Abstract
A large number of tiny noncoding RNAs have been cloned and named microRNAs (miRs). Recently, we have reported that miR-15a and miR-16a, located at 13q14, are frequently deleted and/or down-regulated in patients with B cell chronic lymphocytic leukemia, a disorder characterized by increased survival. To further investigate the possible involvement of miRs in human cancers on a genome-wide basis, we have mapped 186 miRs and compared their location to the location of previous reported nonrandom genetic alterations. Here, we show that miR genes are frequently located at fragile sites, as well as in minimal regions of loss of heterozygosity, minimal regions of amplification (minimal amplicons), or common breakpoint regions. Overall, 98 of 186 (52.5%) of miR genes are in cancer-associated genomic regions or in fragile sites. Moreover, by Northern blotting, we have shown that several miRs located in deleted regions have low levels of expression in cancer samples. These data provide a catalog of miR genes that may have roles in cancer and argue that the full complement of miRs in a genome may be extensively involved in cancers.
Naturally occurring microRNAs (miRs) are 19- to 25-nt transcripts cleaved from 70- to 100-nt hairpin precursors, and are encoded in the genomes of invertebrates, vertebrates and plants (1, 2). Many miRs are conserved in sequence between distantly related organisms, suggesting that these molecules participate in essential processes (3). The biological functions of miRs are not yet fully understood. Several groups have uncovered roles for miRs in the coordination of cell proliferation and cell death during development, and in stress resistance and fat metabolism (4). For example, in Drosophila, miR-14 suppresses cell death and is required for normal fat metabolism (5), whereas bantam encodes a developmentally regulated miR that controls cell proliferation and regulates the proapoptotic gene hid (6). It is believed that miRs could direct positive or negative regulation at a variety of levels, depending on the specific miR and target base pair interaction and the cofactors that recognize them. lin-4 and let-7 are two members of the miR family known as small temporally regulated RNA, because regulate timing of gene expression during development of the nematode Caenorhabditis elegans. Both act as repressors of their respective target genes, lin-14, lin-28, and lin-41, containing 3′ UTRs with sites complementary to the ≈22-nt RNAs (7). Whereas perfect complementarity with targets was found in plants, in animals, the identification of putative targets is much more complicated, because bulges and loops are not only tolerated, but seem to be the rule. Despite the overall imperfect complementarity, a large subset of Drosophila miRs were shown to be precisely complementary to the K box, Brd box, and GY box motifs in the 3′ UTR, motifs found to significantly affect both transcript stability and translational efficiency (8).
At present time there are several indications that miRs might be a new class of genes involved in human tumorigenesis. We previously reported that miR-15a and miR-16a are located at chromosome 13q14, a region deleted in more than half of B cell chronic lymphocytic leukemias (B-CLLs). Detailed deletion and expression analysis shows that miR-15a and miR-16a are located within a 30-kb region of loss in CLL and that both genes are deleted or down-regulated in the majority (≈68%) of CLL cases (9). Hemizygous and/or homozygous loss at 13q14 occur in more than half of cases and constitute the most frequent chromosomal abnormality in CLL (10). Chromosome 13q14 deletions also occur in ≈ 50% of mantle cell lymphoma, in 16-40% of multiple myeloma, and in 60% of prostate cancers, suggesting that one or more tumor suppressor (TS) genes at 13q14 are involved in the pathogenesis of these human tumors (10). However, detailed genetic analysis, including extensive loss of heterozygosity (LOH), mutation, and expression studies have failed to demonstrate the consistent involvement of any of the genes located in the deleted region (9). Here, we present a systematic search for the identification of possible correlations between the genomic position of a large number of miRs and the location of cancer-associated genomic regions.
Methods
The miR Database. The set of 186 miR genes was comprised of 153 miRs identified in the miR registry at www.sanger.ac.uk/Software/Rfam, and 36 other miRs were manually curated from published papers (11-18), or were found in GenBank at www.ncbi.nlm.nih.gov. Nineteen human miRs (10%) were found by homology with cloned miRs from other species (mainly mouse). For all these miRs, we have found the sequence of the precursor by using the M Zucker RNA folding program at www.bioinfo.rpi.edu/applications/mfold/old/rna and selecting the precursor sequence that gave the best score for the hairpin structure.
Genome Analysis. We used the build 33 and build 34 version 1 of the Homo sapiens genome, which is available at www.ncbi.nlm.nih.gov/genome/guide/human. For each miR present in the database, we performed a blast search with the default parameters against the human genome to find the precise location, followed by mapping using the maps available at the Human Genome Resources at the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov). Also, as a confirmation of our data, we have found the human clone corresponding for each miR and mapped it to the human genome (see Table 3, which is published as supporting information on the PNAS web site). perl scripts for the automatic submission of BLAST jobs and for the retrieval of the search results were based on the LPW, HTML, and HTPP perl modules and bioperl modules.
Fragile Site (FRA) Database. We constructed this database by using Virtual Gene Nomenclature Workshop at www.gene.ucl.ac.uk/nomenclature/workshop. For each FRA locus, the literature was screened for papers reporting the cloning. In 10 cases, we found genomic positions for both centromeric and telomeric ends. The total genomic length of these FRA loci was 26.9 Mb. In 29 cases, we were able to identify only one anchoring marker, and we considered, based on the published data, that 3 Mb can be used as the median length for each FRA locus and therefore, we considered for these sites as very close miR located inside this “window” length. The human clones for 17 HPV16 integration sites were precisely mapped on the human genome. We considered by analogy with the length of a FRA that a distance of <2 Mb could define “close” vicinity.
PubMed Database. First, PubMed was screened for papers describing cancer-related abnormalities such as minimal regions of LOH and minimal regions of amplification (minimal amplicons) by using the words “LOH and genome-wide,” “amplification and genome-wide,” and “amplicon and cancer.” The data obtained from 32 papers were used as a screening for the identification of putative cancer-associated genomic regions (CAGRs), based on markers with high frequency of LOH/amplification. As a second step, a literature search was performed to determine the presence or absence of the above three types of alterations and to determine the precise location of miRs in respect with the CAGR (for a detailed list of reference papers see Supporting References, which is published as supporting information on the PNAS web site). We used as searching words, the combinations “minimal regions of LOH and cancer,” and “minimal region amplification and cancer.” We have found a total of 296 papers that were manually curated to find regions defined by both telomeric and centromeric markers. We were able to identify 154 minimally deleted regions (median length 4.14 Mb) and 37 minimally amplified regions (median length 2.45 Mb) with precise genomic mapping for both telomeric and centromeric ends involving all human chromosomes except Y. To identify common breakpoints regions we searched PubMed with the combination “translocation and cloning and breakpoint and cancer,” and we found 308 papers that were manually curated to find a set of 45 translocations with at least one breakpoint precisely mapped.
Statistical Analysis. To test hypotheses related to the incidence of miRs and association with chromosome, FRAs, amplified regions in cancer, deletion regions in cancer, we used random effect Poisson regression models. Under these models, “events” were defined as the number of miRs, and nonoverlapping lengths of the region of interest defined exposure “time” (i.e., FRA versus non-FRA, etc.). The “length” of a region was exactly ± 1 Mb, if known, or estimated as ± 1 Mb if unknown. The random effect used was chromosome, in that data within a chromosome were assumed correlated. The fixed effect in each model consisted of an indicator variable(s) for the type of region. We report the incidence rate ratio (IRR), two-sided 95% confidence interval of the IRR, and two-sided P values for testing the hypothesis that the IRR is 1.0. An IRR significantly >1 indicates an increase in the number of miRs within a region. Each model was repeated considering the distribution of miRs only in the transcriptional active portion of the genome (≈43%, using the published data), rather than the entire chromosome length, with similar results obtained (data not shown). This second model is more conservative and takes into account the phenomenon of clustering that was observed for the miRs genomic location. All computations were completed by using stata v7.0.
Patient Samples and Cell Lines. Patient samples were obtained after informed consent from 12 patients diagnosed with CLL and processed as described (9). We used also seven human lung cancer cell lines: Calu-3, H1299, H522, H460, H23, H1650, and H1573. The cell lines were obtained from the American Type Culture Collection and maintained according to American Type Culture Collection instructions. As normal control, we used normal lung total RNA purchased from Clontech.
Northern Blotting. Total RNA isolation and the blotting were performed as described (9). Blots were stripped by boiling in 0.1% aqueous SDS/0.1 × SSC for 10 min, and were reprobed. As loading control we used 5S rRNA stained with ethidium bromide.
Results
The miRs Are Nonrandomly Distributed in the Human Genome. The present lack of knowledge about the function of miRs highlights the importance of knowing the genomic distribution of miR genes. Frequently, the chromosomal positions of genes have led to important insights into the roles of genes in specific diseases. Therefore, we mapped 186 genes representing known or predicted miRs based on mouse homology or computational methods (refs. 11-18; for more details, see Methods and Table 3). This set of 186 genes is representative of the full human complement of miRs in a genome: experimental and bioinformatics approaches established that the number of miRs is between 188 and 255 (11). The distribution of these genes is nonrandom. Ninety miR genes are located in 36 clusters, usually with two or three genes per cluster (median = 2.5). The largest cluster is composed of six genes at 13q31 (miR-17/miR-18/miR-19a/miR-20/miR-19b1/miR-92-1; see Table 3). We have found a significant association of the incidence of miRs and particular chromosomes: chromosome 4 has a less than expected rate of miRs (IRR = 0.27, P = 0.035), and chromosomes 17 and 19 contain significantly more miR genes than expected, based on chromosome size (IRR = 2.97, P = 0.002 and IRR = 3.39, P = 0.001, respectively). The same results were obtained by using two different models, one considering the random distribution of miRs over the entire chromosome, and the other considering the distribution of miRs only in the transcriptional active portion of the genome (≈43%, using the published, for more details see Methods, and Table 4, which is published as supporting information on the PNAS web site). Six of the 36 clusters (17%) containing 16 of 90 clustered genes (18%) are located on these two small chromosomes, which account for only 5% of the entire genome.
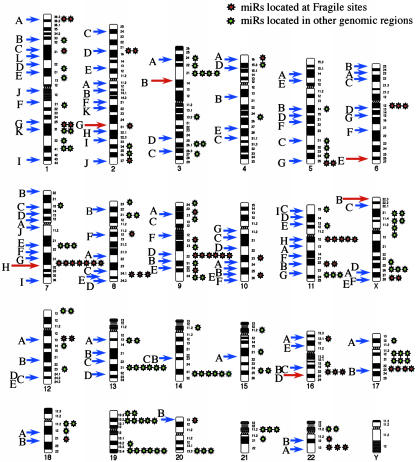
A Significant Number of miRs Are Located in FRAs or Are Close to Human Papilloma Virus (HPV) Integration Sites. Furthermore, 35 of 186 miRs (19%) are located in (13 miRs) or very close (<3Mb; 22 miRs) to cloned FRAs. We used a set of 39 FRAs with available cloning information, for 10 of which we were able to find data about the exact dimension (mean 2.69 Mb) and position (for detailed information, see Methods). The relative incidence of miRs inside FRAs occurred at a rate 9.12 times higher than in non-FRAs (P < 0.001, using random effect Poisson regression models, for detailed information see Methods, Table 1, and Table 4). The same very high statistical significance was also found if we considered only the 13 miRs located exactly inside FRAs or exactly in the vicinity of the “anchoring” marker mapped for an FRA (IRR = 3.22, P < 0.001). Interestingly, among the four most active common FRAs, FRA3B, FRA16D, FRA6E, and FRA7H, we have found seven miRs in (miR-29a and miR-29b) or close (miR-96, miR-182s, miR-182as, miR-183, and miR-129-1) to FRA7H, the only FRA where no candidate TS gene has been found. The other three sites contain known or candidate TS genes: FHIT, WWOX, and PARK2, respectively (19-21). Much more, looking at 113 FRAs scattered in human karyotype we found that 61 miRs are located in the same cytogenetic positions with FRAs (Fig. 1). This result allows us to speculate that more miRs are located in/near FRA and our results represent an underestimation only because the mapping of these unstable regions is not complete. A substantial body of evidence supports the proposal that at least some common chromosomal FRAs predispose to DNA instability in cancer cells (22, 23). Indeed, FRAs are preferential sites of sister chromatid exchange, translocation, deletion, amplification, or integration of plasmid DNA and tumor-associated viruses such as HPV.
Table 1. Mixed effect Poisson regression results for the association between miRs and several types of regions of interest.
| Region of interest | IRR | 95% confidence interval IRR | P |
|---|---|---|---|
| Cloned FRAs vs. non-FRAs | 9.12 | 6.22, 13.38 | <0.001 |
| HPV16 insertion vs. all other | 3.22 | 1.55, 6.68 | 0.002 |
| Deleted region vs. all other | 4.08 | 2.99, 5.56 | <0.001 |
| Amplified region vs. all other | 3.97 | 2.31, 6.83 | <0.001 |
| HOX clusters vs. all other | 15.77 | 7.39, 33.62 | <0.001 |
| HOX genes vs. all other | 2.95 | 1.63, 5.34 | <0.001 |
All other, all the genome except the regions of interest.
Fig. 1.
Correlation between FRAs and miRs. A karyotype showing the position of 113 FRAs and 186 miRs is presented. The 61 miRs located in the same chromosomal band as the FRA are red. We were able to precisely locate 35 miRs inside 12 cloned FRAs. The red arrow shows frequently observed FRAs.
Infection with HPV16 or 18 is the major risk factor for developing cervical cancer, and common FRAs are preferential targets for HPV16 integrations in cervical tumors (24, 25). To understand the significance of miR-FRA correlation, we looked for association between miR gene locations and HPV16 integration site in cervical tumors. Thirteen miR genes (7%) are located near (<2.5 Mb) 7 of 17 (45%) cloned integration sites. The relative incidence of miRs at HPV16 integration sites occurred at a rate 3.22 times higher than in the rest of the genome (P < 0.002; Tables 1 and 3, and Table 5, which is published as supporting information on the PNAS web site). In one cluster of integration sites at chromosome 17q23, with three HPV16 integration events spread over ≈4 Mb of genomic sequence, we found four miR genes (miR-21, miR-301, miR-142s, and miR-142as). Because HPV integration into the host cell genome can cause large deletions, amplification, or complex rearrangements, the expression of cellular genes at or near integration sites may be affected. MiR genes located near the integration sites are possible targets of such genome alterations.
Numerous CAGRs Contain miRs. If the miR-FRA-HPV16 association has significance for cancer pathogenesis, miR genes might be involved in malignancies through other mechanisms, such as deletion, amplification, or epigenetic modifications. Thus, we next searched extensively for reported genomic alterations in human cancers, located in regions containing miRs. PubMed was searched for reports of CAGRs: (i) minimal regions of LOH, suggestive of the presence of TS genes; (ii) minimal regions of amplification, suggestive of the presence of oncogenes (OGs); and (iii) common breakpoint regions in or near possible OGs or TS genes (for detailed information see Methods). Overall, 98 of 186 (52.5%) miR genes are in CAGRs (Table 2, and Table 6, which is published as supporting information on the PNAS web site), including 80 miRs (43%), that are located exactly in minimal regions of LOH or minimal regions of amplification described in a variety of tumors as lung, breast, ovarian, colon, gastric, and hepatocellular carcinoma, as well as leukemias and lymphomas (Tables 2 and 6).
Table 2. Examples of miRs located in minimal deleted regions, minimal amplified regions, and breakpoint regions involved in human cancers.
| Chromosome | Location (defining markers) | Size, Mb | miR | Hystotype | Known OG/TS |
|---|---|---|---|---|---|
| 3p21.1-21.2-D | ARP-DRR1 | 7 | let-7g/miR-135−1 | Lung, breast cancer | — |
| 3p21.3(AP20)-D | GOLGA4-VILL | 0.75 | miR-26a | Epithelial cancer | — |
| 3p23-21.31(MDR2)-D | D3S1768-D3S1767 | 12.32 | miR-26a; miR-138−1 | Nasopharyngeal cancer | — |
| 5q32-D | ADRB2-ATX1 | 2.92 | miR-145/miR-143 | Myelodysplastic syndrome | — |
| 9q22.3-D | D9S280-D9S1809 | 1.46 | miR-24−1/mir-27b/miR-23b; let-7a-1/let-7f-1/let-7d | Urothelial cancer | PTC, FANCC |
| 9q33-D | D9S1826-D9S158 | 0.4 | miR-123 | NSCLC | — |
| 11q23-q24-D | D11S927-D11S1347 | 1.994 | miR-34a-1/miR-34a-2 | Breast, lung cancer | PPP2R1B |
| 11q23-q24-D | D11S1345-D11S1328 | 1.725 | miR-125b-1/let-7a-2/miR-100 | Breast, lung, ovary, cervix cancer | — |
| 13q14.3-D | D13S272-D13S25 | 0.54 | miR-15a/miR-16a | B-CLL | — |
| 13q32-33-A | stSG15303-stSG31624 | 7.15 | miR-17/miR-18/miR-19a/miR-20/miR-19b-1/miR-92−1 | Follicular lymphoma | — |
| 17p13.3-D | D17S1866-D17S1574 | 1.899 | miR-22; miR-132; miR-212 | HCC | — |
| 17p13.3-D | ENO3-TP53 | 2.275 | miR-195 | Lung cancer | TP53 |
| 17q22-t(8; 17) | miR-142s/c-MYC | miR-142s; miR-142as | Prolymphocytic leukemia | c-MYC | |
| 17q23-A | CLTC-PPM1D | 0.97 | miR-21 | Neuroblastoma | — |
| 20q13-A | FLJ33887-ZNF217 | 0.55 | miR-297−3 | Colon cancer | — |
| 21q11.1-D | D21S1911-ANA | 2.84 | miR-99a/let-7c/miR-125b | Lung cancer | — |
D, deleted region; A, amplified region; NSCLC, non-small-cell lung cancer; HCC, hepatocellular carcinoma; PTC, patched homolog (Drosophila); FANCC, Fanconi anemia, complementation group C; PPP2R1B, protein phosphatase 2, regulatory subunit A (PR 65), β isoform, miRs in a cluster are separated by a slash. For references, see Table 6.
Several interesting results came out from this analysis (Table 2). First, on chromosome 9, eight of 15 mapped miRs (including six located in clusters), are inside two regions of deletion on 9q (26): the clusters let-7a-1/let-7f-1/let-7d and miR-23b/miR-27b/miR-24-1 inside the region B at 9q22.3 and miR-181a and miR-199b inside region D at 9q33-34.1. Furthermore, five other miRs are located near (<2 Mb) to the markers with the highest rate of LOH: miR-31 near IFNA, miR-204 near D9S15, miR-181 and miR-147 near GSN, and miR-123 near D9S67. Second, in breast carcinomas, two different regions of loss at 11q23, independent from ATM locus have been studied extensively: the first spans ≈2 Mb between loci D11S1347-D11S927, the second is located between loci D11S1345-D11S1316, and is estimated at ≈1 Mb (27). Candidate TS genes were not found is spite of extensive effort, except the PPP2R1B gene, involved in <10% of cases (9, 28). Both minimal LOH regions contain numerous miRs: the cluster miR-34-a1/miR-34-a2 in the first and the cluster miR-125b1/let-7a-2/miR-100 in the second. Third, high frequency of LOH at 17p13.3 and relatively low frequency of TP53 mutation in cases of hepatocellular carcinomas, lung cancers, and astrocytomas indicate the presence of other TS genes involved in the development of these tumors. We have found that one minimal LOH region described in hepatocellular carcinoma and located telomeric to TP53 between markers D17S1866 and D17S1574 harbor three miRs: miR-22, miR-132, and miR-212 (29), whereas between ENO3 and TP53, we found miR-195 (30). Fourth, homozygous deletions (HDs) in cancer suggest the presence of TS genes (31) and several miR genes are located in homozygously deleted regions without known TS genes. In addition to miR-15a and miR-16a located at 13q14 HD region in B-CLL, the cluster miR-99a/let-7c/miR-125b-2 mapped in a 21p11.1 region of HD in lung cancers and miR-32 at 9q31.2 in a region of HD in various types of cancer. Among the seven regions of LOH and HD on the short arm of chromosome 3, three of them harbor miRs: miR-26a in region AP20, miR-138-1 in region 5 at 3p21.3 and the cluster let-7g/miR-135-1 in region 3 at 3p21.1-p21.2 (32). These locations are very unlikely to be random: overall, we have found that the relative incidence of miRs in both deleted and amplified regions is highly significant (IRR = 4.08, P < 0.001 and IRR = 3.97, P < 0.001, respectively; Table 1). Thus, these miRs expand the spectrum of candidate TS genes.
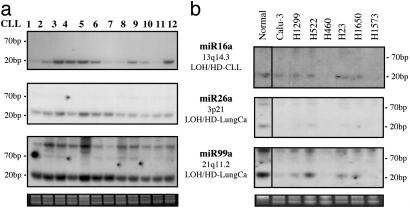
To test whether the genomic location in deleted regions influence the miR expression, we have analyzed a panel of B-CLL samples with known deletions at 13q14 and a set of lung cancer cell lines. Mir-16a expression (located at 13q14) was low or absent in the majority of B-CLL cases although the expression of miR-26a (at 3p21) and miR-99a (at 21q11.2), both regions not involved in B-CLL, was at relatively equal levels in all cases. By contrast, both miR-26a and miR-99a are not expressed or expressed at low levels in lung cancer cell lines, and this finding correlates with their location in regions of LOH/HD in lung tumors/cell lines. On the contrary, the expression of miR-16a was the same in the majority of cell lines as in normal lung (Fig. 2).
Fig. 2.
Expression of miR-16a, miR-26a, and miR-99a in human tumors and cell lines. (a) Northern blot with B-CLL samples. (b) Northern blot with lung cancer cell lines. The genomic location and the type of alteration are presented.
Several miR genes are located near breakpoint regions, including miR-142s at 50 nucleotides from the t(8, 17) break involving chromosome 17 and MYC, and miR-180 at 1 kb from the MN1 gene involved in the t(4, 22) in meningioma (Table 2). The t(8, 17) translocation brings the MYC gene near the miR gene promoter, with consequent MYC overexpression, whereas the t(4, 22) translocation inactivate MN1 gene and possibly the miR gene located in the same position. Other miR genes are located relatively close to breakpoints, for example, the cluster miR 34a-1/34a-2 and miR-153-2 (see Table 6). Another example is the woodchuck homolog of human miR-122a. blast data showed the homologue in sense orientation with the ORF of hcr gene from woodchuck, a noncoding gene involved in a rearrangement with c-myc in a woodchuck hepatocellular carcinoma (33). Further supporting the suggested role of miR-122a in cancer, we found that the human miR-122a is located in the minimal amplicon around MALT1 in aggressive marginal zone lymphoma and ≈160 kb from the breakpoint region of translocation t(11, 18) in mucosa-associated lymphoid tissue lymphoma (34). Apart from miR-122a, several other miR genes are located in regions particularly prone to cancer-specific abnormalities, such as miR-142s and miR-142as at 17q23 close to a t(8, 17) breakpoint in B cell acute leukemia, and also within the minimal amplicon in breast cancer and near the FRA17B site, a target for HPV16 integration in cervical tumors (see Table 6 and Fig. 3).
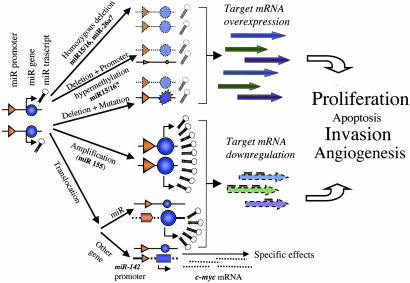
Fig. 3.
MiRs as cancer players. Some of these proposed mechanisms are experimentally proven, like the HD of miR-15a/miR-16a cluster in B-CLL (9), the c-myc overexpression by the reposition near a putative miR promoter, or miR143/miR-145 cluster down-regulation in colon cancers (39).
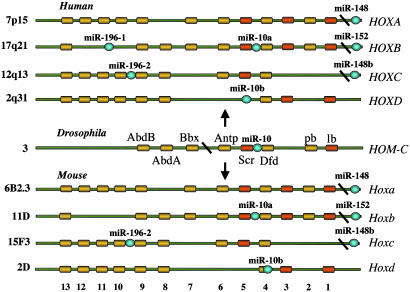
The miRs Are Located Inside or Near Homeobox (HOX) Clusters. Analysis of the genomic location of miR genes provides interesting clues for possible functions. We have found a strong correlation between the location of specific miRs and HOX genes. The miR-10a and miR-196-1 are located within the HOX B cluster on 17q21, whereas miR-196-2 is within the HOX C cluster at 12q13, and miR-10b maps to the HOX D cluster at 2q31. Moreover, three other miRs (miR-148, miR-152, and miR-148b) are close [<1 Mb; we select this distance because some form of long-range coordinated regulation of gene expression was shown to expand up to 1 Mb (35)] to HOX clusters (Fig. 4). Such proximity is unlike to have occurred by chance (IRR = 15.77; P < 0.001; Table 1). HOX-containing genes are a family of encoding transcription factor genes that play crucial roles during normal development and in oncogenesis. HOXB4, HOXB5, HOXC9, HOXC10, HOXD4, and HOXD8, all with miR neighbors, are deregulated in a variety of solid and hematopoietic cancers (36, 37). Because collinear expression of and cooperation between HOX genes is well demonstrated, these data suggest that miRs may be altered along with the HOX genes in human cancers. Class I HOX genes include the Hox genes, whereas class II include Hox-related genes with the conserved HOX domain but less-defined functions. We tested the hypotheses that miR genes would be found within class II Hox gene clusters as well. Thus, we analyzed 14 other human HOX gene clusters (38) and found that seven miRs (miR-129-1, miR-153-2, let-7a-1, let-7f-1, let-7d, miR-202, and miR-139) are located close (<0.5 Mb) to class II homeotic genes, a result which highly improbably occurs by chance (IRR = 2.95, P < 0.001; Table 1 and data not shown). These data strongly support a suggested role of miR genes in development and give hints for possible targets for miRs regulatory effects.
Fig. 4.
MiR location and the HOX gene clusters. Drosophila, mouse, and human clusters are presented (for details see text). Each cluster is ≈100 kb; the figure is not drawn to scale.
Discussion
The data of our systematic analysis argue that the full complement of miRs in a genome may be extensively involved in cancers. Recently, it was reported that miR-143 and miR-145 consistently display reduced steady-state levels of the mature miR at the adenomatous and cancer stages of colorectal neoplasia (39). Other examples of miR genes located in sites shown to be involved in human cancers are: miR-142s located at the breakpoint junction of a t(8, 17) translocation, which causes an aggressive B cell leukemia due to up-regulation of a translocated c-MYC gene (16, 40); miR-155 excised from a noncoding RNA, BIC, identified as a gene transcriptionally activated in B cell lymphomas induced by avian leukemia virus, as a consequence of promoter insertion at a common retroviral integration site (41); miR-7-1 in the last intron of the heterogenous nuclear ribonucleoprotein K gene, a gene overexpressed in cancer cells (16, 42, 43). The miR precursor processing reaction requires Dicer RNase III and Argonaute family members (44). Argonaute has been shown to interact both genetically and biochemically with miRs (15). Further stressing the importance of miRs in cancer, it was shown that mutations in genes required for miRs biosynthesis cause developmental defects and cancer (45, 46). A speculative model was drawn where miRs could be contributors for oncogenesis working as classical TS genes (as is the case of miR-15a/miR-16a) or as classical OGs (as is the case of miR-155), whereas some miRs could participate in “posttranscriptional collapse,” a scenario where misexpression of a miR causes a posttranscriptional misregulation of a TSG or OG (47).
Perhaps surprisingly, specific miR genes (such as miR-33b) are in regions involved in both deletions and amplification depending of cancer type. However, the same gene can behave as an OG or suppressor gene, depending on the type of alteration, cell type, or transcriptional/posttranscriptional events (48). For example, it was shown that within the superfamily of Ras OGs, Kras2 could inhibit lung carcinogenesis in mice, illustrating a TS role of this gene in lung tumorigenesis (49). In Fig. 3 we present a model of possible mechanisms explaining the miR involvement in cancers. We propose that miRs can act both as TS genes and OGs. MiRs are participating in normal cells homeostasy by interfering with mRNA translation and/or stability. Because of the small size, the accumulation of loss-of-function or gain-of-function point mutations may be rare events. HDs (as is the case for miR-15a/miR-16a cluster or probably miR-26a), the combination mutation plus promoter hypermethylation or gene amplification seems to be the main mechanisms of inactivation or activation, respectively. MiRs activity can be influenced either by the reposition of other genes close to miRs promoters/regulatory regions (as is the case of miR-142s - c-myc translocation) or by the relocalization of an miR near other regulatory elements. The overall effects in the case of miRs inactivation is the overexpression of yet unidentified target miRs, whereas the miRs activation will lead to down-regulation of target miRs, supposed to participate in apoptosis, cell cycle, invasivity, or angiogenesis.
In conclusion, our data represent a genome-wide systematic search for correlations between the genomic positions of miR genes and specific cancer-associated abnormalities, and provide a catalog of such data for each miR gene. In the absence of powerful bioinformatic tools for finding miR targets, genomic proximity may be very useful to screen for such interactions.
Note Added in Proof. Recently, it was reported that the precursor miR155/Bic is up-regulated in children with Burkitt's lymphoma (50).
Supplementary Material
Acknowledgments
We thank Dr. Kay Huebner for critical reading of the manuscript, and Ayumi Matsuyama for the assistance with Fig. 1. G.A.C. thanks Prof. Riccardo Fodde (Erasmus University, Rotterdam, The Netherlands) for his continuing support and for his critical review of this work. This work was supported by National Cancer Institute Grants P01CA76259, P01CA56036, and P01CA81534; and Ministero dell'Istruzione, dell'Università e della Ricerca cofin2003 and Ministero della Salute Ricerca Finalizzata 2003.
Abbreviations: miR, microRNA; B-CLL, B cell chronic lymphocytic leukemias; LOH, loss of heterozygosity; CAGR, cancer-associated genomic region; IRR, incidence rate ratio; TS, tumor suppressor; FRA, fragile site; HPV, human papilloma virus; OG, oncogene; HD, homozygous deletion; HOX, homeobox.
References
- 1.Ke, X.-S., Liu, C.-M., Liu, D.-P. & Liang, C.-C. (2003) Curr. Opin. Chem. Biol. 7, 516-523. [DOI] [PubMed] [Google Scholar]
- 2.Moss, E. G. (2003) MicroRNAs in Noncoding RNAs: Molecular Biology and Molecular Medicine (Eurekah.com), pp. 98-114.
- 3.Pasquinelli, A. E., Reinhart, B. J., Slack F., Martindale, M. Q., Kuroda, M. I., Maller, B., Hayward, D. C., Ball, E. E., Degnan, B., Muller, P., et al. (2000) Nature 408, 86-89. [DOI] [PubMed] [Google Scholar]
- 4.Ambros, V. (2003) Cell 113, 673-676. [DOI] [PubMed] [Google Scholar]
- 5.Xu, P., Vernooy, S. Y., Guo, M. & Hay, B. A. (2003) Curr. Biol. 13, 790-795. [DOI] [PubMed] [Google Scholar]
- 6.Brennecke, J., Hipfner, D. R., Stark, A., Russell, R. B. & Cohen, S. M. (2003) Cell 113, 25-36. [DOI] [PubMed] [Google Scholar]
- 7.Pasquinelli, A. E. (2002) Trends Genet. 18, 171-173. [DOI] [PubMed] [Google Scholar]
- 8.Lai, E. C. (2002) Nat. Genet. 30, 363-364. [DOI] [PubMed] [Google Scholar]
- 9.Calin, G. A., Dumitru, C. D., Shimizu, M., Bichi, R., Zupo, S., Noch, E., Aldler, H., Rattan, S., Keating, M., Rai, K., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 15524-15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bullrich, F. & Croce, C. M. (2001) Chronic Lymphoid Leukemia (Dekker, New York).
- 11.Lim, L. P., Glasner, M. E., Yekta, S., Burge, C. B. & Bartel, D. P. (2003) Science 299, 1540. [DOI] [PubMed] [Google Scholar]
- 12.Lagos-Quintana, M., Rauhut, R., Lendeckel, W. & Tuschl, T. (2001) Science 294, 853-858. [DOI] [PubMed] [Google Scholar]
- 13.Lau, N. C., Lim, L. P., Weinstein, E. G. & Bartel, D. P. (2001) Science 294, 858-862. [DOI] [PubMed] [Google Scholar]
- 14.Lee, R. C. & Ambros, V. (2001) Science 294, 862-864. [DOI] [PubMed] [Google Scholar]
- 15.Mourelatos, Z., Dostie, J., Paushkin, S., Sharma, A., Charroux, B., Abel, L., Rappsilber, J., Mann, M. & Dreyfuss, G. (2002) Genes Dev. 16, 720-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W. & Tuschl, T. (2002) Curr. Biol. 12, 735-739. [DOI] [PubMed] [Google Scholar]
- 17.Dostie, J., Mourelatos, Z., Yang, M., Sharma, A. & Dreyfuss, G. (2003) RNA 9, 180-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Houbaviy, H. B., Murray, M. F. & Sharp, P. A. (2003) Dev. Cell 5, 351-358. [DOI] [PubMed] [Google Scholar]
- 19.Ohta, M., Inoue, H., Cotticelli, M., Kastury, K., Baffa, R., Palazzo, J., Siprashvili, Z., Mori, M., McCue, P. & Druck, T. (1996) Cell 84, 587-597. [DOI] [PubMed] [Google Scholar]
- 20.Paige, A. J., Taylor, K. J., Taylor, C., Hillier, S. G., Farrington, S., Scott, D., Porteous, D. J., Smyth, J. F., Gabra, H. & Watson, J. E. (2001) Proc. Natl. Acad. Sci. USA 98, 11417-11422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cesari, R., Martin, E. S., Calin, G. A., Pentimalli, F., Bichi, R., McAdams, H., Trapasso, F., Drusco, A., Shimizu, M., Masciullo, V., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 5956-5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arlt, M. F., Casper, A. M. & Glover, T. W. (2003) Cytogenet. Genome Res. 100, 92-100. [DOI] [PubMed] [Google Scholar]
- 23.Richards, R. I. (2001) Trends Genet. 17, 339-345. [DOI] [PubMed] [Google Scholar]
- 24.Thorland, E. C., Myers, S. L., Persing, D. H., Sarkar, G., McGovern, R. M., Gostout, B. S. & Smith, D. I. (2000) Cancer Res. 60, 5916-5921. [PubMed] [Google Scholar]
- 25.Thorland, E. C., Myers, S. L., Gostout, B. S. & Smith, D. I. (2003) Oncogene 22, 1225-1237. [DOI] [PubMed] [Google Scholar]
- 26.Simoneau, M., Aboulkassim, T. O., LaRue, H., Rousseau, F. & Fradet, Y. (1999) Oncogene 7, 157-163. [DOI] [PubMed] [Google Scholar]
- 27.di Iasio, M. G., Calin, G. A., Tibiletti, M. G., Vorechovsky, I., Benediktsson, K. P., Taramelli, R., Barbanti-Brodano, G. & Negrini, M. (1999) Oncogene 25, 1635-1638. [DOI] [PubMed] [Google Scholar]
- 28.Wang, S. S., Esplin, E. D., Li, J. L., Huang, L., Gazdar, A., Minna, J. & Evans, G. A. (1998) Science 282, 284-287. [DOI] [PubMed] [Google Scholar]
- 29.Zhao, X., He, M., Wan, D., Ye, Y., He, Y., Han, L., Guo, M., Huang, Y., Qin, W., Wang, M. W., et al. (2003) Cancer Lett. 190, 221-232. [DOI] [PubMed] [Google Scholar]
- 30.Tsuchiya, E., Tanigami, A., Ishikawa, Y., Nishida, K., Hayashi, M., Tokuchi, Y., Hashimoto, T., Okumura, S., Tsuchiya, S. & Nakagawa, K. (2000) Jpn. J. Cancer Res. 91, 589-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huebner, K., Garrison, P. N., Barnes, L. D. & Croce, C. M. (1998) Annu. Rev. Genet. 32, 7-31. [DOI] [PubMed] [Google Scholar]
- 32.Zabarovsky, E. R., Lerman, M. I. & Minna, J. D. (2002) Oncogene 21, 6915-6935. [DOI] [PubMed] [Google Scholar]
- 33.Etiemble, J., Moroy, T., Jacquemin, E., Tiollais, P. & Buendia, M. A. (1989) Oncogene 4, 51-57. [PubMed] [Google Scholar]
- 34.Sanchez-Izquierdo, D., Buchonnet, G., Siebert, R., Gascoyne, R. D., Climent, J., Karran, L., Marin, M., Blesa, D., Horsman, D., Rosenwald, A., et al. (2003) Blood 101, 4539-4546. [DOI] [PubMed] [Google Scholar]
- 35.Kamath, R. S., Fraser, A. G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M., et al. (2003) Nature 421, 231-237. [DOI] [PubMed] [Google Scholar]
- 36.Cillo, C., Faiella, A., Cantile, M. & Boncinelli, E. (1999) Exp. Cell Res. 248, 1-9. [DOI] [PubMed] [Google Scholar]
- 37.Owens, B. M. & Hawley, R. G. (2002) Stem Cells 20, 364-379. [DOI] [PubMed] [Google Scholar]
- 38.Pollard, S. L. & Holland, P. W. H. (2000) Curr. Biol. 10, 1059-1062. [DOI] [PubMed] [Google Scholar]
- 39.Michael, M. Z., O' Connor, S. M., van Holst Pellekaan, N. G., Young, G. P. & James, R. J. (2003) Mol. Cancer Res. 1, 882-891. [PubMed] [Google Scholar]
- 40.Gauwerky, C. E., Huebner, K., Isobe, M., Nowell, P. C. & Croce, C. M. (1989) Proc. Natl. Acad. Sci. USA 86, 8867-8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tam, W. (2001) Gene 274, 157-167. [DOI] [PubMed] [Google Scholar]
- 42.Bomsztyk, K., Van Seuningen, I., Suzuki, H., Denisenko, O. & Ostrowski, J. (1997) FEBS Lett. 400, 113-115. [DOI] [PubMed] [Google Scholar]
- 43.Dejgaard, K., Leffers, H., Rasmussen, H. H., Madsen, P., Kruse, T. A., Gesser, B., Nielsen, H. & Celis, J. E. (1994) J. Mol. Biol. 236, 33-48. [DOI] [PubMed] [Google Scholar]
- 44.Sasaki, T., Shiohama, A., Minoshima, S. & Shimizu, N. (2003) Genomics 82, 323-330. [DOI] [PubMed] [Google Scholar]
- 45.Hutvagner, G. & Zamore, P. D. (2002) Science 297, 2056-2060. [DOI] [PubMed] [Google Scholar]
- 46.Carmell, M. A., ZXuan, Z., Zhang, M. Q. & Hannon, G. J. (2002) Genes Dev. 16, 2733-2742. [DOI] [PubMed] [Google Scholar]
- 47.McManus, M. T. (2003) Semin. Cancer Biol. 13, 253-258. [DOI] [PubMed] [Google Scholar]
- 48.Calin, G. (1994) Oncol. Rep. 1, 987-991. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, Z., Wang, Y., Vikis, H. G., Johnson, L., Liu, G., Li, J., Anderson, M. W., Sills, R. C., Hong, H. L., Devereux, T. R., et al. (2001) Nat. Genet. 29, 25-33. [DOI] [PubMed] [Google Scholar]
- 50.Metzler, M., Wilda, M., Busch, K., Viehmann, S. & Borkhardt, A. (2004) Genes Chromosomes Cancer 39, 167-169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.