Abstract
Replication-defective human adenovirus (Ad) group C transducing vectors, most of which have the E1A, E1B, and E3 genes deleted, are highly inflammatory despite the fact that the parental viruses typically cause subclinical or mild infections. To investigate this paradox, the roles that the E1A, E1B, and E3 genes play in inflammation were tested by using replication-incompetent viruses carrying a deletion of the preterminal protein gene. The viruses were injected into BALB/c mouse ears, and edema was monitored as a sensitive surrogate marker of inflammation. A virus deleted for the E1A 289R (transcription activating) protein was noninflammatory, and inhibited edema induced by empty virus particles. The E1A 243R and E1B 55-kDa (p53 binding) proteins play the most important roles in inhibition of inflammation by the noninflammatory virus. The E1B 19-kDa antiapoptotic protein inhibited edema when both the E1A 243R and E1B 55-kDa proteins were expressed but strongly induced edema when only one was expressed. E3 proteins had their greatest effect on the inhibition of edema induced by the E1A 289R protein. The results support a model in which inflammation is countered through a mechanism that involves complex genetic interactions between Ad early region proteins and offer promise for the design and construction of noninflammatory Ad gene therapy vectors that are relatively easy to grow and purify.
The human group C adenoviruses (Ads), particularly Ad serotype 5 (Ad5), have been popular vectors for gene therapy studies. However, these vectors induce strong inflammatory responses that lead to loss of transduced cells and transduced gene expression and a strong adaptive immune response that limits effective readministration of the vector (1).
One promising approach to deal with the induction of inflammation has been the use of helper virus-dependent vectors devoid of all adenoviral genes (2, 3). However, such vectors are considerably more difficult than replication-defective viruses to grow and purify. An alternative approach is to take advantage of the immune modulatory effects of Ad proteins in the design of gene therapy vectors.
Although inflammation induced by replication-defective human group C Ad vectors has come to be accepted as an inherent shortcoming, it is actually surprising given the natural behavior of the parental viruses. Approximately 50% of Ad5 infections are subclinical, and of the infections that are symptomatic, most are associated with mild sore throat and fever. After initial infection of the pharyngeal epithelium, Ad5 moves to the small intestine where it causes persistent and asymptomatic infections with infectious virus shed fecally for days to years (4). Although it is not clear that the amount of virus produced is similar to the amounts of vectors injected to obtain efficient transduction, the ability of Ad5 to cause asymptomatic infections raises the question of why Ad5 vectors made replication defective or replication incompetent induce such strong innate reactions.
Ad vectors are generally made replication defective by deletion of the E1A and E1B genes, and most are also deleted for the E3 gene. E3 proteins are generally considered the most important defense of the virus against host immune responses. Overexpression of E3 or specific E3 ORFs leads to enhanced vector function (5-7) and reduced immune response (5), although contradictory results have also been reported (8, 9). E3 proteins inhibit inflammatory and adaptive responses induced by viral gene expression but appear to have little potential for inhibition of the earliest steps in the induction of inflammation by Ad, those induced by virion binding and uptake. Instead, E1A is the most promising candidate for playing a critical role in the earliest steps.
E1A is the first viral gene expressed after infection and encodes two major proteins, 289 and 243 residues in length in Ad5, that have well-characterized roles in transcription and cell cycle regulation (10, 11). Transcriptional regulation by E1A proteins includes both activation, primarily a function of the 289R protein, and repression, a function of both the 289R and 243R proteins. It is through its transcription repression functions that E1A inhibits host innate immune responses that occur early after infection. Ad particle binding and uptake induce a type I IFN response (12) and expression of inflammatory chemokines (13, 14). E1A inhibits the type I IFN response by repressing transcription of IFN-induced genes (12). Although E1A expression sensitizes certain cells to inflammatory chemokine induction (15), it is likely that E1A also inhibits inflammatory chemokine gene transcription (16, 17). Because E1A encodes the only viral functions known to inhibit the earliest steps in the innate immune response to Ad, we hypothesized that inclusion of E1A is essential for the controlling the deleterious inflammation that occurs in response to Ad vectors.
E1B encodes two major proteins, a 55-kDa protein that, among many functions, binds to and regulates p53 activity (18), and a 19-kDa protein that inhibits apoptosis (19). The E1B 55-kDa protein plays a proinflammatory role in infections by the WT virus (20), whereas a clear role in the regulation of inflammation of the 19-kDa protein has not been shown. In addition, E1B encodes a 17-kDa protein (21) of unknown function (22).
In this study, we have used injection of BALB/c mouse ears and measurement of edema to study the roles of certain Ad early region proteins in the induction and inhibition of inflammation. We demonstrate that E1A 243R inhibits Ad-induced inflammation, but that E1A functions through complex genetic interactions with other Ad early region proteins, particularly those encoded by E1B.
Materials and Methods
Cells. HEK 293 cells are transformed by and express high levels of Ad5 E1 proteins (23), and 293-preterminal protein (pTP) cells were derived from 293 cells by stable transfection with the pTP gene (ref. 24 and J.S., unpublished work).
Virus Growth and Purification. Viruses used in this study were derived from Ad5dl308ΔpTP (25), which has the pTP gene deleted. pTP deletion mutant viruses were grown in 293-pTP cells, and other viruses were grown in 293 cells. Virus stocks were purified as reported (26). Viral DNA-terminal protein (TP) complexes were purified by banding isolated virions in 2.8 M CsCl containing 4 M guanidinium-HCl (27) followed by extensive dialysis vs. 10 mM Tris·HCl, pH 8.0/0.1 mM EDTA.
Plasmids. The Ad5dl308ΔpTP chromosome was introduced into the plasmid p3601 through homologous recombination in Escherichia coli strain BJ5183 (28). To introduce the pTP deletion mutant allele into the backbone of the otherwise WT virus, the fragment from 19550 to 31090 produced by digestion with NdeI was inserted in place of the analogous fragment in p3601-308ΔpTP. To introduce the majority of the Ad chromosomes (excluding the left inverted terminal repeat through the middle of the E1B coding sequence) of Ad5dl300ΔpTP (virus A; viruses tested in this study are listed in Table 1) and Ad5dl327ΔpTP (virus J) into the plasmid pAdEasy1 (29), pAdEasy-1 was digested with XhoI and used with Ad5dl300ΔpT P or Ad5dl327ΔpTP DNA to coelectropoate E. coli strain BJ5183.
Table 1. Viruses analyzed in this study.
| Virus | Genotype |
|---|---|
| A | pTP− |
| B | pTP−, E1A 243R− |
| C | pTP−, E1A 289R− |
| D | pTP−, E1A−, E1B− |
| E | pTP−, E1A− |
| F | pTP−, E1A 289R−, E1B− |
| G | pTP−, E1A 289R−, E1B 55-kDa− |
| H | pTP−, E1A 289R−, E1B 19-kDa− |
| I | pTP−, E1−, LacZ+ |
| J | pTP−, E3− |
| K | pTP−, E3−, E1A 243R− |
| L | pTP−, E3−, E1A 289R− |
| M | E1A 289R− |
The viruses used in this study are indicated by letter designations, and genotypes are presented. The letter designation is used in identification of the viruses in the figures and text. Virus D retains the E1A promoter. Virus I encodes LacZ under the control of the cytomegalovirus major immediate early promoter.
Sequencing of the E1B 55-kDa Gene. Sequence analysis of the Ad5dl308ΔpTP E1B 55-kDa coding region demonstrated deletion of G2760 leading to frameshifting and a C-terminally truncated protein. The E1A and E1B regions of Ad5dl308ΔpTP and all of the viruses derived from it were replaced with the WT E1 region of Ad5dl308 viral DNA by homologous recombination in 293-pTP cells. All of the E1B regions tested in this study were derived directly from viral DNAs to avoid selection of mutated E1B 55-kDa genes that occurs in E. coli.
Construction of New Viruses. To construct a pTP deletion mutant virus deleted for the E3 gene, Ad5dl327-TP complex and Ad5dl308ΔpTP-TP complex that had been digested with SalI (which cleaves uniquely at position 16747 in Ad5dl308ΔpTP and positions 9463, 9842, and 16747 in Ad5dl327) were ligated. The ligation mixture was used to transfect 293-pTP cells, and Ad5dl327ΔpTP was plaque-purified.
pAdEasy plasmids containing the majority of the chromosomes of the pTP deletion mutants were digested with PacI (which cleaves at the right, or E4, end) and used with XhoI-digested Ad-TP complexes to cotransfect 293-pTP cells. The Ad-TP complexes used were derived from Ad5dl308 [WT E1 region (30)], Ad5dl347 [Ad2 E1A 12S cDNA (31)], Ad5dl348 [Ad2 E1A 13S cDNA (31)], Ad5dl312 [deleted for the E1A gene (32)], A5dl337/E1A 243R [E1A coding sequence replaced by the E1A 12S cDNA (this study) and deleted for E1B 19 kDa (33)], and Ad5dl338/E1A 243R [E1A coding sequence replaced by the E1A 12S cDNA (this study) and deleted for E1B 55- and 17-kDa proteins (34)]. The viruses Ad5dl337/E1A 243R and Ad5dl338/E1A 243R were constructed by digesting Ad5dl337 and Ad5dl338 DNAs with XbaI. The large (4.8-100 map unit) fragments were isolated on 10-40% sucrose step gradients and ligated with the purified 0-4.8 map unit fragment of Ad5dl300ΔpTP E1A 243R resulting from digestion with XbaI. The ligation mixtures were used to transfect 293-pTP cells.
The Ad2 E1A 12S cDNA was synthesized by PCR using the primers 5′-AAT TGG TAC CTT GAG TGC CAG CGA GTA G-3′ (homologous to base pairs 502-520) and 5′-AAT TGG TAC CCA TTT AAC ACG CCA TGC AAG-3′ (homologous to base pairs 1635-1654) and Ad5dl347 template DNA and cloned under the control of the E1A promoter into the KpnI site of pShuttle-E1A (35). A clone containing the cDNA with the expected sequence and in the sense orientation, called pShuttle-E1A 243R, was used.
Viruses that did not encode E1B were constructed by using the modified AdEasy system as described (36). E. coli strain BJ5183 that had been transformed with pAdEasy-300ΔpTP was electroporated with 20 ng of PmeI-digested shuttle vector. Recombinant AdEasy plasmids containing the modified left viral ends were digested with PacI to release the Ad chromosomes and used to transfect 293-pTP cells. Resultant viruses were plaque-purified, grown in large scale, and purified as indicated above.
Purification of Empty Ad Particles. The pTP deletion mutant viruses produce large amounts of empty particles during growth in 293-pTP cells for reasons that are not clear. These empty particles have very similar protein profiles when compared to full particles (data not shown). Empty particles produced during growth of virus C (Table 1) were purified by one step and two isopycnic CsCl gradient centrifugations. The concentration of particles was estimated by comparison of OD caused by light scattering at 420 nm with that of virus particles whose concentration had been determined from OD at 260 nm as indicated above. Dilutions of the empty particles used to infect 293-pTP cells demonstrated that the empty particle stock had fewer than one infectious particle per 105 particles.
Confirmation of Virus Genotype and Phenotype. Viruses were tested by PCR for the presence of the expected E1A allele, by restriction digestion of the viral DNAs for the presence of the expected DNA structure in plaque formation assays in 293-pTP and HeLa-pTP cells as a genetic test of viral gene function, and by Western analysis for synthesis of E1B 55-kDa (Fig. 6, which is published as supporting information on the PNAS web site) and E1A 289R and 243R proteins. The results demonstrated that each of the viruses was of the expected genotype and phenotype (Table 1). All of the stocks used in these studies had fewer than one infectious unit in 109 particles when grown in 293 cells and fewer than one infectious unit in 1010 particles (the limit of detection) when grown in HeLa cells.
Analysis of Infectious Activity of Virus Stocks. Virus stocks were tested for plaque titers by using 293-pTP cells. Certain of the virus stocks formed plaques slowly and formed small plaques, making it difficult to effectively compare plaque titers between stocks. Therefore, particle titers were used to standardize virus stocks. To ensure that the stocks contained virus particles of roughly equal infectivity, 293-pTP cells were infected with 104 particles per cell, and cytopathic effect was examined as a function of time. All stocks produced cytopathic effect at a similar rate.
Injections of Mice and Analysis of Edema. All procedures involving animals followed the guidelines of, and were approved by, the University of Colorado Institutional Animal Care and Use Committee. Female, 6-week-old BALB/c mice were anesthetized by subdermal injection of avertin before all procedures. Virus stocks were diluted with PBS such that the glycerol concentration was 5%. Particles (2 × 1010), except as noted, were injected subdermally into ears. Ear thickness was measured immediately before injection and every 24 h through 72 or 96 h after injection by using an engineer's micrometer (24). Ears that developed hematomas were excluded. Data presented represent the average values from 6 to 24 ears, as noted, ± the SEM.
Results
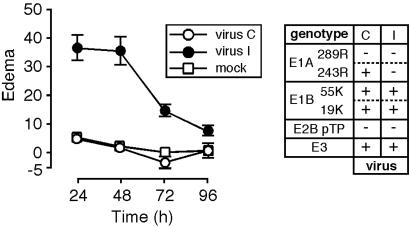
A Virus Deleted for E1A 289R and the pTP Gene Is Not Inflammatory. To test the hypothesis that expression of E1A was required to inhibit Ad-induced inflammation, it was necessary to make the virus replication incompetent, which was accomplished by deleting the pTP gene. Because transcriptional activation by the E1A 289R protein was expected to be inflammatory, the coding region for E1A was replaced with a cDNA encoding only the E1A 243R protein. The resultant virus, virus C (viruses tested in this study are listed in Table 1), was tested by subdermal injection in mouse ears. The results are presented together with those from injection of virus I, which is deleted for E1 and encodes LacZ under the control of the cytomegalovirus major immediate early promoter, to indicate the level of edema induced by an inflammatory vector (Fig. 1). Injection of buffer alone induced modest edema, presumably because of the mild injury from the injection, and virus I induced significant edema, but virus C did not induce edema above the background from injection of buffer alone.
Fig. 1.
Virus deleted for pTP and E1A 289R does not induce edema. Mice were injected in the ears with virus deleted for E1A 289R and pTP (virus C; n = 17 ears) or virus deleted for E1 and pTP and encoding LacZ under the control of the cytomegalovirus major immediate early promoter (virus I; n = 6 ears) or were injected with buffer alone (mock; n = 16 ears). Virus genotypes are indicated to the right of the graph along with single-letter designations of the viruses (see Table 1). Edema is plotted as a function of time after injection.
The inflammatory effects of virus C and virus I were also examined after transduction of mouse liver (Fig. 7, which is published as supporting information on the PNAS web site). Neither virus induced inflammation as monitored by cell infiltration 3-7 days after transduction. The lack of inflammation induced by virus I in the liver reinforces the sensitivity of the mouse ear assay, where edema induced by this virus was readily apparent (Fig. 1).
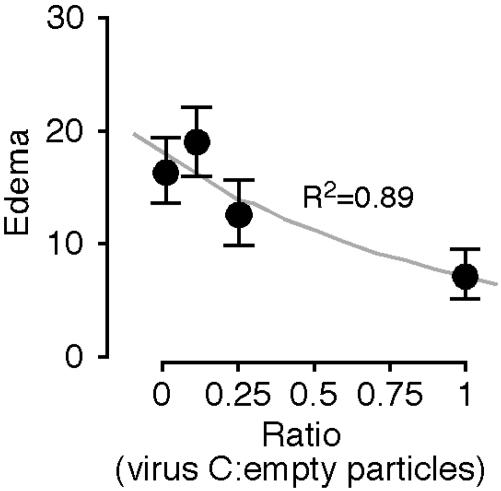
A Virus Deleted for E1A 289R and the pTP Gene Inhibits Edema Induced by the Viral Particle. To test whether the lack of induction of edema by virus C reflected active inhibition rather than the lack of activation of inflammatory processes, varying amounts of virus C were coinjected with a constant amount of empty Ad particles. Injection of empty particles alone induced edema that was inhibited in a dose-dependent manner by virus C (Fig. 2). Analysis of the data with the two-tailed t test demonstrated that P <0.05 for coinjection of equal numbers of virus C and empty particles compared with injection of empty particles alone. The differences for injection of lesser amounts of virus C were not statistically significant.
Fig. 2.
Virus deleted for pTP and E1A 289R inhibits edema induced by empty Ad particles. Mice were injected in the ears with empty Ad particles alone or with increasing doses of virus C (n = 8 ears for injection of empty particles alone and 6 ears for the other points). Edema is plotted as a function of the ratio of virus C particles/empty particles after subtraction of edema resulting from injection of buffer alone. The data are fitted to an exponential curve.
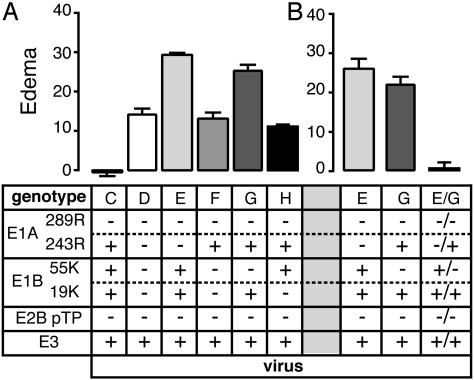
Genetic Interactions Between E1A and E1B Proteins in the Inhibition of Edema. A virus encoding E1A 243R and deleted for the pTP gene, the E1A 289R coding sequence, and the E1B region (virus F) induced moderate edema similar to that induced by a virus deleted for all E1 coding regions (virus D; Fig. 3). This finding suggests that E1A 243R function in the inhibition of inflammation depends on activities of the E1B proteins. However, it is likely that E1A 243R independently both inhibits and induces edema in addition to requiring genetic interactions with E1B proteins for maximal inhibition of inflammation. E1A 243R inhibition of type 1 IFN responses is antiinflammatory, and E1A 243R induction of susceptibility to tumor necrosis factor α (37, 38) is likely to be proinflammatory.
Fig. 3.
Edema as a function of E1A and E1B genes. (A) Mice were injected in the ears with viruses deleted for the pTP gene and mutated in the E1A and/or E1B genes. (B) Mice were injected with 2 × 1010 particles of the strongly inflammatory viruses E (deleted for the E1A gene) or G (deleted for E1B 55-kDa) or were coinjected with 1010 particles of each virus (keeping the total number of particles constant with respect to injection of the individual viruses). Virus genotypes are indicated below the graphs along with single-letter designations of the viruses (see Table 1). For coinjection, the genotypes of both viruses are listed. The numbers of ears analyzed for each virus in A are: C, 17 (data are from Fig. 1); D and E, 6; F, 12; G, 26; and H, 18. The numbers of ears analyzed for each virus in B are: E and G, 6; and coinjection of G+E, 8. Edema is presented as a function of viral genotype after subtraction of edema resulting from injection of buffer alone.
E1B protein function in the inhibition of edema strongly depends on expression of the E1A 243R protein. A virus that had the WT E1B gene but the E1A gene deleted induced high levels of edema (virus D; Fig. 3), suggesting that E1B is proinflammatory in the absence of E1A 243R.
To dissect the roles of E1B proteins on induction of edema, pTP deletion mutant viruses encoding E1A 243R and either the E1B 19-kDa antiapoptotic protein (virus G) or the 55-kDa p53 regulatory protein (virus H) were tested. Virus H induced edema similar to that induced by virus F (which encodes no E1B proteins). In contrast, virus G induced high-level edema similar to that induced by virus E (deleted for E1A and encoding both E1B proteins; Fig. 3). These data lead to the surprising conclusion that expression of the E1B 19-kDa antiapoptotic protein is antiinflammatory when both the E1A 243R and E1B 55-kDa proteins are expressed, but is strongly inflammatory when only one of the E1A 243R and E1B 55-kDa proteins is expressed.
The conclusion that E1B proteins inhibit and activate inflammation as a function of the genetic status of the E1A gene depends on the expression of E1B proteins in the absence of transcriptional activation by E1A 289R. To test whether the E1B promoter is active after transduction at high multiplicity in the absence of the E1A 289R protein, E1B mRNA and protein expression were examined after transduction of human HeLa and KB cells and primary mouse embryo fibroblasts (MEFs) (39). Efficient E1B 55-kDa protein expression occurred after transduction of KB cells with viruses encoding E1B 55-kDa in the absence of E1A 289R expression (viruses C, E, and H, Fig. 7). E1B mRNA expression in HeLa cells and MEF was similar when the E1A gene encoded 289R + 243R (virus A) or only 243R (virus G), whereas E1B mRNA expression was reduced when the E1A gene was deleted (virus E; Fig. 8, which is published as supporting information on the PNAS web site). Virus E was derived from Ad5dl312, which has a deletion extending into the E1A promoter. The reduced E1B mRNA accumulation in cells transduced by virus E is likely explained by the reduced E1B promoter activity in the absence of transcription directed by the E1A promoter during the early phase of infection (40).
As a genetic test of the expression of the E1B proteins in the absence of E1A 289R expression, ears were singly injected and coinjected with the highly inflammatory viruses E (deleted for the E1A coding region) and G (deleted for the E1B 55-kDa protein gene). If both the E1A and E1B proteins are expressed and contribute to the induction and inhibition of edema, the viruses should complement each other, resulting in a substantial reduction in the level of edema. Injection of the viruses individually again led to high-level edema, whereas coinjection led to edema that was only slightly above background (Fig. 3B), offering genetic support for activity of the E1B promoter in the absence of E1A 289R expression. The background edema from injection of buffer in this experiment was ≈3 units higher than the average background edema from all experiments, so the levels of edema are likely slightly understated. The absolute difference between coinjection and single injection is not affected by the modest increase in background.
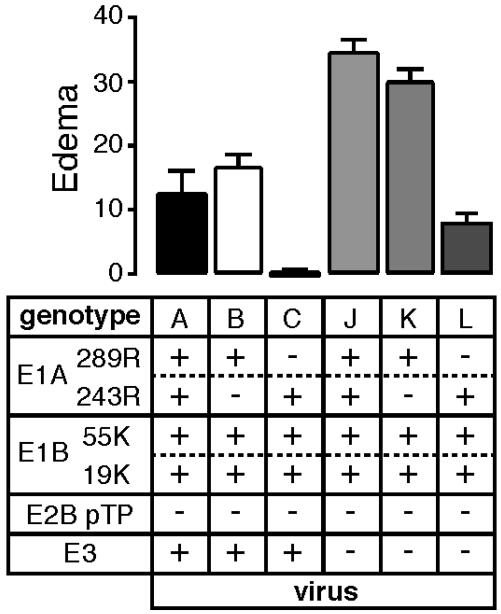
The E1A 289R Protein Induces Edema That Is Inhibited by E3. To test the roles of the E1A proteins in the induction and inhibition of edema, viruses encoding both E1A proteins (virus A) or only the 289R protein (virus B) were compared with the virus encoding only the 243R protein (virus C). Edema was induced to relatively high levels by both the virus encoding only the 289R protein and the virus encoding both E1A proteins (Fig. 4).
Fig. 4.
Edema as a function of E1A and E3 gene products. Mice were injected in the ears with viruses deleted for the pTP gene and mutated in the E1A and/or E3 genes. Virus genotypes are indicated below the graph along with single-letter designations of the viruses (see Table 1). The numbers of ears analyzed for each virus were: A, 11; B and J, 6; C, 17 (data are from Fig. 1); K, 8; and L, 12. Edema is presented as a function of viral genotype after subtraction of edema resulting from injection of buffer alone.
The role of E3 in the inhibition of inflammation was tested by assaying edema induced by viruses encoding both E1A proteins (virus J), the E1A 289R protein alone (virus K), or the E1A 243R protein alone (virus L) with the E3 region deleted (Fig. 4). A moderate level of edema was induced in the absence of E3 protein expression when only E1A 243R was expressed. In contrast, significantly increased edema was induced when E3 was deleted and both E1A proteins or only E1A 289R were expressed.
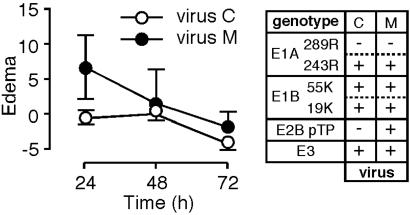
The Role of the pTP Gene in the Induction of Edema. All of the viruses tested to this point were deleted for the pTP gene. To assay the role of pTP in the induction of edema, a virus encoding pTP in which the E1A coding region was replaced with a cDNA encoding the E1A 243R protein was tested. This virus should replicate its DNA, albeit at greatly reduced efficiency relative to the WT virus, in human cells (41) and, because Ad5 DNA replication occurs efficiently in mouse cells in vivo (42) and in vitro (43), after injection into mouse ears. The presence of the pTP gene led to modest edema (Fig. 5), raising the possibility that expression of pTP is directly proinflammatory. Alternatively, the effect of pTP may be indirect, occurring through low-level DNA replication and subsequent increased expression of Ad early region genes.
Fig. 5.
Edema as a function of the pTP gene. Mice were injected in the ears with viruses isogenic except for the presence or absence of the pTP gene. Virus genotypes are indicated to the right of the graph along with single-letter designations of the viruses (see Table 1). Eighteen ears were analyzed for M, and 17 were analyzed for C (data are from Fig. 1). Edema is plotted as a function of time after injection after subtraction of background caused by injection of buffer alone.
Discussion
The majority of research into Ad gene therapy vectors has been in their application, with more modest effort in developing less inflammatory vectors and still less effort in understanding the mechanisms by which Ad vectors induce and inhibit inflammation. Despite the absence of understanding of the mechanism of induction of inflammation, Ad gene therapy vectors have been used in a number of human trials with poor outcomes that have dampened enthusiasm and support for continued use of Ad for gene therapy. However, understanding the mechanisms by which Ad induces and inhibits inflammation offers promise for the development of useful, noninflammatory vectors that are relatively easy to grow and purify.
Phases of Inflammation Induced by Ad. The induction of inflammation by Ad5 occurs in at least two distinguishable steps (44). Binding and uptake of Ad particles induces signal transduction, IFN responses (12), and expression of inflammatory chemokines (13, 14) that initiate the first phase of inflammation (Fig. 2). E1A acts to counter this initial induction of inflammation through transcriptional repression of inflammatory genes (12, 16, 17). Capsid binding and uptake induce the initial inflammatory events (12-14), so all Ad gene therapy vectors induce this phase of inflammation. Because helper-dependent vectors effectively transduce tissues in vivo for prolonged periods (45-47), the first phase does not necessarily limit the effectiveness of Ad vectors. However, it is likely that, given the close juxtaposition of the first and second phases, the first phase contributes to the induction of a strong adaptive immune response by replication-defective and replication-incompetent Ad vectors.
The second phase of inflammation (44), kinetically distinguishable from the first by the expression of Ad genes, is inhibited through the coordinated and balanced expression of E1A, E1B, and E3 proteins, and likely E4 proteins. This phase is critical in the limitation of the effectiveness of first-generation Ad vectors.
Genetic Complexity in the Inhibition of Inflammation by Ad. Comparison of the effects of the genes tested in this study on inflammation demonstrates the genetic complexity involved (Table 2). E1A 243R and E1B 55-kDa had from virtually no effect to a dramatic inhibition of edema. The effect of E1B 19-kDa expression ranged from a substantial increase to a substantial decrease. The comparative analysis also demonstrates the antiinflammatory role of E1A 289R, because addition of its coding sequence to the E1A deletion mutant virus E led to a substantial reduction in edema (compare with virus B, Fig. 4). E3 protein expression led to reduced edema for all viruses tested, although the magnitude of the effect depended on the genetic status of the E1A gene. E3 proteins clearly play an important role in the inhibition of edema by virus C, but their role, as assayed by comparison of viruses C and L, is much less than when E1A 289R is expressed. The reduced role of E3 in inhibiting inflammation induced by virus C relative to its role in productive infection underscores the importance of analyzing the mechanism(s) by which Ad vectors induce and inhibit inflammation.
Table 2. Range of Ad early gene effects on inhibition of edema.
| Inhibition of edema
|
||||
|---|---|---|---|---|
| Gene | Effect | Viruses compared | Effect | Viruses compared |
| E1A 243R | ++++ | C and E | 0 | D and F |
| E1A 289R | +++ | B and E | −−− | A and C |
| E1B 55-kDa | ++++ | C and G | 0 | F and H |
| E1B 19-kDa | +++ | C and H | −−− | F and G |
| E3 | ++++ | A and J | ++ | C and L |
| pTP | −− | C and M | ||
The most divergent effects on the inhibition of edema of the Ad proteins as determined by comparing the levels of edema of viruses isogenic except for the gene/coding sequence (Figs. 3, 4, 5) are summarized. 0, not statistically different; ++, inhibition of 5-10 units; +++, inhibition of 10-20 units; ++++, inhibition of 20-30 units, −−, induction of 5-10 units; and −−−, induction of 10-20 units.
Mechanistic Implications. The E1A 243R and E1B 55-kDa proteins share a variety of properties. Both act as transcriptional repressors and cell cycle regulators and have cell-transforming activities, functions that are at least partially overlapping. Determination of which, if any, of these functions is involved in inhibition of edema will require analysis of viruses with mutations in the E1A 243R and E1B 55-kDa coding sequences that specifically affect either transcription or cell cycle regulatory functions. However, because the same proteins regulate both cell cycle progression and inflammation, it is possible that cell cycle regulation plays an important role in inhibition of inflammation by Ad.
The inflammatory and antiinflammatory behavior of E1B 19-kDa (Table 2 and Fig. 3) could reflect an as-yet-unidentified function, but it is reasonable to assume that the well-defined antiapoptotic activity of the protein (19) is responsible. This assumption suggests that, in the absence of either E1A 243R or E1B 55-kDa protein expression, Ad induces apoptotic signals in certain cells and that inhibition of apoptosis by E1B 19-kDa in these cells is inflammatory. In contrast, when both E1A 243R and E1B 55-kDa are expressed, apoptosis is inflammatory and E1B 19-kDa expression is antiinflammatory.
Expression in vitro from the E1B gene was reduced when the E1A promoter was partially deleted (virus E, Fig. 8). Because virus E is strongly inflammatory, it is possible that only very low levels of E1B proteins are required to induce inflammation in the absence of E1A proteins. Alternatively, it may be that the E1B promoter is activated by inflammatory stimuli, in a manner analogous to activation of the E3 promoter by tumor necrosis factor-α (48), in vivo.
Conservation of the Innate Immune Response Functions Targeted by Ad. Our data all have been accumulated in mice and not humans, the natural host of Ad5. It is clear that murine and human adaptive immune responses differ in important aspects relative to Ad. For example, whereas the E3 19-kDa glycoprotein binds all human HLA types with high affinity, it has variable affinity for different murine MHC molecules, with moderate affinity for H-2d, the class I MHC expressed in BALB/c mice (8). However, the ability of appropriately modified Ad5 to inhibit the early steps in the inflammatory response in mice suggests that the innate functions that the virus targets are highly conserved between mice and humans. Such conservation will make Ad5 an excellent tool for the study of innate immune functions that lead to inflammation and adaptive immune responses.
Antiinflammatory Functions in the Development of Ad Gene Therapy Vectors. The findings presented here hold promise for the development of optimal replication-incompetent and helper-dependent Ad gene therapy vectors. Helper virus contamination likely contributes to the inflammation that occurs in the use of helper-dependent vectors. The use of virus C, or a similarly modified virus, as helper could inhibit the inflammation induced by both the contaminating helper virus and the helper-dependent vector. Although inclusion of transforming genes in a virus vector raises concerns, it should be noted Ad5 is not only not tumorigenic, but that Ad5 E1A, and especially E1A 243R, inhibits tumor growth through a variety of mechanisms (49-53). Thus, it is likely that inclusion of the E1 region will be protective against tumorigenesis.
Supplementary Material
Acknowledgments
We dedicate this work to H. Ginsberg, whose efforts have influenced all aspects of Ad research. We thank E. Hamlin and B. Turner for assistance in image capturing; K. Tardif and A. Siddiqui for mouse embryo fibroblasts; L. Pizer for scientific discussions; and A. Ruiz, M.-C. Kuo, and H. Oginsky for technical support. This work was supported by National Institutes of Health Grants PO1HD38129 and RO1CA77342, the Schweppe Foundation, and the Research Institute at The Children's Hospital.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Ad, adenovirus; TP, terminal protein; pTP, preterminal protein.
References
- 1.Smith, T. A., Mehaffey, M. G., Kayda, D. B., Saunders, J. M., Yei, S., Trapnell, B. C., McClelland, A. & Kaleko, M. (1993) Nat. Genet. 5, 397-402. [DOI] [PubMed] [Google Scholar]
- 2.Mitani, K., Graham, F. L., Caskey, C. T. & Kochanek, S. (1995) Proc. Natl. Acad. Sci. USA 92, 3854-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fisher, K. J., Choi, H., Burda, J., Chen, S. J. & Wilson, J. M. (1996) Virology 217, 11-22. [DOI] [PubMed] [Google Scholar]
- 4.Straus, S. E. (1984) in The Adenoviruses, ed. Ginsberg, H. S. (Plenum, New York), pp. 451-496.
- 5.Ilan, Y., Droguett, G., Chowdhury, N. R., Li, Y., Sengupta, K., Thummala, N. R., Davidson, A., Chowdhury, J. R. & Horwitz, M. S. (1997) Proc. Natl. Acad. Sci. USA 94, 2587-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrod, K. S., Hermiston, T. W., Trapnell, B. C., Wold, W. S. & Whitsett, J. A. (1998) Hum. Gene Ther. 9, 1885-1898. [DOI] [PubMed] [Google Scholar]
- 7.Wen, S., Driscoll, R. M., Schneider, D. B. & Dichek, D. A. (2001) Arterioscler. Thromb. Vasc. Biol. 21, 1777-1782. [DOI] [PubMed] [Google Scholar]
- 8.Schowalter, D. B., Tubb, J. C., Liu, M., Wilson, C. B. & Kay, M. A. (1997) Gene Ther. 4, 351-360. [DOI] [PubMed] [Google Scholar]
- 9.Gantzer, M., Spitz, E., Accard, N. & Rooke, R. (2002) Hum. Gene Ther. 13, 921-933. [DOI] [PubMed] [Google Scholar]
- 10.Moran, E. & Mathews, M. B. (1987) Cell 48, 177-178. [DOI] [PubMed] [Google Scholar]
- 11.Flint, J. & Shenk, T. (1997) Annu. Rev. Genet. 31, 177-212. [DOI] [PubMed] [Google Scholar]
- 12.Reich, N., Pine, R., Levy, D. & Darnell, J. E., Jr. (1988) J. Virol. 62, 114-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higginbotham, J. N., Seth, P., Blaese, R. M. & Ramsey, W. J. (2002) Hum. Gene Ther. 13, 129-141. [DOI] [PubMed] [Google Scholar]
- 14.Liu, Q. & Muruve, D. A. (2003) Gene Ther. 10, 935-940. [DOI] [PubMed] [Google Scholar]
- 15.Higashimoto, Y., Elliott, W. M., Behzad, A., Sedgwick, E., Takei, T., Hogg, J. C. & Hayashi, S. (2002) Am. J. Respir. Crit. Care Med. 166, 200-207. [DOI] [PubMed] [Google Scholar]
- 16.Bhattacharya, S., Eckner, R., Grossman, S., Oldread, E., Arany, Z., D'Andrea, A. & Livingston, D. M. (1996) Nature 383, 344-347. [DOI] [PubMed] [Google Scholar]
- 17.Zhao, H., Granberg, F., Elfineh, L., Pettersson, U. & Svensson, C. (2003) J. Virol. 77, 11006-11015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moran, E. (1993) FASEB J. 7, 880-885. [DOI] [PubMed] [Google Scholar]
- 19.Cuconati, A. & White, E. (2002) Genes Dev. 16, 2465-2478. [DOI] [PubMed] [Google Scholar]
- 20.Ginsberg, H. S., Moldawer, L. L. & Prince, G. A. (1999) Proc. Natl. Acad. Sci. USA 96, 10409-11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson, C. W., Schmitt, R. C., Smart, J. E. & Lewis, J. B. (1984) J. Virol. 50, 387-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montell, C., Fisher, E. F., Caruthers, M. H. & Berk, A. J. (1984) Mol. Cell. Biol. 4, 966-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graham, F. L., Smiley, J., Russell, W. C. & Nairn, R. (1977) J. Gen. Virol. 36, 59-72. [DOI] [PubMed] [Google Scholar]
- 24.Moorhead, J. W., Clayton, G. H., Smith, R. L. & Schaack, J. (1999) J. Virol. 73, 1046-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaack, J., Guo, X. & Langer, S. (1996) Proc. Natl. Acad. Sci. USA 93, 14686-14691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fagan, K. A., Rich, T. C., Tolman, S., Schaack, J., Karpen, J. W. & Cooper, D. M. F. (1999) J. Biol. Chem. 274, 12445-12453. [DOI] [PubMed] [Google Scholar]
- 27.Chinnadurai, G., Chinnadurai, S. & Green, M. (1978) J. Virol. 26, 195-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chartier, C., Degryse, E., Gantzer, M., Dieterle, A., Pavirani, A. & Mehtali, M. (1996) J. Virol. 70, 4805-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He, T. C., Zhou, S., da Costa, L. T., Yu, J., Kinzler, K. W. & Vogelstein, B. (1998) Proc. Natl. Acad. Sci. USA 95, 2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones, N. & Shenk, T. (1978) Cell 13, 181-188. [DOI] [PubMed] [Google Scholar]
- 31.Winberg, G. & Shenk, T. (1984) EMBO J. 3, 1907-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones, N. & Shenk, T. (1979) Cell 17, 683-689. [DOI] [PubMed] [Google Scholar]
- 33.Pilder, S., Logan, J. & Shenk, T. (1984) J. Virol. 52, 664-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pilder, S., Moore, M., Logan, J. & Shenk, T. (1986) Mol. Cell. Biol. 6, 470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaack, J., Allen, B., Orlicky, D., Bennett, M., Maxwell, I. & Smith, R. L. (2001) Virology 291, 101-109. [DOI] [PubMed] [Google Scholar]
- 36.Orlicky, D. J. & Schaack, J. (2001) J. Lipid Res. 42, 460-466. [PubMed] [Google Scholar]
- 37.Chen, M. J., Holskin, B., Strickler, J., Gorniak, J., Clark, M. A., Johnson, P. J., Mitcho, M. & Shalloway, D. (1987) Nature 330, 581-583. [DOI] [PubMed] [Google Scholar]
- 38.Duerksen-Hughes, P., Wold, W. S. & Gooding, L. R. (1989) J. Immunol. 143, 4193-4200. [PubMed] [Google Scholar]
- 39.Scheuner, D., Song, B., McEwen, E., Liu, C., Laybutt, R., Gillespie, P., Saunders, T., Bonner-Weir, S. & Kaufman, R. J. (2001) Mol. Cell 7, 1165-1176. [DOI] [PubMed] [Google Scholar]
- 40.Parks, C. L. & Spector, D. J. (1990) J. Virol. 64, 2780-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shenk, T., Jones, N., Colby, W. & Fowlkes, D. (1979) Cold Spring Harbor Symp. Quant. Biol. 44, 367-375. [DOI] [PubMed] [Google Scholar]
- 42.Duncan, S. J., Gordon, F. C., Gregory, D. W., McPhie, J. L., Postlethwaite, R., White, R. & Willcox, H. N. (1978) J. Gen. Virol. 40, 45-61. [DOI] [PubMed] [Google Scholar]
- 43.Younghusband, H. B., Tyndall, C. & Bellett, A. J. (1979) J. Gen. Virol. 45, 455-467. [DOI] [PubMed] [Google Scholar]
- 44.Liu, Q., Zaiss, A. K., Colarusso, P., Patel, K., Haljan, G., Wickham, T. J. & Muruve, D. A. (2003) Hum. Gene Ther. 14, 627-643. [DOI] [PubMed] [Google Scholar]
- 45.Morsy, M. A., Gu, M., Motzel, S., Zhao, J., Lin, J., Su, Q., Allen, H., Franlin, L., Parks, R. J., Graham, F. L., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 7866-7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morral, N., O'Neal, W., Rice, K., Leland, M., Kaplan, J., Piedra, P. A., Zhou, H., Parks, R. J., Velji, R., Aguilar-Cordova, E., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 12816-12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas, C. E., Schiedner, G., Kochanek, S., Castro, M. G. & Lowenstein, P. R. (2000) Proc. Natl. Acad. Sci. USA 97, 7482-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deryckere, F. & Burgert, H. G. (1996) J. Biol. Chem. 271, 30249-30255. [DOI] [PubMed] [Google Scholar]
- 49.Frisch, S. M. & Mymryk, J. S. (2002) Nat. Rev. Mol. Cell. Biol. 3, 441-452. [DOI] [PubMed] [Google Scholar]
- 50.Sawada, Y., Fohring, B., Shenk, T. E. & Raska, K. J. (1985) Virology 147, 413-421. [DOI] [PubMed] [Google Scholar]
- 51.Cook, J. L., Routes, B. A., Walker, T. A., Colvin, K. L. & Routes, J. M. (1999) Exp. Cell Res. 251, 414-423. [DOI] [PubMed] [Google Scholar]
- 52.Shao, R., Xia, W. & Hung, M. C. (2000) Cancer Res. 60, 3123-3126. [PubMed] [Google Scholar]
- 53.Perez, D. & White, E. (2003) J. Virol. 77, 2651-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.