has potentially significant implications for the blockade of pathological cardiac hypertrophy. In this regard, it is interesting to note that ketanserin has been shown to reduce left ventricular hypertrophy while preserving cardiac function in hypertensive patients (26), and the 5-HT antagonist sarpogrelate attenuates the ability of the hypertrophic agonists angiotensin II and endothelin-1 to stimulate protein synthesis in cardiomyocytes (27).
Based on the effects of 5-HT2 antagonists on the prohypertrophic activity of PAMH and on ligand-receptor-binding data, we conclude that PAMH is selective for 5-HT2A/2B receptors. Signaling from the 5-HT2B receptor influences cardiac growth and development (reviewed in ref. 24). Knockout mice lacking the receptor die from a thin myocardium (22), and overexpression of the receptor in the adult heart results in hypertrophy (23). Remarkably, serotonin, which also binds 5-HT2 receptors, did not promote hypertrophy of cardiac myocytes. This disparity in actions of serotonin and PAMH could be explained if serotonin is unstable and rapidly degraded. Alternatively, it might activate opposing pro- and anti-hypertrophic signaling pathways, whereas PAMH activates only a prohypertrophic pathway. 5-HT2A/2B receptors might also exist in multiple states (for a review, see ref. 28), providing for the possibility that PAMH has a receptor-state specific effect not accessible by serotonin.
Downstream Signaling by PAMH. The 5-HT2 receptor subtypes (2A, 2B, and 2C) couple to Gαq/G11 and phospholipase C, which trigger intracellular calcium release via inositol triphosphate and consequent activation of PKC (Fig. 6). The strong inhibition of PAMH activity by staurosporin, but not by inhibitors of PKC or other kinases implicated in hypertrophic signaling, suggests that a calcium-dependent kinase yet to be identified may mediate the effects of PAMH. Tyrosine kinase inhibitors also suppressed PAMH actions. Several tyrosine kinases, including src, JAK kinase, and Erb-B, have been implicated in 5-HT receptor signaling (24). The tyrosine kinases and their potential targets in the PAMH signaling pathway remain to be determined.
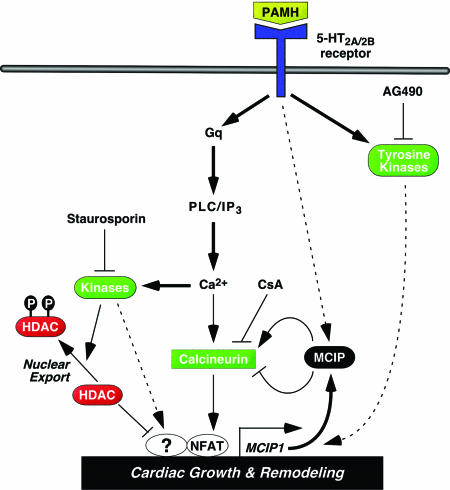
Fig. 6.
Possible signaling pathways controlled by PAMH. The activity of PAMH is blocked by staurosporin and the tyrosine kinase inhibitor AG490, implicating serine, threonine kinases, and tyrosine kinases, respectively, in the signaling pathway controlled by PAMH. PAMH drives nuclear export of HDAC5 and nuclear import of NFATc and stimulates the expression of MCIP1 and other genes involved in cardiac growth and remodeling. Signaling events that appear to play dominant roles in PAMH signaling are shown in bold. Those that are documented, but for which the relative importance remains to be established, are shown by thin lines, and those that remain hypothetical are shown by dashed lines. Prohypertrophic effectors are in green, and antihypertrophic effectors are in red.
In contrast to the complete inhibition of PAMH activity by staurosporin and tyrosine kinase inhibitors, CsA inhibited PAMH activity only partially. Thus, we speculate that PAMH acts primarily through kinase-dependent pathways augmented by calcineurin (Fig. 6). Alternatively, the incomplete inhibition of PAMH activity by CsA could reflect the potency in which PAMH activates the calcineurin pathway. Consistent with the notion that PAMH activates calcineurin signaling, it stimulates nuclear import of NFAT and activation of the NFAT-responsive MCIP1 promoter. However, we also note that MCIP1 protein is up-regulated more dramatically than MCIP1 mRNA by PAMH. Thus, although PAMH was identified on the basis of its ability to stimulate the calcineurin-responsive exon 4 promoter of MCIP1, posttranslational mechanisms may also contribute to the dramatic increase in MCIP1 protein in the presence of PAMH.
5-HT2 receptors couple to calcineurin activation in neurons (29). Similarly, serotonin signaling controls egg laying in Caenorhabditis elegans by activating the calcineurin pathway (30), and transgenic overexpression of MCIP1 mimics the calcineurin loss-of-function phenotype (31), consistent with MCIP functioning as a repressor of calcineurin signaling. Although MCIP1 and CsA can suppress hypertrophy due to calcineurin signaling (18), they may fail to do so in the presence of PAMH, because parallel pathways controlled by PAMH-dependent kinases bypass the requirement of calcineurin.
In addition to its effect on NFAT nuclear import, the ability of PAMH to promote nuclear export of HDAC5 and of a signal-resistant HDAC5 mutant to block hypertrophy in response to PAMH suggests that PAMH stimulates the activity of an HDAC kinase that inactivates HDAC5, a conclusion supported by the complete blockade to PAMH activity by staurosporin. How mutant HDACs suppress the activity of PAMH remains to be determined, but we favor the possibility that they interact with myocyte enhancer factor-2 or other prohypertrophic transcription factors such as NFAT or GATA4 (32).
Implications. The biological activity of PAMH suggests interesting possibilities for pharmacological modulation of muscle growth and function. In addition to influencing cardiac growth and development, 5-HT2B receptor signaling promotes myocyte survival and mitochondrial biogenesis (33). Recent studies have also revealed a profound stimulatory effect of PAMH on skeletal muscle development (unpublished results). Thus, the PAMH signaling pathway provides therapeutic possibilities for manipulating striated muscle function in the settings of pathological hypertrophy, heart failure, or skeletal muscle wasting disorders.
Supplementary Material
Acknowledgments
We are grateful to R. Gorcyznski, B. Rothermel, R. Vega, L. Avery, and R. Bassel-Duby for advice; A. Tizenor for assistance with graphics; and J. Page for editorial assistance. J.F. is a postdoctoral fellow of the Charite, Campus Virchow Klinikum. E.N.O. is supported by grants from the National Institutes of Health, the D. W. Reynolds Center for Clinical Cardiovascular Research, the Robert A. Welch Foundation, and the Texas Advanced Technology Program.
Abbreviations: ANF, atrial natriuretic factor; PAMH, pyridine activator of myocyte hypertrophy; A-PAMH, antagonist of PAMH; CsA, cyclosporine A; HDAC, histone deacetylase; 5-HT, 5-hydroxytryptamine; HTS, high-throughput screen; NFAT, nuclear factor of activated T cells; NRVM, neonatal rat ventricular myocyte; PE, phenylephrine.
References
- 1.Frey, N. & Olson, E. N. (2003) Annu. Rev. Physiol. 65, 45-79. [DOI] [PubMed] [Google Scholar]
- 2.MacLellan, W. R. & Schneider, M. D. (2000) Annu. Rev. Physiol. 62, 289-319. [DOI] [PubMed] [Google Scholar]
- 3.Olson, E. N. & Williams, R. S. (2000) Cell 101, 689-692. [DOI] [PubMed] [Google Scholar]
- 4.Kannel, W. B. & Cobb, J. (1992) Cardiology 81, 291-298. [DOI] [PubMed] [Google Scholar]
- 5.Hogan, P. G., Chen, L., Nardone, J. & Rao, A. (2003) Genes Dev. 17, 2205-2232. [DOI] [PubMed] [Google Scholar]
- 6.McKinsey, T. A., Zhang, C. L. & Olson, E. N. (2002) Trends Biochem. Sci. 27, 40-47. [DOI] [PubMed] [Google Scholar]
- 7.Zhang, C. L., McKinsey, T. A., Chang, S., Antos, C. L., Hill, J. A. & Olson, E. N. (2002) Cell 110, 479-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKinsey, T. A., Zhang, C. L., Lu, J. & Olson, E. N. (2000) Nature 408, 106-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grozinger, C. M. & Schreiber, S. L. (2000) Proc. Natl. Acad. Sci. USA 97, 7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rothermel, B. A., Vega, R. B. & Williams, R. S. (2003) Trends Cardiovasc. Med. 13, 15-21. [DOI] [PubMed] [Google Scholar]
- 11.Kingsbury, T. J. & Cunningham, K. W. (2000) Genes Dev. 14, 1595-1604. [PMC free article] [PubMed] [Google Scholar]
- 12.Rothermel, B., Vega, R. B., Yang, J., Wu, H., Bassel-Duby, R. & Williams, R. S. (2000) J. Biol. Chem. 275, 8719-8725. [DOI] [PubMed] [Google Scholar]
- 13.Vega, R. B., Rothermel, B. A., Weinheimer, C. J., Kovacs, A., Naseem, R. H., Bassel-Duby, R., Williams, R. S. & Olson, E. N. (2003) Proc. Natl. Acad. Sci. USA 100, 669-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryeom, S., Greenwald, R. J., Sharpe, A. H. & McKeon, F. (2003) Nat. Immunol. 4, 874-881. [DOI] [PubMed] [Google Scholar]
- 15.Yang, J., Rothermel, B., Vega, R. B., Frey, N., McKinsey, T. A., Olson, E. N., Bassel-Duby, R. & Williams, R. S. (2000) Circ. Res. 87, E61-E68. [DOI] [PubMed] [Google Scholar]
- 16.Molkentin, J. D., Lu, J. R., Antos, C. L., Markham, B., Richardson, J., Robbins, J., Grant, S. R. & Olson, E. N. (1998) Cell 93, 215-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimes, B. W. & Brandt, B. L. (1976) Exp. Cell Res. 98, 367-381. [DOI] [PubMed] [Google Scholar]
- 18.Rothermel, B. A., McKinsey, T. A., Vega, R. B., Nicol, R. L., Mammen, P., Yang, J., Antos, C. L., Shelton, J. M., Bassel-Duby, R., Olson, E. N., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 3328-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill, J. A., Rothermel, B., Yoo, K. D., Cabuay, B., Demetroulis, E., Weiss, R. M., Kutschke, W., Bassel-Duby, R. & Williams, R. S. (2002) J. Biol. Chem. 277, 10251-10255. [DOI] [PubMed] [Google Scholar]
- 20.Genesca, L., Aubareda, A., Fuentes, J. J., Estivill, X., De La Luna, S. & Perez-Riba, M. (2003) Biochem. J. 374, 567-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang, Y., De Keulenaer, G. W., Weinberg, E. O., Muangman, S., Gualberto, A., Landschulz, K. T., Turi, T. G., Thompson, J. F. & Lee, R. T. (2002) Am. J. Physiol. 283, H533-H539. [DOI] [PubMed] [Google Scholar]
- 22.Nebigil, C. G., Choi, D. S., Dierich, A., Hickel, P., Le Meur, M., Messaddeq, N., Launay, J. M. & Maroteaux, L. (2000) Proc. Natl. Acad. Sci. USA 97, 9508-9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nebigil, C. G., Jaffre, F., Messaddeq, N., Hickel, P., Monassier, L., Launay, J. M. & Maroteaux, L. (2003) Circulation 107, 3223-3229. [DOI] [PubMed] [Google Scholar]
- 24.Nebigil, C. G. & Maroteaux, L. (2003) Circulation 108, 902-908. [DOI] [PubMed] [Google Scholar]
- 25.Thompson, M., Forbes, I. T. & Johnson, C. N. (1992) U. S. Patent 5,093,493.
- 26.Cobo, C., Alcocer, L. & Chavez, A. (1990) Cardiovasc. Drugs Ther. 4, 73-36. [DOI] [PubMed] [Google Scholar]
- 27.Ikeda, K., Tojo, K., Tokudome, G., Hosoya, T., Harada, M. & Nakao, K. (2000) Life Sci. 67, 2991-2996. [DOI] [PubMed] [Google Scholar]
- 28.Strange, P. G. (1999) Biochem. Pharmacol. 58, 1081-1088. [DOI] [PubMed] [Google Scholar]
- 29.Day, M., Olson, P. A., Platzer, J., Striessnig, J. & Surmeier, D. J. (2002) J. Neurophysiol. 87, 2490-2504. [DOI] [PubMed] [Google Scholar]
- 30.Lee, M. H., Han, S. M., Han, J. W., Kim, Y. M., Ahnn, J. & Koo, H. S. (2003) FEBS Lett. 555, 250-256. [DOI] [PubMed] [Google Scholar]
- 31.Bandyopadhyay, J., Lee, J., Lee, J. I., Yu, J. R., Jee, C., Cho, J. H., Jung, S., Lee, M. H., Zannoni, S., Singson, A., et al. (2002) Mol. Biol. Cell 13, 3281-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki, Y. J., Day, R. M., Tan, C. C., Sandven, T. H., Liang, Q., Molkentin, J. D. & Fanburg, B. L. (2003) J. Biol. Chem. 278, 17525-17531. [DOI] [PubMed] [Google Scholar]
- 33.Nebigil, C. G., Etienne, N., Messaddeq, N. & Maroteaux, L. (2003) FASEB J. 17, 1373-1375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.