Abstract
Although the evolution process and ecological benefits of symbiotic species with small genomes are well understood, these issues remain poorly elucidated for free-living species with large genomes. We have compared 115 completed prokaryotic genomes by using the Clusters of Orthologous Groups database to determine whether there are changes with genome size in the proportion of the genome attributable to particular cellular processes, because this may reflect both cellular and ecological strategies associated with genome expansion. We found that large genomes are disproportionately enriched in regulation and secondary metabolism genes and depleted in protein translation, DNA replication, cell division, and nucleotide metabolism genes compared to medium- and small-sized genomes. Furthermore, large genomes do not accumulate noncoding DNA or hypothetical ORFs, because the portion of the genome devoted to these functions remained constant with genome size. Traits other than genome size or strain-specific processes are reflected by the dispersion around the mean for cell functions that showed no correlation with genome size. For example, Archaea had significantly more genes in energy production, coenzyme metabolism, and the poorly characterized category, and fewer in cell membrane biogenesis and carbohydrate metabolism than Bacteria. The trends we noted with genome size by using Clusters of Orthologous Groups were confirmed by our independent analysis with The Institute for Genomic Research's Comprehensive Microbial Resource and Kyoto Encyclopedia of Genes and Genomes' Orthology annotation databases. These trends suggest that larger genome-sized species may dominate in environments where resources are scarce but diverse and where there is little penalty for slow growth, such as soil.
The genome sequences of the smallest genome-sized prokaryotic species, the obligate endocellular parasites, have provided insight into the interrelationship between the ecology and genome evolution of these species (1-3). For instance, when compared their free-living relatives, these reduced genomes have preferentially lost genes underlying the biosynthesis of compounds that can be easily taken up from the host, such as amino acids, nucleotides, and vitamins. Furthermore, regulatory elements, including σ factors, have commonly been eliminated from such symbiotic bacteria, presumably due to the rather stable environment inside host cells, which renders extensive gene regulation useless (4-6). It is not yet clear whether there may also be trends in gene allocation for the larger genome-sized free-living bacteria. If such trends do exist, they could reveal strategies of genome expansion, provide insight into the upper limit of genome size, reveal whether there is more centrally coordinated regulation, and most important, suggest what ecological benefits accrue for such species.
There is currently an increasing amount of evidence that favors the existence of universal trends between functional gene content and genome size. For instance, Jordan et al.'s (7) analysis of 21 genomes showed that lineage-specific gene expansion is positively correlated with genome size and may account for up to 33% of the coding capacities in the genome. Furthermore, comparative genomic studies of Pseudomonas aeruginosa PAO1 and Streptomyces coelicolor A3, two larger genome species, noted a disproportionate increase relative to smaller genome-sized species in regulatory and transport genes and in genes involved in secondary metabolism, respectively (8, 9). However, only a limited number of species were analyzed in both of these studies, and the analysis was restricted to specific functional processes. Furthermore, in the former study, no other species in the panel of strains evaluated had a genome size comparable to strain PA01, a moderately large (6.3-Mb) genome-sized strain; thus, the significance of these findings for other large prokaryotic genomes is unknown.
We sought to more comprehensively evaluate how the relative usage of the genome changes with genome size, using all sequenced genomes and evaluating all functional classes of genes.
Materials and Methods
We undertook the functional characterization of 115 completed genomes deposited in the GenBank database as of May 2003 (the list of genomes is presented as Table 3, which is published as supporting information on the PNAS web site) using the Clusters of Orthologous Groups (COG) database (10, 11). At the time of this study, the COG database was comprised of 144,320 protein sequences from 66 completed genomes forming 4,873 groups of orthologous proteins (COG). Individual COG are clustered in 20 individual functional categories, which are further grouped in four major classes (see Table 1).
Table 1. COG functional categories and category correlation with total number of ORFs.
| Functional class | Individual functional categories | Correlation* | Normalized species† | >2,000 ORFs | All species‡ |
|---|---|---|---|---|---|
| Information | J: Translation, ribosomal structure and biogenesis | − | 0.99, <0.001 | 0.95, <0.001 | 0.98, 0.001 |
| K: Transcription | + | 0.44, <0.001 | 0.18, 0.001 | 0.37, <0.001 | |
| L: DNA replication, recombination, and repair | − | 0.21, <0.001 | 0.19, 0.002 | 0.24, 0.004 | |
| Cellular processes | D: Cell division and chromosome partitioning | − | 0.41, <0.001 | 0.55, <0.001 | 0.37, <0.001 |
| V: Defense mechanisms | No | −0.096 | 0.20 0.001 | −0.38 | |
| O: Posttranslational modification, protein turnover | No | 0.13, 0.002 | −0.66 | 0.29 <0.001 | |
| M: Cell envelope biogenesis, outer membrane | No | −0.19 | −0.26 | −0.40 | |
| P: Inorganic ion transport and metabolism | No | 0.18, <0.001 | −0.60 | 0.1 0.001 | |
| U: Intracellular trafficking, secretion | No | 0.15, 0.001 | −0.36 | 0.27 <0.001 | |
| N: Cell motility | + | 0.1, 0.004 | 0.16, 0.001 | −0.64 | |
| T: Signal transduction mechanisms | + | 0.55, <0.001 | 0.20, <0.001 | 0.46, <0.001 | |
| Metabolism | F: Nucleotide transport and metabolism | − | 0.44, <0.001 | 0.57, <0.001 | 0.53, <0.001 |
| G: Carbohydrate transport and metabolism | No | −0.015 | −0.44 | −0.023 | |
| E: Amino acid transport and metabolism | No | 0.29, <0.001 | −0.09 | 0.07 0.005 | |
| H: Coenzyme metabolism | No | −0.23 | −0.05 | 0.11 0.0006 | |
| I: Lipid metabolism | No | −0.04 | 0.15, 0.002 | −0.14 | |
| C: Energy production and conversion | + | 0.1, 0.004 | 0.15 0.002 | −0.29 | |
| Q: Secondary metabolites transport and metabolism | + | 0.30, <0.001 | 0.12, 0.005 | 0.31, 0.001 | |
| Poorly characterized | R: General function prediction only | No | −0.012 | −0.48 | 0.06, 0.008 |
| S: Function unknown | No | 0.4, <0.001 | − <0.86 | 0.24 <0.001 |
The 20 functional categories (second column) are grouped in four major classes (first column) (adapted from the COG web site).
The genomic fraction attributable to a functional category showed universal positive (+), negative (−), or no (No) correlation with total ORFs in the genome when both correlations for normalized genomes (fourth column), and normalized genomes with >2,000 ORFs (fifth column) were significant at a P value threshold of 0.01 (P value denotes the confidence level that the correlation observed is significantly different from the null hypothesis, e.g., no correlation).
Power correlation R2 and P values for each set are shown. The power correlation gave among the highest R2 values from the types of correlations tested for most functional categories. It should be mentioned, however, that there were typically very small differences among different models (e.g., linear, power, logarithmic, etc.) in their ability to describe the trends with total ORFs in the genome (data not shown). Thus, no assumptions can be made about the mechanisms underlying the relationship between functional gene content and total ORFs in the genome.
The sixth column shows correlations for all 99 bacterial genomes used in this study. The 16 archaeal genomes were not included in the analysis, because Archaea had significantly different genomic fractions from Bacteria in many functional categories.
All possible ORFs from the 115 genomes were assigned to a functional category according to the category where their best COG homolog is classified. Homologs were identified by using the blast local alignment algorithm (12) and a cut-off of at least 30% identity at the amino acid level over 70% of the length of the query protein in pair-wise sequence comparisons. This cut-off is above the twilight zone of similarity searches where inference of homology is error-prone due to low similarity between aligned sequences; thus query proteins were presumably homologous to their COG match (13, 14). Homologous proteins can be either orthologs (homology through speciation) or paralogs (homology through lineage specific gene duplication), and both paralogs and orthologs are assumed to retain the same biochemical function, whereas paralogs have usually diverged in specificity (15, 16). Therefore, ORFs are expected to share at least the same general function with their COG matches. PERL scripts were used to edit ORF assignments where necessary; formatting databases for blast searches and automatically parsing blast outputs.
We further tested our findings from the COG database by using the publicly available data from the ortholog group table database at the Kyoto Encyclopedia of Genes and Genomes (KEGG) and the Comprehensive Microbial Resource database (CMR) supported by The Institute for Genomic Research (TIGR). The KEGG database classifies orthologous genes from all sequenced species into 24 functional categories (17). An identical strategy as previously mentioned for COG was used to assign each ORF from 75 fully sequenced genomes (the same genomes used for TIGR data below) to a KEGG functional category. TIGR performs an automated whole-genome annotation on any published microbial genome, which classifies genes in 19 redundant Role Categories (or functional categories), i.e., a single protein can be assigned in more than one category (18). The number of proteins devoted to a Role Category for each of the 75 genomes incorporated in CMR as of July 2002 was obtained from the Multi Genome Query Tool at the CMR web site (www.spacetransportation.org/Detailed/44108.html).
The amount of noncoding DNA in any genome was calculated by subtracting the sum of the lengths of the coding sequences annotated in the GenBank files from the estimated size of the genome.
Results and Discussion
With the previously described strategy, we were able to assign, on average, 70.3% of the ORFs in any genome to a COG functional category. If one considers that a significant amount of predicted genes (≈15-20%) is species-specific in every genome sequenced so far (19), we have characterized the large majority of the repertoire of each cell.
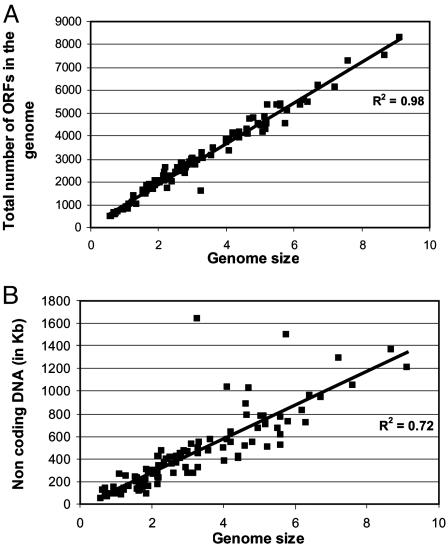
Data Normalization. Our main objective was to study the relationship between the total ORFs in the genome and the genomic fraction devoted to a functional category. To normalize the effect of the different degrees of representation in the database, genomes with too many or too few genes homologous to the database were not included in inferring patterns with genome size, i.e., genomes in which the percentage of genes homologous to the database fell within one standard deviation from the mean (x̄ 70.3%, SD 11.2) are represented by solid squares (87 of the 115 genomes), whereas the rest are represented by open squares (Fig. 1). Functional categories showed similar trends with total ORFs in the genome both when the normalized set and all genomes were considered (Table 1). However, trends with the normalized set should be more accurate because this set minimizes the bias in database representation. The use of genome size instead of total ORFs in the genome gave identical results due to the high correlation (R2 = 0.98) between these two parameters of the genome (Fig. 2A). Therefore, total ORFs in the genome and genome size are used interchangeably in the following text.
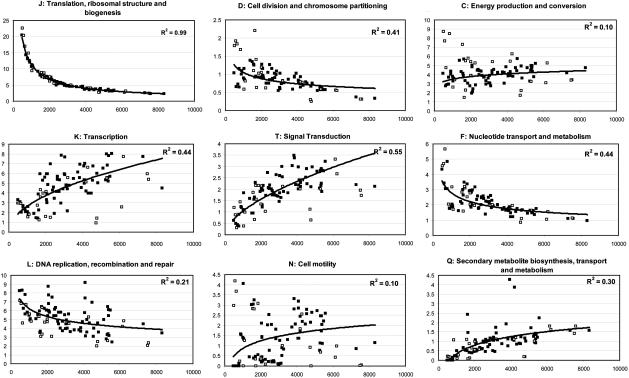
Fig. 1.
COG functional categories that showed universal correlation with total ORFs in the genome. y axes are the percent of ORFs in the genome attributable to a specific COG category (graph title), and x axes are the total ORFs in the genome for each of the 99 fully sequenced bacterial genomes. Solid squares represent genomes that had a reasonable number of genes with homologs in the COG database, whereas open squares represent genomes that had either too many or too few genes with homologs in the database (outliers). Trendlines and R2 shown are for the solid squares. Archaeal genomes were not included because Archaea had significantly different genomic fractions from Bacteria in many functional categories.
Fig. 2.
Correlation among total number of ORFs in the genome, noncoding DNA, and genome size for prokaryotic genomes. (A) The total number of ORFs in the genome vs. the genome size for 115 completed prokaryotic genomes. (B) The total amount of noncoding DNA in the genome vs. genome size.
Major Trends with Genome Size. To identify major universal trends, as opposed to ones that are attributable to the preferential gene loss in the reduced genomes, the analysis was repeated including only normalized genomes that had at least 2,000 ORFs annotated in their genomic sequences. COG functional categories that showed correlation with genome size for both sets tested (i.e., all solid squares and solid squares with 2,000 ORFs) were considered cases of major trends, and these categories are shown in Fig. 1. Categories that showed correlation with genome size (at a P value threshold of 0.01) for only one of the two sets of genomes tested were considered cases of minor trends and are not shown for simplicity (but presented as Fig. 6, which is published as supporting information on the PNAS web site). All findings are summarized in Table 1.
The COG functional categories that showed universal correlation with genome size were: informational categories of translation, ribosomal structure and biogenesis, and DNA replication recombination and repair. These categories showed a strong negative correlation with genome size, whereas transcription (transcription apparatus and transcription control genes) showed a strong positive correlation (Fig. 1 Left). Of the cellular processing categories, the percent of genes related to cell division and chromosome partitioning category showed a small decrease with genome size (≈1-2%), whereas the percent of genes related to signal transduction mechanisms and cell motility strongly and moderately increased with genome size, respectively (Fig. 1 Center). Among the individual metabolism categories, nucleotide transport and metabolism showed a strong negative correlation with genome size, whereas energy production and conversion and secondary metabolite biosynthesis, transport, and catabolism showed a moderate and strong positive correlation with genome size, respectively (Fig. 1 Right). Notably, genomes with <2,000 ORFs have almost no secondary metabolism-related genes (Fig. 1 Right).
Minor Trends with Genome Size. Categories of posttranslational modification and protein turnover, inorganic ion transport and metabolism, intracellular trafficking and secretion, amino acid transport and metabolism, and function unknown categories showed correlation only when all solid squares were considered, i.e., no correlation for solid squares with >2,000 ORFs (Table 1). Therefore, these trends are attributable to the preferential gene loss in the reduced genomes. Furthermore, several categories that were universally correlated with total ORFs in the genome showed stronger correlation with all solid squares compared to solid squares with >2,000 ORFs. Thus, such categories like transcription, signal transduction, and secondary metabolite biosynthesis are also affected by preferential gene loss in the reduced genomes. These results are in good agreement with the current knowledge of which functional categories are more likely to have been reduced in the symbiotic genomes.
On the other hand, categories of defense mechanisms and lipid metabolism showed correlation only when solid squares with >2,000 ORFs were considered. These trends, however, are more likely a database artifact due to the underrepresentation of large genomes than a real preferential accumulation of such genes by the large genomes. The fact that there were several small genomes with high percentages of ORFs devoted to these categories (which accounted for the lack of correlation when all solid squares were considered) supports the former interpretation. Last, it should be mentioned that most minor trends involved weak correlations and small changes (≈1-2%) in the fraction of the genome devoted to the corresponding functional categories.
Noncoding DNA and Hypothetical ORFs. Interestingly, the genomic fraction assigned to hypothetical ORFs (i.e., poorly characterized categories) remained constant for genomes with >2,000 ORFs. Moreover, the fraction of noncoding DNA was also invariable (at ≈12-14% of the genome) for all 115 genomes evaluated (Fig. 2B), which confirmed previous results that analyzed a smaller set of species (20). Therefore, the large prokaryotic genomes overall are not explained by disproportionate accumulation of junk DNA, i.e., hypothetical genes or noncoding sequence.
In contrast, genomes with <2,000 ORFs have a smaller percent of function unknown (or conserved hypothetical) ORFs compared to larger genome-sized species. This suggests that some of these genes, if they indeed code for proteins, have dispensable functions in the larger genome-sized bacteria. If these genes follow the trends of the other functional categories, then these unknown genes may be involved in regulation or secondary metabolism rather than in informational processes. Nonetheless, a significant fraction (≈3%) of the genes in the reduced genomes remains attributable to the function unknown category. Their retention suggests that at least some of the conserved hypothetical genes encode for functional proteins.
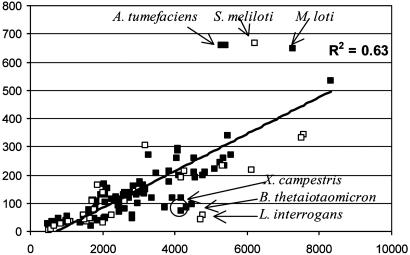
Factors Other than Genome Size. The correlation R2 values indicate that genome size can only partially explain some of the shifts in gene content. Strain-specific traits are assumed to be responsible for datapoint dispersion around the mean, which is pronounced for several functional categories. For example, by examining individual COG, we conclude that the number of the prevalent ABC transporter genes (and transport genes in general) was proportionately increased (i.e., the genomic fraction devoted to them remained constant) with genome size, and there was little dispersion around the mean suggesting a universal relationship with genome size (Fig. 3). However, specific bacterial groups like the ecologically versatile α-Proteobacteria Agrobacterium and Mesorhizobium sp. had a disproportionately increased number of ABC transporters, whereas the more habitat-specific bacteria like the γ-Proteobacteria Xanthomonas sp. had fewer than the average ABC transporters.
Fig. 3.
ABC transporter genes proportionately increase with genome size. y axis is the number of genes attributable to ABC transporter functions, and x axis is the total ORFs in the genome for each of the 99 fully sequenced bacterial genomes. Genomes that have disproportionately increased or decreased their number of ABC transporter genes are denoted on the graph.
As far as traits other than total ORFs in the genome are concerned, we evaluated whether the ribosomal rRNA (rrn) copy number could explain some of the shifts in functional gene content. The rrn copy number had, typically, a small effect on functional gene content compared to the total ORFs in the genome. However, in the case of carbohydrate transport and metabolism, the correlation was stronger for rrn copy number (R2 = 0.4, P < 0.001) than for total ORFs in the genome (correlation not significant at P = 0.01). The rrn copy number is positively associated with the rate at which phylogenetically diverse bacteria respond to resource availability (21), thus the strong correlation between carbohydrate metabolism and transport and rrn copy number is not surprising.
Last, the higher variability observed for data points representing small genomes is partially attributable to the fact that a small genome will show a dramatic change in functional patterns with a small change in the number of genes for a cellular process. Thus, while analyzing the percent of genes in a functional category can reveal major changes, it is less sensitive for detecting changes among large genome-sized prokaryotes.
Results from KEGG and TIGR Annotation Databases. Results using COG, KEGG, and TIGR databases are not always directly comparable because of database-specific characteristics. Although the KEGG Orthology database performs high-quality annotation, it has incorporated a limited (only the well-described) number of pathways and processes (17). Thus, more orthologous groups can be found in COG than in the KEGG database. With respect to TIGR annotation, although assignment of correct function is usually satisfactory (≈90%), ≈50% of the genes in a genome remain unassigned or are assigned to poorly characterized categories (vs. ≈40% for COG) (18). Moreover, as noted on the CMR web site, all Role Category data were generated at the time each genome was entered into the CMR; thus newer genomes may have more genes assigned to Role Categories than older ones. Despite these limitations, there are several categories that are comparable among the three databases and hence can be used to test the validity of the trends revealed with COG. Our results for these categories were congruent (a selected set of KEGG and TIGR's functional categories is presented as Fig. 7, which is published as supporting information on the PNAS web site). For example, KEGG and TIGR informational categories of protein translation and DNA replication were negatively correlated with genome size (R2 > 0.4 for all categories), whereas regulation category was positively correlated with genome size (R2 > 0.52), similar to the COG data.
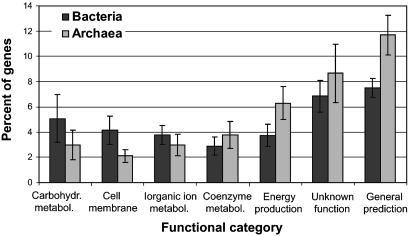
Bacteria vs. Archaea. Our analysis also revealed that there were some notable but small differences between Bacteria and Archaea in the relative usage of the genome for the different cell functions (Fig. 4). Archaea appeared to have a higher genomic portion devoted to energy production and conversion, coenzyme metabolism, and poorly characterized categories than their bacterial counterparts of the same genome size. On the other hand, Archaea had relatively fewer genes involved in carbohydrate transport and metabolism, cell envelope and membrane biogenesis, and inorganic ion transport and metabolism. Some of the differences, like those concerning energy production, cell envelope, and general prediction-only categories were more strongly supported by the data (compare errors bars in Fig. 4).
Fig. 4.
Differences between Archaea and Bacteria in the relative usage of the genome. Bars represent the average from 34 bacterial and 12 archaeal genomes, which have between 1,500 and 3,500 ORFs (to avoid any genome size effect on the data). Only normalized genomes have been included (see text). Averages are statistically different by two-tailed t test, assuming unequal variances and 0.05 confidence level. Functional categories that had <2% of the genes in the genome are not shown.
A set of archaeal specific proteins in addition to the standard proteins encountered in a typical prokaryotic cell would explain the higher genomic fraction in the above categories for Archaea. In agreement with this hypothesis, Graham et al. (22), in an attempt to define an archaeal genomic signature, concluded that genes with no detectable bacterial or eukaryotic homologs mostly involve energetic systems and cofactor biosynthesis, e.g., genes involved in methanogenesis. On the other hand, the fewer genes for cell-wall biogenesis are probably attributable to the fact that Archaea possess a different cell wall from Bacteria. Archaea lack peptidoglycan in their cell wall, and peptidoglycan biosynthesis requires a battery of enzymes in bacteria (23). Furthermore, the archaeal cell wall components and metabolism have not been studied to the same extent as those for Bacteria and hence are missing from the database.
Joint Genome Institute (JGI)'s Species Sequenced to High Draft. We also analyzed the 39 partially sequenced genomes in the JGI database in the same way. This is a collection of exclusively environmental strains, which includes seven strains with genome sizes >6 Mb (average genome size, 3.83 vs. 3.23 Mb in the closed set). Although trends between gene functional categories and total ORFs in the genome for JGI genomes were very similar to those for the fully sequenced genomes (data not shown), only 59.8% (vs. 70.3% for the closed set) of the ORFs in the JGI set were assignable to a COG category. This may indicate that this genome set samples more of the uncharacterized genes in nature, although some of the difference is likely due to the lack of manual curation of the annotation.
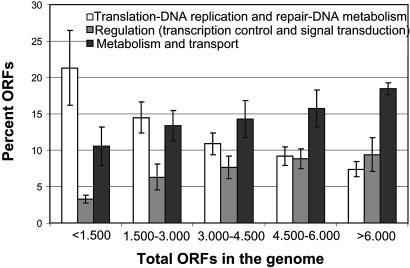
What Is Gained in a Large Genome? Our analysis showed that larger genomes preferentially accumulate regulation, secondary metabolism, and, to a smaller degree, energy conversion-related genes as opposed to informational ones, judging from the inverse pattern for these classes with genome size (Fig. 5). We performed the same analysis in May of 2002, using the 75 genomes available at that time and a database of 3,852 COG groups (vs. 4,873 COG currently). The results between this set and the expanded set of 115 genomes presented herein were very consistent, and correlations were often more significant in the latter set. This consistency gives higher confidence in the trends reported.
Fig. 5.
Summary of the shifts in gene content with genome size in prokaryotic genomes. The bars represent the sum of the COG functional categories, which showed strong correlation with genome size and are involved in the same major cellular processes. Only normalized genomes (represented by solid squares in Fig. 1) have been included. Errors bars represent the standard deviation from the mean except for the last genome size class, where error bars represent data range due to a small number of normalized genomes in this class (three genomes).
These data suggest that secondary metabolism and energy conversion rather than general metabolism are disproportionately expanded in larger genomes and thus should explain a large part of the broad metabolic diversity that characterizes large genome-sized species. The expansion involved both expansions of specific COG and de novo acquisitions of new COG (or pathways), with the latter case being roughly twice as frequent as the former one (data not shown). On the other hand, the genes assignable to the remaining metabolism, except nucleotide metabolism, and several cellular processes categories are only proportionally increased with genome size (similar to the example of ABC transporter genes mentioned previously).
Regardless of a proportional or disproportional increase in metabolic or cellular pathways, large genome-sized species would need increased regulation to successfully control the extensive metabolic repertoire they apparently possess under different growth conditions. Thus, it is not surprising that regulatory genes, i.e., transcription control, and signal transduction, dominated the genes that are disproportionately increased in larger genomes. In addition, many regulation systems are expected to cross talk, because their genes share high sequence similarity (paralogous genes of expanded gene families), which suggests increased complexity in regulation as well. In agreement with these interpretations, all species with genome sizes >6 Mb in our set are free-living bacteria that can grow in very diverse environments, several using alternative electron acceptors and a great range of substrates for energy production (Table 2).
Table 2. Genomic information and ecological niche(s) of species with a genome size > 6 Mb.
| Species | Genome size | Percent in COG | Ecological niche |
|---|---|---|---|
| Bacteroides thetaiotaomicron* | 6.26 | 33.5 | Human gut, metabolically versatile |
| Bradyrhizobium japonicum | 9.11 | 60.4 | Soil, rhizosphere; N2 fixing symbiont of legumes |
| Mesorhizobium loti | 7.59 | 69 | Soil, rhizosphere; N2 fixing symbiont of legumes |
| Nostoc sp. | 7.2 | 58.2 | Cyanobacteria, ubiquitous in nature; photosynthetic |
| Pseudomonas sp. (average of three strains) | 6.2-6.4 | 69-80 | Soil, water; opportunistic pathogens of plants and humans |
| Sinorhizobium meliloti | 6.7 | 63 | Soil, mizosphere; N2 fixing symbiont of legumes |
| Streptomyces avermitilis | 9.03 | 48.8 | Ubiquitous in soil; very versatile metabolically |
| Streptomyces coelicolor | 8.67 | 40 | Ubiquitous in soil; very versatile metabolically |
All environmental and nonproteobacteria strains (bold) have <58.2% (vs. an average of 70.3%) of their genes homologous to COG proteins (third column). This indicates that the overrepresentation of specific lineages (e.g., proteobacteria) and clinical strains in the database has possibly biased our knowledge of microbial functional gene content.
The negative correlation with genome size of informational and DNA metabolism categories is equally interesting (Figs. 1 and 5). This trend suggests that a similar number of informational and DNA metabolism related proteins is able to cope with an increased number of genes. For instance, there is a relatively small increase in the absolute number of genes (of ≈20%) in the translation category between 2- and 8-Mb-sized genomes. This may be attributable to there being sufficient informational processes present and active at any time in the cell. Thus, when there is an unusual demand for informational proteins because of a larger genome, their transcription or posttranslational modification can be regulated accordingly to yield sufficient more active proteins.
A Hypothesis for Large Genomes. Presumably the interactions between the organism and particular habitat(s) have selected for genome expansion. Large genomes do not appear to be uncommon in nature (Table 2 and JGI genomes), and hence they must have value. As noted above, all overamplified gene families are associated directly or indirectly (regulation) with metabolism. However, the lack of knowledge of the population sizes and activities of such species in natural environments does not allow specific inferences about which environmental factors may have fostered genome expansion. In contrast, the genome evolution in endosymbiotic bacteria is much better understood. The relief from selection for specific pathways and regulation systems along with population bottlenecks that allow more rapid fixation of mutations are proposed to determine their genome evolution (1, 20, 24). Also, the higher number of bacterial generations in these nonnutrient-limiting environments probably facilitates loss of DNA through spontaneous recombination events at repeated or mobile sequences (1, 24).
One hypothesis for large genomes consistent with the above data is that Bacteria with such genomes are more ecologically successful in environments where resources are scarce but diverse and where there is little penalty for slow growth. These are characteristics of soil. In support of this, Mitsui et al. (25) and Klappenbach et al. (21) found slow-growing oligotrophic α-Proteobacteria to be more dominant in soil. In the former study, many of these isolates were nonsymbiotic members of the Rhizobiaceae and Bradyrhizobiaceae (25, 26), families that have genomes >6-8 Mb. Generation times in soil are thought to be low, with mean generations measured at three per year (27).
Although this study shows some clear trends between gene content and genome size, the dispersion around the mean for many categories suggests that features other than genome size likely explain what is gained in larger genomes. These traits need to be explored for a fuller understanding of the interactions between ecology and genome evolution. This study also draws attention to the limited number of large genomes sequenced to date. The possibility that large genomes represent a significant fraction of the extant microbial world and that they may possess unique traits missed in the current annotation knowledge is a major challenge for microbiologists.
Supplementary Material
Acknowledgments
We thank Tom Schmidt, Rebecca Grumet, Joel Kapplenbach, Frank Larimer, and an anonymous reviewer for helpful discussions regarding the manuscript. This work was supported by the Bouyoukos Fellowship Program (K.T.K.), the U.S. Department of Energy's Microbial Genome Program, and the Center for Microbial Ecology.
Abbreviations: COG, Clusters of Orthologous Groups; KEGG, Kyoto Encyclopedia of Genes and Genomes; CMR, Comprehensive Microbial Resource; TIGR, The Institute for Genomic Research; rrn, ribosomal rRNA; JGI, Joint Genome Institute.
References
- 1.Andersson, S. & Kurland, C. (1998) Trends Microbiol. 6, 263-268. [DOI] [PubMed] [Google Scholar]
- 2.Galperin, M. & Koonin, E. (1999) Genetica 106, 159-170. [DOI] [PubMed] [Google Scholar]
- 3.Moran, N. (2002) Cell 108, 583-586. [DOI] [PubMed] [Google Scholar]
- 4.Andersson, S., Zomorodipour, A., Andersson, J., Sicheritz-Pontent, T., Alsmark, U., Podowski, R., Naslund, A., Eriksson, A., Winkler, H. & Kurland, C. (1998) Nature 396, 109-110. [DOI] [PubMed] [Google Scholar]
- 5.Fraser, C, Gocanye, J., White, O., Adams, M., Clayton, R., Fleischmann, R., Bult, D., Kerlavage, A., Sutton, G., Kelly, J., et al. (1995) Science 270, 397-403. [DOI] [PubMed] [Google Scholar]
- 6.Shigenobu, S., Watanabe, H., Hattori, M., Sakaki, K. & Ishikawa, H. (2000) Nature 7, 81-86. [DOI] [PubMed] [Google Scholar]
- 7.Jordan, I., Makarova, K., Spouge, J., Wolf, Y. & Koonin, E. (2001) Genome Res. 11, 555-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bentley, S., Chater, K., Cerdeno-Tarraga, A.-M., Challis, G., Thompson, N., James, K., Harris, D., Quail, M., Kieser, H., Harper, D., et al. (2002) Nature 9, 141-147. [DOI] [PubMed] [Google Scholar]
- 9.Stover, C., Pham, X., Erwin, A., Mizoguchi, S., Warrener, P., Hickey, M., Brinkman, F., Hufnagle, W., Kowalik, D., Lagrou, M., et al. (2000) Nature 406, 959-964. [DOI] [PubMed] [Google Scholar]
- 10.Tatusov, R., Koonin, E. & Lipman, D. (1997) Science 278, 631-637. [DOI] [PubMed] [Google Scholar]
- 11.Tatusov, R., Fedorova, N., Jackson, J., Jacobs, A., Kiryutin, B., Koonin, E., Krylov, D., Mazumder, R., Mekhedov, S., Nikolskaya, A., et al. (2003) BMC Bioinformatics 4, 41-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altschul, S., Madden, T., Schäffer, A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sander, C. & Schneider, R. (1991) Proteins 9, 56-58. [DOI] [PubMed] [Google Scholar]
- 14.Rost, B. (1999) Protein Eng. 12, 85-94. [DOI] [PubMed] [Google Scholar]
- 15.Eisen, J. A. (1998) Genome Res. 8, 163-167. [DOI] [PubMed] [Google Scholar]
- 16.Gerlt, J. & Babbitt, P. (2001) Annu. Rev. Biochem. 70, 209-246. [DOI] [PubMed] [Google Scholar]
- 17.Kanehisa, M. & Goto, S. (2000) Nucleic Acids Res. 28, 27-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peterson, J., Umayam, L., Dicknson, T., Hickey, E. & White, O. (2001) Nucleic Acids Res. 29, 123-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson, K., Paulsen, I., Heidelberg, J. & Fraser, C. (2000) Nat. Biotechnol. 18, 1049-1054. [DOI] [PubMed] [Google Scholar]
- 20.Mira, A., Ochman, H. & Moran, N. (2001) Trends Genet. 17, 589-596. [DOI] [PubMed] [Google Scholar]
- 21.Klappenbach, J., Dunbar, J. & Schmidt, T. (2000) Appl. Env. Microbiol. 66, 1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graham, D., Overbeek, R., Olsen, G. & Woese, C. (1999) Proc. Natl. Acad. Sci. USA 97, 3304-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konig, H. (1988) Can. J. Microbiol. 34, 395-406. [Google Scholar]
- 24.Frank, C. A., Amiri, H. & Andersson, S. (2002) Genetica 115, 1-12. [DOI] [PubMed] [Google Scholar]
- 25.Mitsui, H., Gorlach, K., Lee, H., Hattori, R. & Hattori, T. (1997) J. Microbiol. Methods 30, 103-110. [Google Scholar]
- 26.Saito, A., Mitsui, H., Hattori, R., Minamisawa, K. & Hattori, T. (1998) FEMS Microbiol. Ecol. 25, 277-286. [Google Scholar]
- 27.Grey, T. & Willimas, S. (1971) Symp. Soc. Gen. Microbiol. 21, 255-286. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.