Recent advancements in technology have led to the development of new techniques that hold promise for improved diagnosis and management of infectious diseases. Here, we review new assays that help better identify pathogens and tailor antibiotic therapy to patients' needs.
Keywords: procalcitonin, gene expression profiling, mass spectrometry, MALDI-TOF MS, sepsis
Abstract
In the first decade of the 21st century, we have seen the completion of the human genome project and marked progress in the human microbiome project. The vast amount of data generated from these efforts combined with advances in molecular and biomedical technologies have led to the development of a multitude of assays and technologies that may be useful in the diagnosis and management of infectious diseases. Here, we identify several new assays and technologies that have recently come into clinical use or have potential for clinical use in the near future. The scope of this review is broad and includes topics such as the serum marker procalcitonin, gene expression profiling, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), and nucleic acid aptamers. Principles that underlie each assay or technology, their clinical applications, and potential strengths and limitations are addressed.
Recent advancements in the fields of immunology, molecular biology, bioinformatics, and biomedical engineering have led to the development of new assays that hold promise for the improved diagnosis and management of infectious diseases. For the clinician faced with a patient suffering from possible infection, a key challenge is knowing when to provide and when to withhold antimicrobial medications. Initiating effective antimicrobial therapy early has proven to decrease morbidity and mortality in severe infection [1, 2], while the overuse of antimicrobials leads to antimicrobial resistance. Antimicrobial resistance is a major public health threat that has substantial economic impact on our healthcare system [3, 4]. Judicious use of antibiotics and effective patient care rely on the ability to quickly and accurately (1) distinguish infectious from noninfectious etiologies of illness, (2) define either the class of pathogen or specific pathogen responsible for that illness, (3) assess disease severity, (4) assess response to therapy, and (5) define the length of a treatment course. Here, we review 4 new up-and-coming assays and technologies that offer improvement in each of these areas. The intent of this review is to provide an overview of the principles that underlie each assay or technique, the clinical or preclinical setting in which they have been used, and the potential limitations of each modality. The assays and technologies reviewed here have either recently come into clinical use or are expected to come into clinical use in the near future.
Procalcitonin
A novel approach to estimating the likelihood of a patient having a bacterial infection, and monitoring response to antimicrobial therapy, is to assess host response. Traditionally, this was done by monitoring clinical signs and symptoms, such as the systemic inflammatory response syndrome (SIRS). However, these criteria have low sensitivity and/or specificity for bacterial infections, and lack standardization in clinical practice. A more recent approach is to measure serum markers of infection. One such marker that has generated recent interest is procalcitonin (PCT).
PCT is released in multiple body tissues in response to bacterial infections via direct stimulation of bacterial cytokines, such as interleukin1β, tumor necrosis factor α, and interleukin 6 [5]. Interferon γ, a cytokine released in response to viral infections, blocks the upregulation of PCT, resulting in a higher specificity of PCT for distinguishing bacterial from viral infection compared to other inflammatory markers such as C-reactive protein. Quantitatively, PCT helps estimate the risk of severe bacterial infections or milder viral illnesses [6]. PCT promptly increases within 6 to 12 hours of bacterial infection. PCT decreases daily by around 50% if the patient responds to therapy and the infection is well controlled [7]. Unlike C-reactive protein and other markers, PCT appears not to be influenced by systemic corticosteroid treatments.
Recent research has focused on the impact of PCT testing on patient management and outcomes. Studies were done in different clinical settings ranging from low acuity in primary care, to intermediate acuity in emergency departments and hospital wards, to high acuity in intensive care units. PCT protocols have been adapted according to these settings with different PCT cutoff ranges (reviewed in [8]). In low-acuity settings, PCT was used to guide initial prescription of antimicrobials, while in higher-acuity settings PCT was used to guide duration of treatment: antibiotics were recommended to be stopped once patients showed clinical response and PCT dropped to normal values.
Fifteen randomized controlled trials including >4000 patients have evaluated the efficacy and safety of using PCT for antibiotic decision making [9]. PCT protocols proved to be effective in reducing antibiotic exposure compared to standard groups in all trials. PCT guidance lowered antibiotic prescription rates by 65% in primary care patients, 35% in the emergency department setting, and almost 30% in the critical care setting (see detailed results in Table 1). Such a strategy has also been found cost-effective in a North American healthcare system assuming PCT costs of around CA$40 [10]. No other biomarker has been evaluated within such rigorous trial designs. Still, concern about low adherence to PCT protocols remains, especially in critical care settings. Studies from “real life” have shown that adherence is a crucial factor when such protocols are implemented [11, 12]. Also, studies that have used PCT to escalate antibiotic therapy when values did not drop were disappointing [13].
Table 1.
Efficacy and Safety of Procalcitonin Protocols in Previous Randomized Controlled Trials
| Efficacy |
Safety |
||||||
|---|---|---|---|---|---|---|---|
| Initiation of Antibiotics, % | Duration of Antibiotics, Median (IQR) | Total Antibiotic Exposure, Median (IQR) | Mortality, Adjusted OR (95% CI) | P Value | Treatment Failurea, Adjusted OR(95% CI) | P Value | |
| Setting | |||||||
| Overall | 64% vs 84% | 7 (4–10) vs 10 (7–13) | 4 (0–8) vs 8 (5–12) | 0.94 (.71–1.23) | .754 | 0.82 (.71–.97) | .02 |
| Primary care | 23% vs 63% | 7 (5–8) vs 7 (6–8) | 0 (0–0) vs 6 (0–7) | … | … | 0.95 (.73–1.24) | .687 |
| Emergency department | 73% vs 88% | 7 (4–10) vs 10 (7–12) | 5 (0–8) vs 9 (5–12) | 1.03 (.7–1.5) | .895 | 0.76 (.61–.95) | .01 |
| ICU | 100% vs 100% | 8 (5–15) vs 12 (8–18) | 8 (5–15) vs 12 (8–18) | 0.84 (.54–1.31) | .443 | ||
| Diagnoses | |||||||
| Upper ARI | 15% vs 48% | 7 (5–8) vs 7 (6–7) | 0 (0–0) vs 0 (0–7) | … | … | 0.95 (.73–1.24) | .687 |
| Community- acquired pneumonia | 90% vs 99% | 7 (5–10) vs 10 (8–14) | 6 (4–10) vs 10 (8–14) | 0.89 (.64–1.23) | .471 | 0.77 (.62–.96) | .02 |
| Ventilator- associated pneumonia | 99% vs 100% | 11 (6–17) vs 14 (9–19.5) | 11 (6–17) vs 14 (9–19.5) | 0.69 (.25–1.94) | .486 | 0.69 (.25–1.94) | .486 |
| Acute bronchitis | 24% vs 66% | 7 (4–9) vs 7 (5–8) | 0 (0–0) vs 5 (0–7) | … | … | 1.09 (.70–1.70) | .71 |
| Exacerbation of COPD | 48% vs 73% | 6 (3–9) vs 8 (6–10) | 0 (0–6) vs 7 (0–10) | 1.15 (.43–3.09) | .774 | 0.75 (.46–1.22) | .25 |
Abbreviations: ARI, acute respiratory infection; COPD, chronic pulmonary obstructive disease; ICU, intensive care unit.
a Treatment failure was defined according to clinical setting: primary care (death, hospitalization, ARI-specific complications, recurrent or worsening infection, and discomfort at 30 days), emergency department (mortality, ICU admission, rehospitalization, complications, recurrent or worsening infection within 30 days), intensive care unit (all-cause mortality within 30 days).
Source: Adapted from Schuetz et al [9].
Traditional culture methods, such as blood cultures, focus on identification and characterization of pathogens. Yet, they have low sensitivity and, thus, if negative, may not influence clinical decision making. A blood marker, such as PCT, mirrors a patient's response to infection and indirectly the severity of infection. The marker may not be able to identify the etiology of infection, but the likelihood of a relevant bacterial infection increases with increasing marker levels. The marker then may help rule out infection and provides information about patient recovery. With new microbiological methods becoming available that rapidly identify microorganisms with higher sensitivity as discussed below, PCT may help to increase specificity by providing information about the “relevance” of microbiological results in individual patients.
Gene Expression Profiling of Peripheral Blood Leukocytes
Gene expression profiling of peripheral blood cells is an emerging strategy for diagnosing and monitoring infection. Like procalcitonin, gene expression profiles use host response to pathogens as a means of diagnosing infection rather than direct pathogen detection. Unlike PCT, gene expression profiles simultaneously measure the expression of a large number of genes to generate a snapshot of host immune cell function. Pattern-recognition receptors on immune cells are activated by different pathogen-derived ligands. This results in the initiation of distinct sets of transcriptional programs. The resultant pattern of gene expression may be viewed as a transcriptional signature that represents or is diagnostic of a specific pathogen.
The first step in gene expression profiling requires isolation of RNA transcripts from cells of interest, most commonly peripheral blood cells. Microarrays are the most frequently used gene expression platform in clinical work and allow for analysis on a genome-wide scale. A microarray is a solid support to which an array of oligonucleotide probes representing specific gene coding regions is affixed. Isolated RNA is typically transcribed into complementary DNA (cDNA), fluorescently labeled and hybridized to the array. The strength of the fluorescent signal generated from each probe represents the relative abundance of the corresponding RNA transcript. The differential expression of genes between 2 states, such as infection versus health, or bacterial versus viral infection, can then be compared. While microarrays have been the mainstay for gene expression profiling in the recent past, they may soon be superseded by newer technologies such as high-throughput cDNA sequencing, which has a greater dynamic range and also provides sequence information on each RNA transcript (reviewed in [14]) or by direct RNA sequencing, which eliminates some of the artifact introduced with cDNA synthesis (reviewed in [14, 15]).
Some of the initial work using gene expression profiling of peripheral blood leukocytes to discriminate bacterial from viral infections was recently described [16, 17]. Using genome-wide expression profiles obtained from 131 pediatric patients with acute infections, Ramilo et al [16] identified a set of 35 genes that distinguished influenza A infection from bacterial infections with 87%–95% accuracy. Using sets of either 30 or 45 genes, they were able to differentiate Escherichia coli, Staphylococcus aureus, and Streptococcus pneumoniae infections with 83%–95% accuracy. On the basis of this data, they generated a microarray of 137 genes that had the greatest power to discriminate among pathogens. This array was able to distinguish the expression profiles of 27 patients with acute respiratory infection compared to 7 healthy controls. In subsequent work, Zaas et al developed a 30-gene transcriptional signature of acute respiratory viral infections in adults experimentally infected with common respiratory viruses [17]. Using this 30-gene signature, reanalysis of the pediatric datasets from Ramilo's work was able to distinguish children with viral infection from healthy controls with 100% accuracy. Influenza A was distinguished from any bacterial infection with 80% accuracy and influenza A from streptococcal pneumonia infection with 93% accuracy. The work of these 2 groups also highlights a shortcoming of gene expression profiling. In the setting of polymicrobial or mixed viral and bacterial infections, indistinct profiles can be generated, thus limiting the utility of this potential new diagnostic.
Multiple groups have used gene expression profiling of peripheral blood leukocytes to distinguish sepsis from sterile SIRS. Tang et al generated a 50-gene transcriptional profile of sepsis that distinguished sepsis from sterile SIRS with 88%–91% accuracy [18]. Notably, the majority of these studies have shown an upregulation of pattern recognition receptors such as Toll-like receptors and CD14, as well as signal transduction pathways including nuclear factor kappa B, mitogen-activated protein kinase, and Janus kinase, that are involved in coordinating host immune responses [19]. By contrast, the expression levels of inflammatory cytokines such as tumor necrosis factor and members of the interleukin family, vary between studies and no consistent pattern has been discerned [19]. This interstudy variability highlights some of the challenges in bringing gene expression profiling into the clinical arena.
Many of these studies are limited by small sample size and, ultimately, large, prospective clinical trials will be needed for validation of transcriptional profiles in diagnosing and monitoring infection. Significant challenges in standardizing technique and data analysis also remain before expression profiling in infectious diseases is brought into clinical use. However, similar challenges have been overcome in the fields of oncology, where transcriptional profiling is used to help predict outcomes in breast cancer patients [20, 21]. As the technology improves, cost declines, and knowledge of leukocyte biology grows, it becomes increasingly likely that peripheral blood transcriptional profiling will become a clinically valuable diagnostic.
Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), a method with broad applications in biochemistry, proteomics, and polymer chemistry, has recently been adapted for the identification of whole microorganisms [22–26]. The development of user-friendly, integrated commercial MALDI-TOF MS platforms has brought this technology into many European, and some US, clinical microbiology laboratories, where it has been used to identify organisms from colonies on solid media as well as directly from positive blood and urine cultures [22, 27–31]. The technique has proved capable of accurately identifying mycobacteria and nonfermenting bacteria, organisms that have posed difficulties for conventional methods [33, 34]. Although the US Food and Drug Administration (FDA) has yet to approve any commercial MALDI-TOF MS system for clinical use, this technology offers improved turnaround time and will complement, if not someday supplant, conventional microbiologic identification methods.
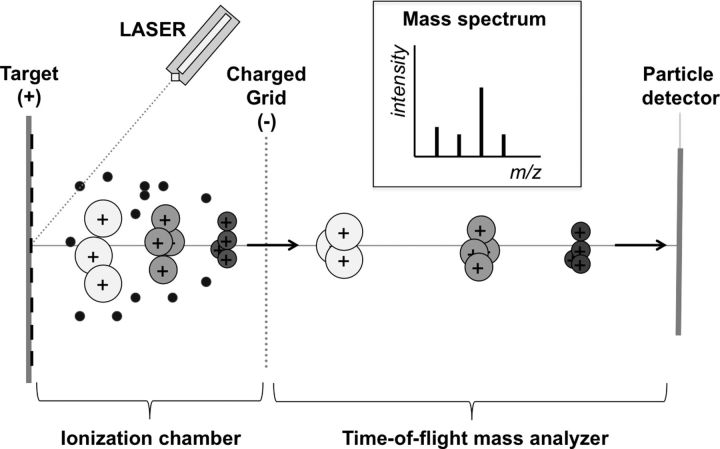
MALDI-TOF MS instruments have 3 components: a specimen ionization chamber, a time-of-flight mass analyzer, and a particle detector [32] (Figure 1). Sample preparation is simple, and involves transferring a portion of an isolated colony onto a target plate. The deposited colony is then covered with a chemical matrix and the target plate is loaded into the instrument. The sample-matrix mixture is pulsed by a laser, converting the sample into an ionic gas composed of small proteins and peptides and other molecules. In the ionization chamber, positively charged molecules are accelerated through an electric field to velocities that depend on their mass-to-charge (m/z) ratios. The particles then leave the electric field and enter the time-of-flight mass analyzer. The time it takes a particle to traverse the mass analyzer (“flight time”) depends on the velocity developed in the ionization chamber, and hence, on the m/z ratio. Flight times of individual particles are measured by a particle detector at the end of the mass analyzer, and are converted into m/z values that are plotted on a mass spectrogram. The spectrogram is then compared to a library by a proprietary algorithm to identify the organism [22].
Figure 1.
Schematic diagram of a matrix-assisted laser desorption/ionization time-of-flight mass spectrometry instrument. The specimen is irradiated with a laser and converted into an ionic gas in the ionization chamber. Positively charged particles are accelerated in an electric field and then traverse the time-of-flight mass analyzer before colliding with a particle detector. The mass spectrogram is calculated from the measured flight times.
Principal advantages of MALDI-TOF MS technology include ease of use, potential to automate, rapid turnaround time, and low reagent costs [22]. The simplicity of setup and the ability to run large numbers of isolates per batch readily lend this technique to high-throughput workflow and potential automation. Once the instrument is loaded, identifications can typically be performed in <1 minute, compared with hours to days for conventional methods. This improvement in turnaround time may carry substantial clinical benefit. Although purchase of a MALDI-TOF MS instrument involves a significant capital commitment with recurrent annual service contract fees, the reagents and disposables required consist primarily of target plates, microliter quantities of inexpensive organic compounds, inoculation loops, and pipette tips with estimated costs of as little as US$0.10–$0.40 per identification when optimized [22]. In some cases, this operational cost amounts to one-tenth that of conventional identification with automated biochemical testing platforms.
Multiple studies comparing MALDI-TOF with conventional techniques have been performed in Europe, where at least 1 commercial MALDI-TOF instrument has CE approval [22]. A Swiss study compared a MALDI-TOF MS system to conventional methods for the identification of 1371 routine bacteria and yeast isolates in a clinical microbiology laboratory [27]. MALDI-TOF MS provided identifications for 98.5% of the isolates, including 93.2% at the species level and 5.3% at the genus level. Of the species level identifications, 95.1% matched conventional identifications. Eleven percent of discordant results were due to errors in conventional identification, whereas the remaining 89% of discrepancies were due to errors in MALDI-TOF identification.
Important deficiencies in present MALDI-TOF MS platforms include misclassification of Shigella as Escherichia coli, and misclassification of Streptococcus pneumoniae as Streptococcus mitis with one instrument [22]. Additionally, present MALDI-TOF MS instruments have demonstrated poor performance with polymicrobial samples. In some cases, instruments have identified only 1 organism without indicating the presence of others [22].
Commercial MALDI-TOF instruments are expected to evolve rapidly under intense competition for US market share, pending FDA approval. Additionally, instruments that perform microbial identifications by combining polymerase chain reaction with electrospray ionization time-of-flight mass spectrometry are likely to enter the competition. While there is room for improvement, the fast turnaround times, ease of use, and potential operational cost savings are likely to make mass spectrometry technology popular in US clinical laboratories in the near future.
Nucleic Acid Aptamers
Nucleic acid aptamers are short, single-stranded oligonucleotides that bind to a broad range of targets with high affinity and specificity. The use of oligonucleotides as affinity probes for proteins or other molecules was first described by Tuerk and Gold [35] and Ellington and Szostak [36] in 1990. A nucleic acid aptamer may be composed of either DNA or RNA and is typically 15–90 bases in length. Aptamers with high affinity and specificity for their targets are selected in an iterative process called systematic evolution of ligands by exponential enrichment (SELEX) [35–37] (Figure 2). The binding affinity of aptamers rival and often surpass that of antibodies. Aptamers also have great discriminatory power. As an example, an aptamer developed for the detection of theophylline is 14 000-fold more specific for theophylline than caffeine, a molecule that differs from theophylline by the presence of a single methyl group [38].
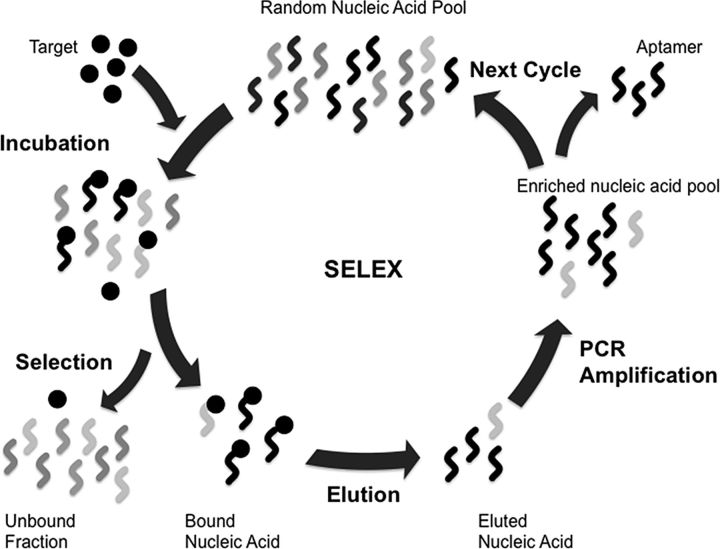
Figure 2.
In systematic evolution of ligands by exponential enrichment (SELEX), a large pool of random oligonucleotides is incubated with a target molecule. Upon binding, the short nucleic acid sequence folds into a 3-dimensional structure that binds to its target. Bound nucleic acids are then eluted and amplified by polymerase chain reaction. This cycle is repeated until oligonucleotides with suitably high specificity and binding affinity are isolated [47]. Through this process aptamers with low picomolar to nanomolar dissociation constants (Kd) can be obtained [48]. The SELEX process involves 5 main steps: (1) incubation of a random pool of nucleic acids with a target molecule or cell; (2) separating bound nucleic acids from unbound nucleic acids; (3) eluting the bound nucleic acid from the target; (4) amplifying the eluted nucleic acids to generate a refined pool of nucleic acids; and (5) using amplicons as new nucleic acid pools in subsequent rounds of selection. This process is repeated until “aptamers” with high specificity and affinity for the target are isolated. Abbreviations: PCR, polymerase chain reaction; SELEX, systematic evolution of ligands by exponential enrichment.
Conventionally, nucleic acid aptamers were selected for purified target molecules. In 2008, Hamula et al modified the SELEX process using live bacterial cells in suspension as targets [39]. Using this technique, this group was able to develop an aptamer to Lactobacillus acidophilus that distinguished this bacterium from other bacterial and fungal genera. Their work highlights one of the major strengths of aptamer technology—namely, that a specific protein target does not need to be isolated prior to selection. The use of whole bacterium as a target also allows for the detection selection of aptamers that target proteins in their native conformation. Subsequently, this group used the 10 most prevalent strains of group A streptococcus as targets in the SELEX process and were able to derive aptamers with high affinity and specificity compared to other streptococcal species [40]. Whole cell SELEX has also been used to derive aptamers capable of distinguishing Escherichia coli O157:H7 from a nonpathogenic strain of Escherichia coli [41]. This technique has also been used in vitro to identify a panel of 5 aptamers capable of distinguishing Staphylococcus aureus from Staphylococcus epidermidis. The same aptamer panel was then used to identify Staphylococcus aureus in the fluid of an infected wound [42]. Similar whole bacterium SELEX methods have been used to develop aptamers for Campylobacter jejuni [43], Vibrio parahaemolyticus [44], and multiple Salmonella species [45, 46]. Further refinement is needed before aptamer-based assays for pathogen detection come into clinical use; however, given their high specificity and the ease with which they can be modified, aptamers may become ideal reagents for use in point-of-care tests.
Because of their high discriminatory power, aptamers have potential for genotyping and serotyping viruses. Gopinath et al developed an RNA aptamer capable of distinguishing a single-subtype H3N2 influenza from other influenza strains, including other H3N2 viruses [49]. Notably, the aptamer had a 15-fold greater affinity for the virus when compared to a commercially available antibody that targeted the same H3N2 strain.
Aptamer manufacturing has many advantages. The oligonucleotides are readily synthesized at scale. They are stable and robust, with long shelf lives and an ability to withstand fluctuations in temperature. Aptamers may be easily attached to a wide variety of detection moieties. But important disadvantages include the cost associated with chemical modification, the difficulty designing aptamers against hydrophobic nonpolar targets, and the limited library of available commercial aptamers at present. Another potential deficiency is that nucleic acid aptamers can be degraded by serum nucleases that may limit their applicability in ex vivo diagnostics. Despite these drawbacks, aptamers are versatile recognition molecules that can be designed to target virtually any pathogen and are expected to find application in many future diagnostic techniques.
CONCLUSIONS
Antimicrobial resistance is a growing global public health problem that has substantial impact on morbidity, mortality, and healthcare costs. Diagnostic uncertainty has been identified as a driving factor in the misuse and overuse of antimicrobials, which can result in selection of resistant microbes. Diagnostic modalities that help distinguish infectious from noninfectious causes of illness are much needed. We have highlighted 2 approaches, one on the early and accurate identification of the pathogen, and the second on the patients' host response to the pathogen. This review identifies several novel technologies that have the potential to dramatically improve the assessment of patients with presumed infectious diseases, and help monitor the course of infection and response to therapy. These technologies are advancing, but additional research and clinical trials are needed to establish their safe, appropriate, and cost-effective use in everyday practice.
Notes
Disclaimer. No commercial sponsor had any involvement in design and conduct of this study, namely collection, management, analysis, or interpretation of the data; and preparation, decision to submit, review, or approval of the manuscript.
Financial support. M. K. M. is supported by NIAID NIH T32-AI007061-35.
Potential conflicts of interest. P. S. has received support from BRAHMS and bioMérieux to attend meetings and has fulfilled speaking engagements. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Kumar A, Roberts D, Wood KE, et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med. 2006;34:1589–96. doi: 10.1097/01.CCM.0000217961.75225.E9. [DOI] [PubMed] [Google Scholar]
- 2.Proulx N, Frechette D, Toye B, Chan J, Kravcik S. Delays in the administration of antibiotics are associated with mortality from adult acute bacterial meningitis. QJM. 2005;98:291–8. doi: 10.1093/qjmed/hci047. [DOI] [PubMed] [Google Scholar]
- 3.Spellberg B, Guidos R, Gilbert D, et al. The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin Infect Dis. 2008;46:155–64. doi: 10.1086/524891. [DOI] [PubMed] [Google Scholar]
- 4.Roberts RR, Hota B, Ahmad I, et al. Hospital and societal costs of antimicrobial-resistant infections in a Chicago teaching hospital: implications for antibiotic stewardship. Clin Infect Dis. 2009;49:1175–84. doi: 10.1086/605630. [DOI] [PubMed] [Google Scholar]
- 5.Linscheid P, Seboek D, Schaer DJ, Zulewski H, Keller U, Muller B. Expression and secretion of procalcitonin and calcitonin gene-related peptide by adherent monocytes and by macrophage-activated adipocytes. Crit Care Med. 2004;32:1715–21. doi: 10.1097/01.ccm.0000134404.63292.71. [DOI] [PubMed] [Google Scholar]
- 6.Cuquemelle E, Soulis F, Villers D, et al. Can procalcitonin help identify associated bacterial infection in patients with severe influenza pneumonia? A multicentre study. Intensive Care Med. 2011;37:796–800. doi: 10.1007/s00134-011-2189-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker KL. Procalcitonin and the calcitonin gene family of peptides in inflammation, infection, and sepsis: a journey from calcitonin back to its precursors. J Clin Endocrinol Metab. 2004;89:1512–25. doi: 10.1210/jc.2002-021444. [DOI] [PubMed] [Google Scholar]
- 8.Schuetz P, Chiappa V, Briel M, Greenwald JL. Procalcitonin algorithms for antibiotic therapy decisions: a systematic review of randomized controlled trials and recommendations for clinical algorithms. Arch Intern Med. 2011;171:1322–31. doi: 10.1001/archinternmed.2011.318. [DOI] [PubMed] [Google Scholar]
- 9.Schuetz P, Briel M, Christ-Crain M, et al. Procalcitonin to guide initiation and duration of antibiotic treatment in acute respiratory infections: an individual patient data meta-analysis. Clin Infect Dis. 2012;55:651–62. doi: 10.1093/cid/cis464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heyland DK, Johnson AP, Reynolds SC, Muscedere J. Procalcitonin for reduced antibiotic exposure in the critical care setting: a systematic review and an economic evaluation. Crit Care Med. 2011;39:1792–9. doi: 10.1097/CCM.0b013e31821201a5. [DOI] [PubMed] [Google Scholar]
- 11.Albrich WC, Dusemund F, Bucher B, et al. Effectiveness and safety of procalcitonin-guided antibiotic therapy in lower respiratory tract infections in “real life”: an international, multicenter poststudy survey (ProREAL) Arch Intern Med. 2012;172:715–22. doi: 10.1001/archinternmed.2012.770. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert D. Serum procalcitonin levels: comment on “effectiveness and safety of procalcitonin-guided antibiotic therapy in lower respiratory tract infections in ‘real life’”. Arch Intern Med. 2012;172:722–3. doi: 10.1001/archinternmed.2012.1327. [DOI] [PubMed] [Google Scholar]
- 13.Jensen JU, Hein L, Lundgren B, et al. Procalcitonin-guided interventions against infections to increase early appropriate antibiotics and improve survival in the intensive care unit: a randomized trial. Crit Care Med. 2011;39:2048–58. doi: 10.1097/CCM.0b013e31821e8791. [DOI] [PubMed] [Google Scholar]
- 14.Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ozsolak F, Milos PM. Single-molecule direct RNA sequencing without cDNA synthesis. Wiley Interdiscip Rev RNA. 2011;2:565–70. doi: 10.1002/wrna.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramilo O, Allman W, Chung W, et al. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood. 2007;109:2066–77. doi: 10.1182/blood-2006-02-002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaas AK, Chen M, Varkey J, et al. Gene expression signatures diagnose influenza and other symptomatic respiratory viral infections in humans. Cell Host Microbe. 2009;6:207–17. doi: 10.1016/j.chom.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang BM, McLean AS, Dawes IW, Huang SJ, Lin RC. The use of gene-expression profiling to identify candidate genes in human sepsis. Am J Respir Crit Care Med. 2007;176:676–84. doi: 10.1164/rccm.200612-1819OC. [DOI] [PubMed] [Google Scholar]
- 19.Tang BM, Huang SJ, McLean AS. Genome-wide transcription profiling of human sepsis: a systematic review. Crit Care. 2010;14:R237. doi: 10.1186/cc9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamal AH, Loprinzi CL, Reynolds C, et al. Breast medical oncologists' use of standard prognostic factors to predict a 21-gene recurrence score. Oncologist. 2011;16:1359–66. doi: 10.1634/theoncologist.2011-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pham MX, Teuteberg JJ, Kfoury AG, et al. Gene-expression profiling for rejection surveillance after cardiac transplantation. N Engl J Med. 2010;362:1890–900. doi: 10.1056/NEJMoa0912965. [DOI] [PubMed] [Google Scholar]
- 22.Dekker JP, Branda JA. MALDI-TOF mass spectrometry in the clinical microbiology laboratory. Clin Microbiol Newsl. 2011;33:87–93. [Google Scholar]
- 23.Sauer S, Kliem M. Mass spectrometry tools for the classification and identification of bacteria. Nat Rev Microbiol. 2010;8:74–82. doi: 10.1038/nrmicro2243. [DOI] [PubMed] [Google Scholar]
- 24.Sauer S, Freiwald A, Maier T, et al. Classification and identification of bacteria by mass spectrometry and computational analysis. PLoS One. 2008;3:e2843. doi: 10.1371/journal.pone.0002843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freiwald A, Sauer S. Phylogenetic classification and identification of bacteria by mass spectrometry. Nat Protoc. 2009;4:732–42. doi: 10.1038/nprot.2009.37. [DOI] [PubMed] [Google Scholar]
- 26.Ho YP, Reddy PM. Identification of pathogens by mass spectrometry. Clin Chem. 2010;56:525–36. doi: 10.1373/clinchem.2009.138867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bizzini A, Durussel C, Bille J, Greub G, Prod'hom G. Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J Clin Microbiol. 2010;48:1549–54. doi: 10.1128/JCM.01794-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.La Scola B, Raoult D. Direct identification of bacteria in positive blood culture bottles by matrix-assisted laser desorption ionisation time-of-flight mass spectrometry. PLoS One. 2009;4:e8041. doi: 10.1371/journal.pone.0008041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stevenson LG, Drake SK, Murray PR. Rapid identification of bacteria in positive blood culture broths by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2010;48:444–7. doi: 10.1128/JCM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prod'hom G, Bizzini A, Durussel C, Bille J, Greub G. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for direct bacterial identification from positive blood culture pellets. J Clin Microbiol. 2010;48:1481–3. doi: 10.1128/JCM.01780-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferreira L, Sanchez-Juanes F, Gonzalez-Avila M, et al. Direct identification of urinary tract pathogens from urine samples by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2010;48:2110–5. doi: 10.1128/JCM.02215-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horton G. The MALDI-TOF mass spectrometric view of the plasma proteome and peptidome. Clin Chem. 2006;52:1223–37. doi: 10.1373/clinchem.2006.069252. [DOI] [PubMed] [Google Scholar]
- 33.Mellmann A, Cloud J, Maier T, et al. Evaluation of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry in comparison to 16S rRNA gene sequencing for species identification of nonfermenting bacteria. J Clin Microbiol. 2008;46:1946–54. doi: 10.1128/JCM.00157-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pignone M, Greth KM, Cooper J, Emerson D, Tang J. Identification of mycobacteria by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry. J Clin Microbiol. 2006;44:1963–70. doi: 10.1128/JCM.01959-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–10. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 36.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–22. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 37.Dua P, Kim S, Lee DK. Patents on SELEX and therapeutic aptamers. Recent Pat DNA Gene Seq. 2008;2:172–86. doi: 10.2174/187221508786241710. [DOI] [PubMed] [Google Scholar]
- 38.Jenison RD, Gill SC, Pardi A, Polisky B. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–9. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 39.Hamula CL, Zhang H, Guan LL, Li XF, Le XC. Selection of aptamers against live bacterial cells. Anal Chem. 2008;80:7812–9. doi: 10.1021/ac801272s. [DOI] [PubMed] [Google Scholar]
- 40.Hamula CL, Le XC, Li XF. DNA aptamers binding to multiple prevalent M-types of Streptococcus pyogenes. Anal Chem. 2011;83:3640–7. doi: 10.1021/ac200575e. [DOI] [PubMed] [Google Scholar]
- 41.Lee YJ, Han SR, Maeng JS, Cho YJ, Lee SW. In vitro selection of Escherichia coli O157:H7-specific RNA aptamer. Biochem Biophys Res Commun. 2012;417:414–20. doi: 10.1016/j.bbrc.2011.11.130. [DOI] [PubMed] [Google Scholar]
- 42.Cao X, Li S, Chen L, et al. Combining use of a panel of ssDNA aptamers in the detection of Staphylococcus aureus. Nucleic Acids Res. 2009;37:4621–8. doi: 10.1093/nar/gkp489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dwivedi HP, Smiley RD, Jaykus LA. Selection and characterization of DNA aptamers with binding selectivity to Campylobacter jejuni using whole-cell SELEX. Appl Microbiol Biotechnol. 2010;87:2323–34. doi: 10.1007/s00253-010-2728-7. [DOI] [PubMed] [Google Scholar]
- 44.Duan N, Wu S, Chen X, Huang Y, Wang Z. Selection and identification of a DNA aptamer targeted to Vibrio parahemolyticus. J Agric Food Chem. 2012;60:4034–8. doi: 10.1021/jf300395z. [DOI] [PubMed] [Google Scholar]
- 45.Duan N, Wu S, Zhu C, et al. Dual-color upconversion fluorescence and aptamer-functionalized magnetic nanoparticles-based bioassay for the simultaneous detection of Salmonella Typhimurium and Staphylococcus aureus. Anal Chim Acta. 2012;723:1–6. doi: 10.1016/j.aca.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 46.Hyeon JY, Chon JW, Choi IS, Park C, Kim DE, Seo KH. Development of RNA aptamers for detection of Salmonella Enteritidis. J Microbiol Methods. 2012;89:79–82. doi: 10.1016/j.mimet.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 47.Yan AC, Levy M. Aptamers and aptamer targeted delivery. RNA Biol. 2009;6:316–20. doi: 10.4161/rna.6.3.8808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mairal T, Ozalp VC, Lozano Sanchez P, Mir M, Katakis I, O'Sullivan CK. Aptamers: molecular tools for analytical applications. Anal Bioanal Chem. 2008;390:989–1007. doi: 10.1007/s00216-007-1346-4. [DOI] [PubMed] [Google Scholar]
- 49.Gopinath SC, Misono TS, Kawasaki K, et al. An RNA aptamer that distinguishes between closely related human influenza viruses and inhibits haemagglutinin-mediated membrane fusion. J Gen Virol. 2006;87(Pt 3):479–87. doi: 10.1099/vir.0.81508-0. [DOI] [PubMed] [Google Scholar]