Abstract
Active DNA demethylation regulates many vital biological processes, including early development and locus-specific gene expression in plants and animals. In Arabidopsis, bifunctional DNA glycosylases directly excise the 5-methylcytosine base and then cleave the DNA backbone at the abasic site. Recent evidence suggests that mammals utilize DNA glycosylases after 5-methylcytosine is oxidized and/or deaminated. In both cases, the resultant single-nucleotide gap is subsequently filled with an unmodified cytosine through the DNA base excision repair pathway. The enzymatic removal of 5-methylcytosine is tightly integrated with histone modifications and possibly noncoding RNAs. Future research will increase our understanding of the mechanisms and critical roles of active DNA demethylation in various cellular processes as well as inspire novel genetic and chemical therapies for epigenetic disorders.
In combination with histones and nonhistone architectural proteins, eukaryotic genomic DNA is hierarchically packed into complex chromatin structures. Chromatin can be adjusted from a compact status to an easy-to-access condition, and vice versa, through individual or combined epigenetic alterations in DNA methylation patterns and histone modifications as well as by actions of chromatin-remodeling factors and noncoding RNAs.
DNA methylation at the fifth position of cytosine is an important epigenetic modification. Via its effects on chromatin structures, DNA methylation regulates many critical biological processes such as gene imprinting, silencing of transposable elements (TEs), and X-chromosome inactivation. In plants, methylation occurs in all DNA sequence contexts including CG, CHG, and CHH (H represents either A, T, or G), with transposon-rich heterochromatic regions being the major targets (Zhang et al. 2006; Henderson and Jacobsen 2007). In mammals, cytosine in CG context throughout the genome is the predominant methylation target, with the exception of those densely clustered into CG islands near gene promoters (Ehrlich et al. 1982; Suzuki and Bird 2008; Cedar and Bergman 2009). Mammalian DNA can also be methylated in non-CG contexts, as has been observed in embryonic stem cells (Ramsahoye et al. 2000; Lister et al. 2009). On a whole-genome scale, both Arabidopsis and human cytosine methylation shows a preference for nucleosome-bound DNA over flanking sequences (Chodavarapu et al. 2010), indicating an interplay between nucleosomes and DNA methyltransferases or demethylases in determining genomic DNA methylation patterns. Cytosine methylation can be faithfully inherited during DNA replication and is thus often considered as a stable epigenetic mark. Nevertheless, DNA methylation is subject to dynamic regulation of establishment, maintenance, and removal in response to developmental and environmental cues.
Establishment and maintenance of cytosine methylation are mediated by DNA methyltransferases (DNMTs) that use S-adenosyl-l-methionine (SAM) as the methyl donor. Mammalian DNMT3 family members and their plant homolog DOMAIN REARRANGED METHYLTRANSFERASE 2 (DRM2) catalyze de novo DNA methylation (Cao and Jacobsen 2002; Goll and Bestor 2005). The targeting of DRM2 for de novo methylation can be mediated by the RNA-directed DNA methylation (RdDM) pathway, which requires 24-nucleotide small interfering RNAs (siRNAs) (Law and Jacobsen 2010; Haag and Pikaard 2011; Zhang and Zhu 2011). Similarly, mammalian piRNAs, which are small RNAs (25 to 30 nucleotides) that mediate posttranscriptional silencing of transposons, are thought to pair with nascent transcripts and directly or indirectly recruit de novo methyltrasferases (Law and Jacobsen 2010; Watanabe et al. 2011). In addition, evidence has been published in support of a model in which DNMT3A is activated or recruited to target loci via protein interaction with DNMT3L, a noncatalytic DNMT3 paralog that binds unmethylated histone 3 lysine 4 (H3K4) tails (Ooi et al. 2007; Cheng and Blumenthal 2008). Once established, mammalian DNA methylation is maintained by DNMT1, which restores the hemimethylated DNA to a fully methylated status during DNA replication (Goll and Bestor 2005). In plants, DNA methylation in CG and CHG contexts is maintained by MET1 (DNA METHYLTRANSFERASE 1) and CMT3 (CHROMOMETHYLASE 3), respectively (Law and Jacobsen 2010). Cytosine methylation in the asymmetric CHH context cannot be maintained and thus must occur de novo during every cell cycle.
Preexisting DNA methylation can be lost as a consequence of passive or active demethylation processes. The former refers to the failure in maintaining DNA methylation owing to DNA methyltransferase dysfunction or shortage of methyl group supplies, whereas the latter is an outcome of enzymatic action resulting in the replacement of 5-methylcytosine (5-mC) with cytosine. This review focuses on active DNA demethylation in plants and animals. We start by reviewing active DNA demethylation processes and their functional significance. We then consider the molecular players involved in removing methylated cytosines as well as the established and emerging models of enzymatic demethylation. Finally, we highlight the interplay between DNA demethylases and other epigenetic regulators in carrying out a fully functional demethylation pathway.
PRUNING DNA METHYLATION PATTERNS
An extensive literature has documented strong correlations between various diseases and aberrant DNA methylation patterns. Active demethylation is critical for the pruning of DNA methylation patterns that is required for normal developmental reprogramming and proper transcriptional activities. In both plants and animals, enzyme-mediated removal of 5-mC has been observed in specific early developmental stages and in somatic cells as well.
Genome-Wide Demethylation during Development
In some mammals including humans, two waves of global DNA demethylation have been documented at specific times during early development. The first genome-wide demethylation occurs in paternal pronuclei in the zygote (Mayer et al. 2000; Oswald et al. 2000). Detection of 5-mC by immunostaining suggested a global loss of genomic DNA methylation that was observed before the completion of the first round of DNA replication. In consistence, such loss of DNA methylation was not abolished by treatment of zygotes with the replication inhibitor aphidiolin (Mayer et al. 2000). Closer examination revealed that such paternal demethylation bypassed certain genomic loci, including centromeric and pericentromeric heterochromatin, some transposons, and imprint regions (Olek and Walter 1997; Rougier et al. 1998; Santos et al. 2002; Lane et al. 2003). In addition, the maternal genome maintains its methylation patterns despite the ongoing paternal demethylation in the same zygote. Although how such a wave of active demethylation locates its target regions remains to be explored, some proteins that protect DNA methylation from demethylation at specific imprinted loci have been identified (Nakamura et al. 2007; Reese et al. 2007; Li et al. 2008). Several recent studies (Gu et al. 2011; Inoue and Zhang 2011; Iqbal et al. 2011) have suggested that passive demethylation probably accounts, at least in part, for this global demethylation, although this replication-dependent demethylation process is indeed initiated by enzymatic catalysis (discussed later). Nevertheless, active DNA demethylation does occur in the paternal pronucleus before the zygote enters into the two-cell stage, as indicated by the rapid reduction in methylation levels at the two TEs, Line1 and Etn (Okada et al. 2010).
Zygotic paternal demethylation is followed by a replication-dependent passive DNA demethylation in the maternal genome before the formation of the blastocyst, at which stage the de novo DNA methyltransferases reestablish genomic DNA methylation in both parental origins (Wu and Zhang 2010). Most of these newly created DNA methylation patterns are erased during the second wave of global demethylation, which occurs in primordial germ cells (PGCs). Genome-wide loss of 5-mC in PGCs is considered as active demethylation, because this process undergoes several cell cycles in the presence of the maintenance methyltransferase DNMT1 (Morgan et al. 2005). In support of the view that this active process is replication independent, chronological investigation revealed that the profound epigenetic changes occur while the PGCs are in the G2 phase of the cell cycle (Hajkova et al. 2008). Unlike zygotic paternal DNA demethylation, imprinted methylation marks are targets for demethylation in the PGC genome, although some repetitive elements still showed incomplete demethylation (Hajkova et al. 2002).
Following the onset of DNA demethylation in PGCs, extensive chromatin rearrangement occurs (Hajkova et al. 2008), consistent with the notion that this global erasure of DNA methylation allows for epigenetic reprogramming toward totipotency. Similarly, epigenetic reprogramming involving active DNA demethylation is required for the differentiated somatic cell nucleus to become totipotent during nuclear transfer or cloning (Dean et al. 2001; Zhu 2009). Indeed, researchers have postulated that incomplete and aberrant epigenetic reprogramming is the cause of the high frequency of failed cloning and of developmental abnormalities in cloned animals (Morgan et al. 2005; Yang et al. 2007; Zhu 2009). In contrast to its significance in PGCs, the significance of paternal DNA demethylation in zygotes is less clear. Given that TEs and other repeat sequences account for half of the genome in mammals, the question has been raised as to whether global demethylation generally releases silencing of such types of sequences, even though some of them escape paternal demethylation (Wu and Zhang 2010). In fact, such a scenario has been observed in plants.
In Arabidopsis, evidence exists for genome-wide DNA demethylation concurrent with transposon reactivation during gametogenesis. Like most seed plants, Arabidopsis uses a double-fertilization strategy that depends on the multicellular nature of male and female gametophytes (Fig. 1). Mature pollen grains are tricellular, containing two sperm cells and one vegetative nucleus, whereas the female gametophyte contains one egg cell, one diploid central cell, two synergid cells, and three antipodal cells. The two sperm cells fertilize the egg cell and the central cell, thereby producing the embryo and the endosperm, respectively. Genome-wide hypomethylation was observed in the Arabidopsis endosperm (Gehring et al. 2009; Hsieh et al. 2009). Mutation of DEMETER (DME), one of the bifunctional DNA glycosylases known for active DNA demethylation in Arabidopsis, partially prevents the global loss of DNA methylation (Hsieh et al. 2009). Gene expression of DME is detectable in the female gametophyte but quickly disappears after fertilization of the central cell with a sperm cell (Choi et al. 2002), suggesting that the endosperm hypomethylation may result from demethylation in the central cell nucleus. At the same time, MET1 is transcriptionally repressed during female gametogenesis (Jullien et al. 2008). Therefore, endosperm hypomethylation is likely to be a combined outcome of active and passive DNA demethylation.
Figure 1.
Global DNA demethylation during Arabidopsis early development. (A) Multicellular female gametophyte consisting of three antipodal cells (AC), two synergid cells (Syn), one haploid egg cell (EC), and one diploid central cell nucleus (CCN). DME is expressed in the central cell before fertilization and thereby may account for the global demethylation in the maternal endosperm genome (Gehring et al. 2009; Hsieh et al. 2009). (B) Tricellular male gametophyte consisting of two sperm cells (SC) and one vegetative cell nucleus (VCN). Desilencing of transposable elements (TEs) and production of 21-nucleotide siRNA occur only in the VCN (Slotkin et al. 2009). These 21-nt siRNA may travel to SC and reinforce TE silencing. (C) The endosperm and embryo are produced after double fertilization. The endosperm exhibits genome-wide hypomethylation compared with the embryo, with TEs being extensively demethylated. Compared with aerial tissues, both endosperm and embryo show local CHH hypermethylation in TE loci targeted by siRNA, which are likely produced by transcriptional activation of endosperm TEs (Gehring et al. 2009; Hsieh et al. 2009). The 21-nucleotide siRNA originating from the male gametophyte may contribute to posttranscriptional silencing of TE primary transcripts in the endosperm.
In addition to the global hypomethylation patterns in the endosperm, increased CHH methylation relative to adult tissues at siRNA target loci in both the endosperm and the embryo has also been reported (Gehring et al. 2009). Such local hypermethylation was hypothesized to result from augmented RdDM, an enhancement owing to elevated siRNA production that is attributed to transcriptional activation of TEs and other siRNA target loci by the genome-wide demethylation in the endosperm. Although the cell-non-autonomous effects of small RNAs have been reported in other parts of the plant, including male gametocytes (Slotkin et al. 2009; Molnar et al. 2010), transport of siRNAs from the central cell (or endosperm) to the egg (or embryo) has yet to be demonstrated. Nevertheless, this enhanced-silencing hypothesis seems appealing. Arabidopsis endosperm is a transient nutrient reservoir that is lost by the time the seed germinates, whereas the embryo eventually develops into a mature plant. Therefore, genome-wide DNA demethylation in the endosperm and the resultant siRNA burst would not threaten transgenerational genome integrity; instead, it could serve as a “phishing” attempt with the purpose of identifying and silencing potentially mobile DNA elements in the embryo (Fig. 1C).
Demethylation-induced activation of TEs was also observed in the vegetative nucleus of Arabidopsis pollen (Slotkin et al. 2009). Consistent with DNA hypomethylation, the vegetative cell exhibits significant down-regulation of DDM1 (DECREASED DNA METHYLATION 1), a chromatin-remodeling factor necessary for maintaining DNA methylation (Slotkin et al. 2009). Dysfunction of DDM1 reduces global 5-mC levels by more than 70% in Arabidopsis (Vongs et al. 1993). Additionally, DME is expressed in the vegetative cell but not in the sperm cells (Schoft et al. 2011). Therefore, both passive and active demethylation seems to contribute to DNA hypomethylation in the vegetative nucleus. Whereas activation of TEs seems to be restricted to the vegetative cell, 21-nucleotide small RNAs from the activated TEs accumulate in sperm cells (Slotkin et al. 2009). In support of the hypothesis that these 21-nucleotide small RNAs traveled from the vegetative cell to sperm cells, an artificial miRNA from the vegetative cell successfully silenced a reporter gene specifically expressed in sperm cells (Slotkin et al. 2009). In the sperm cells, DDM1 is expressed and TEs are silenced as in adult tissues (Martienssen et al. 2008; Slotkin et al. 2009). Therefore, these mobile 21-nucleotide small RNAs may reinforce TE silencing in sperm cells by posttranscriptional cleavage of the primary TE transcript. In addition, these small RNAs may be carried over during fertilization and thereby contribute to TE silencing in the zygote, for instance, by cleaving retrotransposon transcripts to prevent reverse transcription in the endosperm (Fig. 1C).
Locus-Specific Active DNA Demethylation
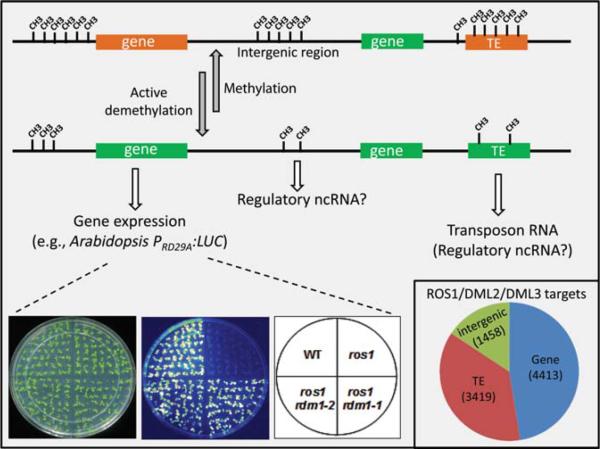
In addition to genome-wide regulation, active DNA demethylation also functions extensively in epigenetic regulation at discrete genetic loci. The Arabidopsis REPRESSOR OF SILENCING 1 (ROS1) is a well-established DNA demethylase that prunes DNA methylation for dynamic transcriptional regulation. Unlike DME, which appears to have little preference for specific DNA sequences during global demethylation (Huh et al. 2008; Gehring et al. 2009; Hsieh et al. 2009), ROS1 seems to preferentially counteract rigorous DNA methylation established by the RdDM pathway (Gong et al. 2002; He et al. 2009; Zhu 2009; Gao et al. 2010). The RdDM pathway effectively silences TEs, repetitive sequences, and foreign DNA (Matzke et al. 2007). However, this de novo methylation process may overexert its function and cause unwanted methylation for several reasons. First, RdDM establishes methylation in a target DNA sequence that matches the 24-nucleotide siRNA(s), but the methylation may spread to neighboring sequences. If an RdDM target locus is near a transcriptional unit that needs to be expressed, prevention of spreading of DNA methylation may become important. Second, RdDM blocks transposition of TEs and thereby protects genome integrity under normal conditions. However, TE transposition when an organism is exposed to harsh environmental stress may be evolutionarily beneficial (Ito et al. 2011a). In addition, transcripts from TE or other DNA repeats may carry regulatory functions and are needed under certain circumstances, as exemplified by the mammalian SINE (short interspersed nuclear element) transcripts that control mRNA production at multiple levels (Ponicsan et al. 2010). Third, siRNAs guide RdDM to their homologous loci. Unwanted silencing may thus occur if an otherwise transcriptionally active locus shows homology to newly generated siRNAs, such as those from foreign DNA. Therefore, active DNA demethylation is required for the proper control of epigenetic regulation (Fig. 2).
Figure 2.
Active DNA demethylation at specific loci. Active DNA demethylation may generate a chromatin environment that permits transcription at loci including genes, TEs, and intergenic regions. As an example, ROS1-mediated demethylation is required for active transcription of the transgene PRD29A:LUC, which is otherwise silenced by RdDM that requires RDM1 (Gao et al. 2010). (Inset) Pie chart indicating categories (numbers) of hypermethylated loci in the Arabidopsis rdd mutant (Qian et al. 2012).
Transcription of RD29A (RESPONSIVE TO DESSICATION 29A) is highly induced by some abiotic stress conditions in wild-type Arabidopsis. In the transgenic line containing the transgene PRD29A:LUC (luciferase under the promoter of RD29A), RD29A is silenced along with the transgene when ROS1 is mutated (Gong et al. 2002). This transcriptional silencing in the ros1 mutant is attributable to DNA hypermethylation in the endogenous and transgenic RD29A promoters, which are located in different chromosomes (Gong et al. 2002). This is evidence that active DNA demethylation can protect genes from being incorrectly silenced. Bisulfite sequencing of the endogenous RD29A promoter in this transgenic plant revealed that methylated cytosines closer to the transcriptional start site are more prone to ROS1-mediated demethylation (Duan and Zhu, unpubl.), supporting the view that active DNA demethylation may restrict over-spreading of DNA methylation. Genetic screening for ros1 suppressors using this transgenic line has identified many known RdDM components (Zhang and Zhu 2011), strongly supporting an antagonistic relationship between active DNA demethylation and RdDM. Intriguingly, the ROS1 transcript level is significantly repressed in RdDM mutants and in met1 as well as by treatment with the DNA methyltransferase inhibitior 5-aza (Huettel et al. 2006; Mathieu et al. 2007; Martínez-Macías et al. 2012), indicating a feedback regulation of ROS1-dependent active DNA demethylation. Recently, an example of DNA demethylation-regulated TE transposition was reported. Active DNA demethylation by DNG701, a rice homolog of AtROS1, promotes transposition of the retrotransposon Tos17 in rice calli (La et al. 2011). Although ROS1 does not change global DNA methylation levels (Penterman et al. 2007), a recent characterization of the DNA methylome has identified nearly 5000 regions in the ros1 mutant that are hypermethylated relative to the wild type (Qian et al. 2012). Among these ROS1 targets, 44% are TEs, 26% are intergenic regions, and 30% are genic loci.
Mammalian cells also use active DNA demethylation for locus-specific gene regulation. One recent example is the transcriptional regulation of a human tumor suppressor gene, p15ink4b (Thillainadesan et al. 2012). In human breast cancer MCF7 cells, p15ink4b is silenced by promoter hypermethylation, which involves the oncogene ZNF217 and a corepressor complex CoREST that collectively recruit the DNA methyltransferase DNMT3A. Treatment with transforming growth factor β (TGF-β) repelled the DNMT3A/ZNF217/CoREST silencing complex and reduced DNA methylation in the p15ink4b promoter by recruiting a demethylation complex containing thymine DNA glycosylase (TDG), one of the mammalian enzymes involved in active DNA demethylation. On the other hand, knockdown of TDG or MBD4 prevents TGF-β-dependent demethylation of p15ink4b (Thillainadesan et al. 2012). Also in the MCF7 cells, the pS2/TFF1 gene promoter undergoes periodic methylation and active demethylation that coincides with cyclical estrogen receptor binding and gene expression of pS2 (Métivier et al. 2008). Studies in mammals have also revealed important roles of active DNA demethylation in neurogenesis and immune responses. The promoter of brain-derived neurotrophic factor (BDNF), an important gene for adult neural plasticity, is methylated under unstimulated conditions. In response to electroconvulsive treatment, the BDNF promoter exhibits active DNA demethylation that involves Gadd45β (growth arrest and DNA-damage-inducible 45 β) (Ma et al. 2009). In T lymphocytes, active DNA demethylation occurs in a small region of the promoter-enhancer of the interleukin-2 gene shortly after stimulation and independently of DNA replication (Bruniquel and Schwartz 2003).
ENZYMATIC REMOVAL OF 5-METHYLCYTOSINES
The importance of active DNA demethylation in epigenetic regulation has motivated extensive research to identify DNA demethylases as well as the underlying mechanisms. Studies in Arabidopsis have established the role of the bifunctional DNA glycosylases in direct removal of 5-mC through a base excision repair (BER) pathway. Although orthologous DNA glycosylases have not been identified in mammals, research to date has suggested several mechanisms involving some DNA glycosylases that mediate mammalian active DNA demethylation.
Direct Excision of 5-mC
DNA demethylation can be achieved by a process that starts with direct excision of 5-mC (Fig. 3). In Arabidopsis, removal of 5-mC is catalyzed by a subfamily of bifunctional DNA glycosylases that include ROS1, DME, DML2 (DMETER like 2), and DML3 (Zhu 2009). Each of these four enzymes excises 5-mC from all sequence contexts in vitro, although discrepancies regarding substrate preference exist (Agius et al. 2006; Gehring et al. 2006; Morales-Ruiz et al. 2006; Penterman et al. 2007). In addition to catalyzing the hydrolysis of a glycosylic bond between the base and the deoxyribose, these DNA demethylases also possess apurinic/apyrimidinic (AP) lyase activity that nicks the DNA backbone at the abasic site (Zhu 2009). During demethylation, these bifunctional enzymes consecutively exert DNA glycosylase and AP lyase activities, generating a single-nucleotide gap that is eventually refilled with an unmethylated cytosine through the DNA damage repair pathway. On the basis of genetic interaction analyses of maternal effects on seed development, Arabidopsis DNA LIGASE 1 (LIG1) has been suggested to participate in DME-mediated demethylation (Andreuzza et al. 2010), whereas the DNA polymerase responsible for repair remains unknown. Recently, a zinc finger DNA 3′ phosphoesterase (ZDP) was shown to function in ROS1-mediated DNA demethylation (Martínez-Macías et al. 2012). Biochemical results demonstrated that ZDP removes the 3′ phosphate from ROS1-nicked DNA, facilitating subsequent repair. ZDP mutation consistently reduces DNA demethylation in vitro and causes DNA hypermethylation at hundreds of loci in vivo. In addition, ZDP interacts with ROS1 in vitro and its transcript level is down-regulated in RdDM pathway mutants. Although ZDP is undoubtedly involved in active DNA demethylation, there appears to be genetic redundancy for ZDP function, given that ZDP target loci represent fewer than one-tenth of ROS1 targets, as determined in a parallel study (Qian et al. 2012).
Figure 3.
Active DNA demethylation pathways in plants and animals. Arabidopsis bifunctional DNA glycosylases, such as ROS1 and DME, can directly excise 5-mC and then cleave the DNA backbone at the abasic site, resulting in a gap that is filled with an unmodified cytosine through the BER pathway. In mammals where no ROS1 ortholog has been identified to date, 5-mC can undergo deamination and/or hydroxylation before being excised by DNA mismatch repair glycosylases, which triggers the BER pathway to complete the active demethylation process. See text for details.
Whereas DME functions during gametogenesis, the other three Arabidopsis demethylases are expressed in vegetative tissues (Penterman et al. 2007; Zhu et al. 2007; Huh et al. 2008). Triple mutation of ROS1, DML2, and DML3 in the rdd mutant does not alter total DNA methylation levels compared with the wild-type background (Penterman et al. 2007), yet these enzymes clearly monitor a broad range of target loci, as revealed by the characterization of the Arabidopsis methylome (Penterman et al. 2007; Lister et al. 2008; Qian et al. 2012). The most recent results showed ~5000 hypermethylated regions in ros1 mutants and ~9000 in rdd mutants (Qian et al. 2012). More than half of the hypermethylated regions in rdd do not appear to be targeted by ROS1 (Qian et al. 2012), indicating that DML2 and DML3 possess unique functions compared with ROS1, although partial redundancy of these three glycosylases has been observed (Penterman et al. 2007). Meanwhile, DME is distinctive for its well-known function in transcriptional activation of the maternal allele of imprinted genes (Huh et al. 2008).
In vertebrates including mammals, TDG and the methyl-CpG-binding protein 4 (MBD4) are reported to mediate active DNA demethylation by functioning as DNA glycosylases (Rai et al. 2008; Wu and Zhang 2010; Cortellino et al. 2011; Thillainadesan et al. 2012). However, the glycosylase activities of TDG and MBD4 against 5-mC are very weak compared with their glycosylase activities in removing thymine in T/G mismatch (Zhu et al. 2000a,b). Excision of 5-mC by TDG and MBD4 can be enhanced by the presence of other cellular components (Frémont et al. 1997; Jost et al. 1999; Rai et al. 2008), suggesting that these two enzymes may have strong 5-mC glycosylase activities in vivo. Alternatively, the role of TDG and MBD4 in active DNA demethylation may be attributed to their T/G mismatch repair activities, if 5-mC is deaminated into T. In addition, these DNA glycosylases may recognize other derivatives of 5-mC for the purpose of demethylation.
Excision after Derivatization of 5-mC
In contrast to plants that directly excise 5-mC, mammals apparently utilize DNA glycosylases after 5-mC is oxidized and/or deaminated (Fig. 3). In both cases, DNA base excision repair follows. Recent advances highlight several key enzyme families in regulating mammalian active demethylation, including the Tet (ten-eleven translocation) family that catalyzes iterative oxidation of 5-mC, the AID/APOBECs (activation-induced cytidine deaminase/apolipoprotein B mRNA-editing catalytic polypeptides) family that deaminates 5-mC and its oxidative derivative 5-hydroxymethylcytosine (5-hmC), and DNA glycosylases (especially TDG) that excise 5-mC derivatives and initiate subsequent DNA repair.
In mammals, 5-mC can be hydroxylated to 5-hmC by the Tet family of dioxygenases, which include TET1, 2, and 3 in humans and mice (Bhutani et al. 2011; Williams et al. 2011). In mouse embryonic stem cells, 5-hmC shows strong enrichment within exons and near transcriptional start sites (Pastor et al. 2011), yet the conversion of 5-mC to 5-hmC alone does not activate genes silenced by DNA methylation (Bhutani et al. 2011; Williams et al. 2011). However, 5-hmC can be either deaminated by AID/APOBEC family proteins to yield 5-hydroxymethyluracil (5-hmU) (Cortellino et al. 2011) or further oxidized by Tet proteins to consecutively generate 5-formylcytosine (5-fC) and 5-carboxylcytosine (5-caC) (He et al. 2011; Ito et al. 2011b). Mammalian DNA glycosylase TDG recognizes and efficiently excises 5-fC and 5-caC, while leaving 5hmC intact (He et al. 2011; Maiti and Drohat 2011; Zhang et al. 2012). In fact, TDG is the only known mammalian DNA glycosylase that cleaves 5-fC and 5-caC, because other tested DNA glycosylases including MBD4 and SMUG1 (single-strand-selective monofunctional U DNA glycosylase 1) showed no significant activity in removing 5-fC and 5-caC (He et al. 2011; Maiti and Drohat 2011). It is now clear that TDG-initiated DNA repair completes the process of active DNA demethylation (Cortellino et al. 2011; He et al. 2011). However, it remains to be determined whether a putative decarboxylase may remove the carboxyl group from 5-caC to regenerate unmodified cytosine (Wu and Zhang 2010; Ito et al. 2011b), because depletion of TDG can only partially restore the 5-caC level to that under normal conditions in mouse embryonic stem cells, in which Tet2 catalyzes 5-caC production with high efficiency (He et al. 2011; Ito et al. 2011b).
The importance of 5-mC oxidation is also indicated by recent findings that active demethylation in the zygotic paternal pronucleus is dependent on TET3-dependent conversion of 5-mC to 5-hmC (Gu et al. 2011; Iqbal et al. 2011). Deficiency of TET3 was also shown to impede demethylation of two embryonic stem cell marker genes and to cause developmental defects (Gu et al. 2011), although it is unknown whether 5-hmC in the paternal pronucleus undergoes deamination or further oxidation before being excised. Although new 5-hmC may be generated while the preexisting 5-hmC is being processed for excision, 5-hmC seems to persist throughout the early embryonic stages (Gu et al. 2011), suggesting that the global loss of 5-mC observed during the one-cell stage (Mayer et al. 2000) may not completely yield in replacement with unmodified cytosines as quickly as previously thought. Because several cycles of cell division occur after the one-cell embryonic stage, the presence of 5-hmC probably contributes to passive DNA demethylation, given that 5-hmC is not recognized by Dnmt1 for methylation maintenance during DNA replication (Valinluck and Sowers 2007). Indeed, a recent study reported that, in the two-cell embryonic stage, only one of the two sister chromatids of sperm-derived chromosomes is enriched for 5-hmC (Inoue and Zhang 2011). Moreover, blastomeres at the four- and eight-cell stages showed a gradual loss of 5-hmC, suggesting a replication-dependent demethylation during preimplantation development (Inoue and Zhang 2011).
Besides 5-fC and 5-caC, T and 5-hmU can be produced from deamination of 5-mC or 5-hmC, respectively (Fig. 3). The AID/APOBEC family of proteins catalyzes these deamination reactions and thereby initiates mismatch repair for active DNA demethylation (Cortellino et al. 2011; Guo et al. 2011). Mammalian AID is originally known for its function in generating antibody diversity in B lymphocytes (Muramatsu et al. 2000). AID belongs to the APOBEC family, which is so named because members other than AID were originally identified as RNA editors (Conticello et al. 2007). The ability of AID and APOBEC1 to mediate 5-mC deamination has been documented in vitro and in Escherichia coli (Morgan et al. 2004), and the first substantiation of AID's role in mediating vertebrate active DNA demethylation came from zebrafish embryos (Rai et al. 2008). Overexpression of AID and MBD4 together in zebrafish embryos caused demethylation of the bulk genome and of the injected methylated DNA fragments. Gadd45a was then shown to enhance the cooperational demethylation by AID and MBD4. On the other hand, depletion of AID or Gadd45a attenuated demethylation of the injected methylated DNA, which was linearized and nonreplicating. It is therefore postulated that MBD4 excision of thymine follows AID deamination of 5-mC, a process that is promoted by Gadd45 (Rai et al. 2008). Indeed, increase in T/G mismatch was observed when AID was expressed with a noncatalytic MBD4 (Rai et al. 2008).
Similar to the proposed deamination → excision → repair demethylation process in zebrafish, a process involving AID, TDG, and Gadd45a was identified in a mouse study that focused on TDG (Cortellino et al. 2011). RNAi targeting TDG in embryonic cells compromised DNA demethylation in the promoter of an invitro-methylated reporter gene. Contribution of passive demethylation was excluded because DNA demethylation occurred within 12 h of transfection and because the reporter plasmid contained no replication origin. TDG was also shown to control several development- and hormone-regulated genes through DNA demethylation of their corresponding promoters, which is consistent with the fact that knockout or catalytic dysfunction of TDG leads to embryonic lethality (Cortellino et al. 2011). In contrast, mice that lack AID are viable and fertile, although their PGCs exhibit genome-wide hypermethylation that is still low (~20%) relative to that in embryonic stem cells or somatic cells (70%–80%) (Popp et al. 2010), pointing to possible functional redundancy in the AID/APOBEC family. Alternatively, the lack of dependency on AID may indicate that the deamination step, on a whole-genome scale, can be largely bypassed through, for instance, iterative oxidation of 5-hmC followed by enzymatic removal of 5-fC and/or 5-caC (Fig. 3). In addition, because this study used bisulfite sequencing, which cannot distinguish between 5-mC and 5-hmC (Huang et al. 2010; Pastor et al. 2011), it remains to be explored to what extent 5-mC or 5-hmC contributes as AID deamination substrates to demethylation. Nevertheless, AID was demonstrated to be necessary for promoter demethylation and concomitant gene induction of OCT4 and NANOG, two human pluripotency genes whose promoters are rapidly demethylated during nuclear reprogramming (Bhutani et al. 2010). AID binds to methylated OTC4 and NANOG promoters in fibroblasts, but not to the demethylated promoters in embryonic stem cells, supporting the direct involvement of AID in active DNA demethylation. In addition to TDG, SMUG1 also efficiently cleaves 5-hmU in double-stranded DNA, yet depletion of TDG alone is lethal in mice (Cortellino et al. 2011).
BER is a well-established pathway that begins with DNA glycosylase activity for nucleotide mismatch repair (Norbury and Hickson 2001). In both plants and mammals, BER can accomplish active DNA demethylation (Zhu 2009; Wu and Zhang 2010; Bhutani et al. 2011). In addition to DNA glycosylases, the proteins in the DNA damage-responsive Gadd45 family have been implicated in active DNA demethylation (reviewed in Niehrs and Schäfer 2012). Moreover, single-stranded DNA breaks and BER were shown to be mechanistically linked to global DNA demethylation in mouse PGCs and in the paternal pronucleus (Hajkova et al. 2010).
INTERPLAY BETWEEN 5-mC REMOVAL AND OTHER EPIGENETIC MODIFICATIONS
Active DNA demethylation is a coordinated process. In addition to proteins that directly mediate enzymatic replacement of 5-mC with unmodified cytosine, other molecular components can also be involved by recruiting the 5-mC removal machinery to its target loci, by establishing a chromatic environment that allows for excision of 5-mC, and conversely, by repelling the demethylation enzymes in order to protect the 5-mC mark.
Recruitment for 5-mC Removal
Although enzymes that excise 5-mC can be targeted to specific loci, the molecular mechanisms underlying the recruitment remain elusive. In theory, target specificity can originate from distinct characteristics of the chromatins or the individual chromatin components including DNA sequences and histones. Enzymes involved in 5-mC excision may carry functional domains that facilitate the recognition of their targets. DME and ROS1 each contain a characterized domain that mediates methylation-independent DNA binding (Mok et al. 2010; Ponferrada-Marín et al. 2010). Such domains may either facilitate target searching and/or the enzymatic removal of 5-mC. Similarly, the mammalian DNA glycosylase MBD4 contains a methyl DNA-binding domain that may help MBD4 identify methylated substrates (Ballestar and Wolffe 2001). Nevertheless, other factors would also be required to precisely target active DNA demethylation to specific loci.
In Arabidopsis, an RNA-binding protein named ROS3 has been implicated in targeting ROS1 to certain loci (Zheng et al. 2008). ROS3 is required for demethylation at a number of loci, some of which overlap with ROS1 targets. ROS3 and ROS1 proteins colocalize in discrete foci dispersed throughout the nucleus in wild-type plants, whereas ROS1 localization was severely disrupted in the ros3 mutant, suggesting that the two proteins may function in a demethylation complex. ROS3 binds single-stranded small RNAs of specific sequences in vivo; therefore, it was proposed that ROS3 may use small RNAs to identify homologous target loci for demethylation (Zheng et al. 2008). However, the general sequence features of ROS3-bound small RNAs are currently obscure except that they appear to be rich in guanine (Zheng et al. 2008). It is also unknown whether ROS3 can bind long, noncoding RNAs (lncRNAs), such as those generated by RNA polymerase II (Pol II) or the plant-specific Pol V. In the RdDM pathway, Pol II- or Pol V-dependent, nascent lncRNAs can pair with their homologous 24-nucleotide siRNAs to recruit DNA methylases (Law and Jacobsen 2010; Zhang and Zhu 2011). Although Pol V mutation usually results in desilencing, it also causes hypermethylation at certain loci where DNA methylation patterns are independent of the siRNAs (Mosher et al. 2008), indicating a possibility that lncRNA might also facilitate the recruitment of DNA demethylases. In fact, the initial hint that RNA might contribute to DNA demethylation was the observation of RNA-dependent 5-mC glycosylase activity against hemimethylated DNA in chicken embryo extracts (Frémont et al. 1997). 5-mC DNA glycosylase activity from chicken embryos or mice G8 myoblasts was blocked by RNase treatment and was restored by the addition of synthetic RNA complementary to the methylated strand of the substrate DNA (Jost et al. 1999; Zhu et al. 2000a). Although these in vitro results may reflect direct assistance during enzymatic removal of 5-mC, a role of RNA in targeting active DNA demethylation is plausible.
DNA demethylation can be induced by hormones and other signals. Recruitment of the demethylases in these cases may be mediated by the signaling components or their downstream effectors. TDG can directly interact with the estrogen receptor a in activating transcription of estrogen-regulated genes (Chen et al. 2003). In addition, TDG interacts with SRC1, NCoA-3, CBP, and p300 (Tini et al. 2002; Lucey et al. 2005; Chiang et al. 2010), all of which are transcriptional coactivators that activate transcription at genes downstream from various signaling pathways (Goodman and Smolik 2000); whether these protein interactions result in active DNA demethylation in the gene promoters remains to be determined. Similarly, the retinoid receptor physically associates with human TDG (Um et al. 1998), which causes demethylation of a retinoic acid–responsive promoter linked to a reporter gene (Zhu et al. 2001). These results suggest that DNA demethylases may be guided to their target loci through physical interaction with certain transcriptional regulators.
Interplay with Histone Modifications
Removal of 5-mC may occur directly in the established chromatic environment. Alternatively, a call for demethylation may also lead to other modifications to the chromatin before or during the removal of 5-mC. Recently, a histone acetyltransferase named IDM1 (INCREASED DNA METHYLATION 1) was identified as a regulator of DNA demethylation in Arabidopsis (Qian et al. 2012). IDM1 recognizes chromatin containing CG methylation and low histone 3 lysine 4 (H3K4) and arginine 2 (H3R2) methylations that are characteristic of IDM1's target loci and subsequently catalyzes H3K18 and H3K23 acetylation. Analysis of genetic interactions revealed that IDM1 and ROS1 function in the same pathway for DNA demethylation, although IDM1 controls only a portion of the ROS1 targets. It is still unclear how ROS1 is recruited to the modified chromatin, but the discovery of IDM1 underscores the contribution of histone modifications to active DNA demethylation. Similarly, maintenance of 5-mC in the CHG context can be impaired by IBM1 (INCREASE IN BONSAI METHYLATION 1), which is a histone demethylase that prevents H3K9 methylation (Saze et al. 2008; Miura et al. 2009). Arabidopsis SSRP1 (STRUCTURE SPECIFIC RECOGNITION PROTEIN 1) is required for DNA demethylation and regulation of many paternally imprinted genes in the central cell (Ikeda et al. 2011). This non-histone chromosomal protein is a component of a heterodimeric histone chaperone complex, which has a conserved function in chromatin remodeling in Arabidopsis, Drosophila, and humans (Orphanides et al. 1999; Shimojima et al. 2003; Duroux et al. 2004). Interestingly, hypomethylation by MET1 dys-function in the ssrp1–3 met1–3 double mutant only partially activates the maternal allele of the imprinted FWAGFP reporter in the central cell; in contrast, FWA-GFP is fully active in dme-1 met1–3. Therefore, mutation of SSRP1 apparently leads to additional repressive chromatin modification besides DNA hypermethylation.
In Arabidopsis and humans, nucleosome-bound DNA is more densely methylated than linker DNA sequences (Chodavarapu et al. 2010). Such a bias may be attributable to preferential methylation of nucleosomal DNA, more frequent demethylation in linker DNA, or both. Each member of the bifunctional DNA glycosylase family in Arabidopsis possesses an amino-terminal domain similar to the linker histone H1 (Zhu 2009; Law and Jacobsen 2010). Although it is unknown whether these H1-like domains are functional in binding to linker DNA, it might be interesting to determine whether nucleosome positioning contributes to genome-wide DNA demethylation patterning. In fact, researchers have speculated that the obvious differences in histone compositions and histone marks between the two parental pronuclei could underlie the paternal-specific demethylation in zygotes (Wu and Zhang 2010). In mouse embryonic stem cells, genome-wide profiling revealed a strong correlation between 5-hmC enrichment and the dual histone mark H3K27 trimethylation plus H3K4 trimethylation in gene promoter regions (Pastor et al. 2011).
Preventing Demethylation
During the genome-wide loss of 5-mC in the zygotic paternal pronucleus, DNA methylation in the maternal genome and in certain paternal loci remains unchanged. Although the docking of demethylation enzymes to DNA substrates can potentially be blocked by repressive histone modifications, several nonhistone proteins have been shown to protect DNA methylation regardless of the global demethylation. Mutation of stella/PGC7, which binds to both unmethylated and methylated DNA in vitro, leads to DNA demethylation in both paternal and maternal pronuclei (Nakamura et al. 2007). MBD3, which is an essential component of a chromatin-remodeling complex, also may block demethylation because demethylation in the promoter of the paternally imprinted H19 gene was observed in MBD3-depleted mouse embryos (Reese et al. 2007). Chromatin immunoprecipitation showed that MBD3 binds to the methylated region in the H19 promoter (Reese et al. 2007). However, it is unknown whether stella and MBD3 repel demethylation enzymes to protect 5-mC or whether they maintain DNA methylation imprint through interactions with methyltransferases as does the zinc finger protein ZFP57 (Li et al. 2008). Another way to prevent active DNA demethylation (in addition to counteracting the activities of DNA demethylases) is via transcriptional repression of DNA demethylases. For instance, DME is nondetectable in the germ cells but is expressed in the central cell (Choi et al. 2002; Schoft et al. 2011), thereby allowing the establishment of gene imprinting in the endosperm.
CONCLUDING REMARKS AND PERSPECTIVES
In both plants and animals, genome-wide active DNA demethylation mediates developmental reprogramming, whereas locus-specific removal of 5-mC ensures proper gene regulation. As exemplified in Arabidopsis, identification of active DNA demethylation targets on a whole-genome scale has revealed thousands of such loci (Qian et al. 2012), indicating that dynamic editing of DNA methylation patterns is a common epigenetic feature. The biochemical mechanism for 5-mC cleavage is well established in plants, but convincing evidence of active DNA demethylation pathways in mammals has been obtained only recently. In particular, the discovery of Tet-dependent 5-hmC has triggered a flood of studies, resulting in significant advances in understanding of mammalian active DNA demethylation processes. Nevertheless, more efforts are needed to understand fully this critical epigenetic regulation. Gene families exist for enzymes known to be involved in 5-mC excision. Do all members of each family mediate active DNA demethylation? How much redundancy is there among their functions? How may they cooperate in shaping the cellular methylome? For instance, array-based data show that ROS1 transcript abundance increases during early embryo development in Arabidopsis seeds (Schmid et al. 2005). This might indicate a temporal coediting of the embryonic methylome by DME and ROS1. In fact, ROS1-dependent demethylation mediates postembryonic remodeling of the pericentromeric 5S rDNA chromatin (Douet et al. 2008). Another interesting question is how the demethylation enzymes might be regulated in accordance with the need of demethylation, as exemplified by the repression of Arabidopsis ROS1 transcript (Huettel et al. 2006; Mathieu et al. 2007; Martínez-Macías et al. 2012).
The whole spectrum of biological processes under regulation by active DNA demethylation has yet to be completely revealed, even though some enzymes have long been known as DNA demethylases. A recent study reported that telomeres were longer in ros1 mutants than in wild-type Arabidopsis, but the underlying mechanism is unknown (Liu et al. 2010). Mechanistic investigations have been focusing on 5-mC excision, and molecular players in the follow-up repairing process have received less attention. Research is needed to determine which BER proteins are involved in active DNA demethylation and how active DNA demethylation interacts with the canonical DNA damage repair pathways. In addition, although recent advances have provided novel insights into mammalian genome-wide demethylation, how such global demethylation is processed remains to be determined. At the same time, the mechanisms underlying locus-specific recruitment or protection from active DNA demethylation are still unclear. As a major means of epigenetic regulation, active DNA demethylation clearly interacts with other epigenetic processes, such as histone modifications, and thereby needs to be evaluated in the context of chromatin. Future research on this topic will greatly increase our understanding of epigenetics and should also lead to the development of novel genetic and chemical therapies for epigenetic disorders.
REFERENCES
- Agius F, Kapoor A, Zhu JK. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc Natl Acad Sci. 2006;103:11796–11801. doi: 10.1073/pnas.0603563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreuzza S, Li J, Guitton AE, Faure JE, Casanova S, Park JS, Choi Y, Chen Z, Berger F. DNA LIGASE I exerts a maternal effect on seed development in Arabidopsis thaliana. Development. 2010;137:73–81. doi: 10.1242/dev.041020. [DOI] [PubMed] [Google Scholar]
- Ballestar E, Wolffe AP. Methyl-CpG-binding proteins. Targeting specific gene repression. Eur J Biochem. 2001;268:1–6. doi: 10.1046/j.1432-1327.2001.01869.x. [DOI] [PubMed] [Google Scholar]
- Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell. 2011;146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol. 2003;4:235–240. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE. Role of the Arabidopsis DRM methyltransferases in denovo DNA methylation and gene silencing. Curr Biol. 2002;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: Patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Chen D, Lucey MJ, Phoenix F, Lopez-Garcia J, Hart SM, Losson R, Buluwela L, Coombes RC, Chambon P, Schär P, et al. T:G mismatch-specific thymine-DNA glycosylase potentiates transcription of estrogen-regulated genes through direct interaction with estrogen receptor alpha. J Biol Chem. 2003;278:38586–38592. doi: 10.1074/jbc.M304286200. [DOI] [PubMed] [Google Scholar]
- Cheng X, Blumenthal RM. Mammalian DNA methyltransferases: A structural perspective. Structure. 2008;16:341–350. doi: 10.1016/j.str.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang S, Burch T, Van Domselaar G, Dick K, Radziwon A, Brusnyk C, Edwards MR, Piper J, Cutts T, Cao J, et al. The interaction between thymine DNA glycosylase and nuclear receptor coactivator 3 is required for the transcriptional activation of nuclear hormone receptors. Mol Cell Biochem. 2010;333:221–232. doi: 10.1007/s11010-009-0223-1. [DOI] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, et al. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Gehring M, Johnson L, Hannon M, Harada JJ, Goldberg RB, Jacobsen SE, Fischer RL. TER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- Conticello SG, Langlois MA, Yang Z, Neuberger MS. DNA deamination in immunity: AID in the context of its APOBEC relatives. Adv Immunol. 2007;94:37–73. doi: 10.1016/S0065-2776(06)94002-4. [DOI] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean W, Santos F, Stojkovic M, Zakhartchenko V, Walter J, Wolf E, Reik W. Conservation of methylation reprogramming in mammalian development: Aberrant reprogramming in cloned embryos. Proc Natl Acad Sci. 2001;98:13734–13738. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douet J, Blanchard B, Cuvillier C, Tourmente S. Interplay of RNA Pol IV and ROS1 during post-embryonic 5S rDNA chromatin remodeling. Plant Cell Physiol. 2008;49:1783–1791. doi: 10.1093/pcp/pcn152. [DOI] [PubMed] [Google Scholar]
- Duroux M, Houben A, Ruzicka K, Friml J, Grasser KD. The chromatin remodelling complex FACT associates with actively transcribed regions of the Arabidopsis genome. Plant J. 2004;40:660–671. doi: 10.1111/j.1365-313X.2004.02242.x. [DOI] [PubMed] [Google Scholar]
- Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10:2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frémont M, Siegmann M, Gaulis S, Matthies R, Hess D, Jost JP. Demethylation of DNA by purified chick embryo 5-methylcytosine-DNA glycosylase requires both protein and RNA. Nucleic Acids Res. 1997;25:2375–2380. doi: 10.1093/nar/25.12.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Liu HL, Daxinger L, Pontes O, He X, Qian W, Lin H, Xie M, Lorkovic ZJ, Zhang S, et al. An RNA polymerase II-and AGO4-associated protein acts in RNA-directed DNA methylation. Nature. 2010;465:106–109. doi: 10.1038/nature09025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Huh JH, Hsieh TF, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Bubb KL, Henikoff S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science. 2009;324:1447–1451. doi: 10.1126/science.1171609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Gong Z, Morales-Ruiz T, Ariza RR, Roldán-Arjona T, David L, Zhu JK. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: Purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol. 2011;12:483–492. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, Surani MA. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–881. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science. 2010;329:78–82. doi: 10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Hsu YF, Zhu S, Wierzbicki AT, Pontes O, Pikaard CS, Liu HL, Wang CS, Jin H, Zhu JK. An effector of RNA-directed DNA methylation in arabidopsis is an ARGONAUTE 4-and RNA-binding protein. Cell. 2009;137:498–508. doi: 10.1016/j.cell.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- Hsieh TF, Ibarra CA, Silva P, Zemach A, Eshed-Williams L, Fischer RL, Zilberman D. Genome-wide demethylation of Arabidopsis endosperm. Science. 2009;324:1451–1454. doi: 10.1126/science.1172417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettel B, Kanno T, Daxinger L, Aufsatz W, Matzke AJ, Matzke M. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J. 2006;25:2828–2836. doi: 10.1038/sj.emboj.7601150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh JH, Bauer MJ, Hsieh TF, Fischer RL. Cellular programming of plant gene imprinting. Cell. 2008;132:735–744. doi: 10.1016/j.cell.2008.02.018. [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Kinoshita Y, Susaki D, Ikeda Y, Iwano M, Takayama S, Higashiyama T, Kakutani T, Kinoshita T. HMG domain containing SSRP1 is required for DNA demethylation and genomic imprinting in Arabidopsis. Dev Cell. 2011;21:589–596. doi: 10.1016/j.devcel.2011.08.013. [DOI] [PubMed] [Google Scholar]
- Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Jin SG, Pfeifer GP, Szabó PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Gaubert H, Bucher E, Mirouze M, Vaillant I, Paszkowski J. An siRNA pathway prevents transgenerational retro-transposition in plants subjected to stress. Nature. 2011a;472:115–119. doi: 10.1038/nature09861. [DOI] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011b;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost JP, Siegmann M, Thiry S, Jost YC, Benjamin D, Schwarz S. A re-investigation of the ribonuclease sensitivity of a DNA demethylation reaction in chicken embryo and G8 mouse myoblasts. FEBS Lett. 1999;449:251–254. doi: 10.1016/s0014-5793(99)00454-8. [DOI] [PubMed] [Google Scholar]
- Jullien PE, Mosquna A, Ingouff M, Sakata T, Ohad N, Berger F. Retinoblastoma and its binding partner MSI1 control imprinting in Arabidopsis. PLoS Biol. 2008;6:e194. doi: 10.1371/journal.pbio.0060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La H, Ding B, Mishra GP, Zhou B, Yang H, Bellizzi Mdel R, Chen S, Meyers BC, Peng Z, Zhu JK, et al. A 5-methylcytosine DNA glycosylase/lyase demethylates the retrotransposon Tos17 and promotes its transposition in rice. Proc Natl Acad Sci. 2011;108:15498–15503. doi: 10.1073/pnas.1112704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J, Reik W. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35:88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, Ferguson-Smith AC. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell. 2008;15:547–557. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O′Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Wang J, Miki D, Xia R, Yu W, He J, Zheng Z, Zhu JK, Gong Z. DNA replication factor C1 mediates genomic stability and transcriptional gene silencing in Arabidopsis. Plant Cell. 2010;22:2336–2352. doi: 10.1105/tpc.110.076349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucey MJ, Chen D, Lopez-Garcia J, Hart SM, Phoenix F, Al-Jehani R, Alao JP, White R, Kindle KB, Losson R, et al. T:G mismatch-specific thymine-DNA glycosylase (TDG) as a coregulator of transcription interacts with SRC1 family members through a novel tyrosine repeat motif. Nucleic Acids Res. 2005;33:6393–6404. doi: 10.1093/nar/gki940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: Potential implications for active demethylation of CpG sites. J Biol Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen RA, Kloc A, Slotkin RK, Tanurdzić M. Epigenetic inheritance and reprogramming in plants and fission yeast. Cold Spring Harb Symp Quant Biol. 2008;73:265–271. doi: 10.1101/sqb.2008.73.062. [DOI] [PubMed] [Google Scholar]
- Martínez-Macías MI, Qian W, Miki D, Pontes O, Liu Y, Tang K, Liu R, Morales-Ruiz T, Ariza RR, Roldán-Arjona T, et al. A DNA 3′ phosphatase functions in active DNA demethylation in Arabidopsis. Mol Cell. 2012;45:357–370. doi: 10.1016/j.molcel.2011.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell. 2007;130:851–862. doi: 10.1016/j.cell.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Matzke M, Kanno T, Huettel B, Daxinger L, Matzke AJ. Targets of RNA-directed DNA methylation. Curr Opin Plant Biol. 2007;10:512–519. doi: 10.1016/j.pbi.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- Métivier R, Gallais R, Tiffoche C, Le Péron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- Miura A, Nakamura M, Inagaki S, Kobayashi A, Saze H, Kakutani T. An Arabidopsis jmjC domain protein protects transcribed genes from DNA methylation at CHG sites. EMBO J. 2009;28:1078–1086. doi: 10.1038/emboj.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok YG, Uzawa R, Lee J, Weiner GM, Eichman BF, Fischer RL, Huh JH. Domain structure of the DEMETER 5-methylcytosine DNA glycosylase. Proc Natl Acad Sci. 2010;107:19225–19230. doi: 10.1073/pnas.1014348107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A, Melnyk CW, Bassett A, Hardcastle TJ, Dunn R, Baulcombe DC. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science. 2010;328:872–875. doi: 10.1126/science.1187959. [DOI] [PubMed] [Google Scholar]
- Morales-Ruiz T, Ortega-Galisteo AP, Ponferrada-Marín MI, Martínez-Macías MI, Ariza RR, Roldán-Arjona T. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc Natl Acad Sci. 2006;103:6853–6858. doi: 10.1073/pnas.0601109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: Implications for epigenetic reprogramming. J Biol Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14(Spec No 1):R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- Mosher RA, Schwach F, Studholme D, Baulcombe DC. PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc Natl Acad Sci. 2008;105:3145–3150. doi: 10.1073/pnas.0709632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol. 2007;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- Niehrs C, Schaäfer A. Active DNA demethylation by Gadd45 and DNA repair. Trends Cell Biol. 2012;22:220–227. doi: 10.1016/j.tcb.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Norbury CJ, Hickson ID. Cellular responses to DNA damage. Annu Rev Pharmacol Toxicol. 2001;41:367–401. doi: 10.1146/annurev.pharmtox.41.1.367. [DOI] [PubMed] [Google Scholar]
- Okada Y, Yamagata K, Hong K, Wakayama T, Zhang Y. A role for the elongator complex in zygotic paternal genome demethylation. Nature. 2010;463:554–558. doi: 10.1038/nature08732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olek A, Walter J. The pre-implantation ontogeny of the H19 methylation imprint. Nat Genet. 1997;17:275–276. doi: 10.1038/ng1197-275. [DOI] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. DNMT3 L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999;400:284–288. doi: 10.1038/22350. [DOI] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL. DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci. 2007;104:6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponferrada-Marín MI, Martínez-Macías MI, Morales-Ruiz T, Roldán-Arjona T, Ariza RR. Methylation-independent DNA binding modulates specificity of Repressor of Silencing 1 (ROS1) and facilitates demethylation in long substrates. J Biol Chem. 2010;285:23032–23039. doi: 10.1074/jbc.M110.124578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponicsan SL, Kugel JF, Goodrich JA. Genomic gems: SINE RNAs regulate mRNA production. Curr Opin Genet Dev. 2010;20:149–155. doi: 10.1016/j.gde.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Miki D, Zhang H, Liu Y, Zhang X, Tang K, Kan Y, La H, Li X, Li S, et al. A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science. 2012;336:1445–1448. doi: 10.1126/science.1219416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese KJ, Lin S, Verona RI, Schultz RM, Bartolomei MS. Maintenance of paternal methylation and repression of the imprinted H19 gene requires MBD3. PLoS Genet. 2007;3:e137. doi: 10.1371/journal.pgen.0030137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougier N, Bourc'his D, Gomes DM, Niveleau A, Plachot M, Pàldi A, Viegas-Péquignot E. Chromosome methylation patterns during mammalian preimplantation development. Genes Dev. 1998;12:2108–2113. doi: 10.1101/gad.12.14.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- Saze H, Shiraishi A, Miura A, Kakutani T. Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science. 2008;319:462–465. doi: 10.1126/science.1150987. [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Schoft VK, Chumak N, Choi Y, Hannon M, Garcia-Aguilar M, Machlicova A, Slusarz L, Mosiolek M, Park JS, Park GT, et al. Function of the DEMETER DNA glycosylase in the Arabidopsis thaliana male gametophyte. Proc Natl Acad Sci. 2011;108:8042–8047. doi: 10.1073/pnas.1105117108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojima T, Okada M, Nakayama T, Ueda H, Okawa K, Iwamatsu A, Handa H, Hirose S. Drosophila FACT contributes to Hox gene expression through physical and functional interactions with GAGA factor. Genes Dev. 2003;17:1605–1616. doi: 10.1101/gad.1086803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, Tanurdzić M, Becker JD, Feijó JA, Martienssen RA. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136:461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: Provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Thillainadesan G, Chitilian JM, Isovic M, Ablack JN, Mymryk JS, Tini M, Torchia J. TGF-β-dependent active demethylation and expression of the p15(ink4b) tumor suppressor are impaired by the ZNF217/CoREST complex. Mol Cell. 2012;46:636–649. doi: 10.1016/j.molcel.2012.03.027. [DOI] [PubMed] [Google Scholar]
- Tini M, Benecke A, Um SJ, Torchia J, Evans RM, Chambon P. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol Cell. 2002;9:265–277. doi: 10.1016/s1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- Um S, Harbers M, Benecke A, Pierrat B, Losson R, Chambon P. Retinoic acid receptors interact physically and functionally with the T:G mismatch-specific thymine-DNA glycosylase. J Biol Chem. 1998;273:20728–20736. doi: 10.1074/jbc.273.33.20728. [DOI] [PubMed] [Google Scholar]
- Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- Vongs A, Kakutani T, Martienssen RA, Richards EJ. Arabidopsis thaliana DNA methylation mutants. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ, Toyoda A, et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science. 2011;332:848–852. doi: 10.1126/science.1203919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2011;13:28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Active DNA demethylation: Many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Smith SL, Tian XC, Lewin HA, Renard JP, Wakayama T. Nuclear reprogramming of cloned embryos and its implications for therapeutic cloning. Nat Genet. 2007;39:295–302. doi: 10.1038/ng1973. [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhu JK. RNA-directed DNA methylation. Curr Opin Plant Biol. 2011;14:142–147. doi: 10.1016/j.pbi.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Zhang L, Lu X, Lu J, Liang H, Dai Q, Xu GL, Luo C, Jiang H, He C. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat Chem Biol. 2012;8:328–330. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Pontes O, Zhu J, Miki D, Zhang F, Li WX, Iida K, Kapoor A, Pikaard CS, Zhu JK. ROS3 is an RNA-binding protein required for DNA demethylation in Arabidopsis. Nature. 2008;455:1259–1262. doi: 10.1038/nature07305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Hess D, Angliker H, Schwarz S, Siegmann M, Thiry S, Jost JP. 5-methylcytosine-DNA glycosylase activity is present in a cloned G/T mismatch DNA glycosylase associated with the chicken embryo DNA demethylation complex. Proc Natl Acad Sci. 2000a;97:5135–5139. doi: 10.1073/pnas.100107597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Angliker H, Schwarz S, Thiry S, Siegmann M, Jost JP. 5-Methylcytosine DNA glycosylase activity is also present in the human MBD4 (G/T mismatch glycosylase) and in a related avian sequence. Nucleic Acids Res. 2000b;28:4157–4165. doi: 10.1093/nar/28.21.4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Benjamin D, Zheng Y, Angliker H, Thiry S, Siegmann M, Jost JP. Overexpression of 5-methylcytosine DNA glycosylase in human embryonic kidney cells EcR293 demethylates the promoter of a hormone-regulated reporter gene. Proc Natl Acad Sci. 2001;98:5031–5036. doi: 10.1073/pnas.091097298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu JK. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr Biol. 2007;17:54–59. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]