Abstract
Leber congenital amaurosis (LCA) is a blinding retinal disease that presents within the first year after birth. Using exome sequencing, we identified mutations in the nicotinamide adenine dinucleotide (NAD) synthase gene NMNAT1 encoding nicotinamide mononucleotide adenylyltransferase 1 in eight families with LCA, including the family in which LCA was originally linked to the LCA9 locus. Notably, all individuals with NMNAT1 mutations also have macular colobomas, which are severe degenerative entities of the central retina (fovea) devoid of tissue and photoreceptors. Functional assays of the proteins encoded by the mutant alleles identified in our study showed that the mutations reduce the enzymatic activity of NMNAT1 in NAD biosynthesis and affect protein folding. Of note, recent characterization of the slow Wallerian degeneration (Wlds) mouse model, in which prolonged axonal survival after injury is observed, identified NMNAT1 as a neuroprotective protein when ectopically expressed. Our findings identify a new disease mechanism underlying LCA and provide the first link between endogenous NMNAT1 dysfunction and a human nervous system disorder.
The molecular mechanisms underlying neurodegeneration are highly complex, and strategies to prevent this process are under intense investigation1,2. Important insights into protection from neurodegeneration come from studies of the Wlds mouse model, which harbors a spontaneous mutation and has a marked reduction in the rate of axonal degeneration after injury3–7. This protective effect is due to the expression of a unique chimeric protein, named the Wlds fusion protein, that is composed of ubiquitination factor E4B (Ube4b) and full-length Nmnat1 (refs. 8,9), a highly conserved protein that is present throughout evolution from archaebacteria to humans. Recent work has shown that the Nmnat1 portion of the Wlds fusion protein is responsible for the observed delay in axonal degeneration10. NMNAT1 is an essential enzyme in NAD biosynthesis. Despite the intense interest in and importance of NMNAT1, the mechanism by which NMNAT1 protects against neurodegeneration remains controversial. Although the enzymatic activity does not seem to be necessary for neuroprotection in Drosophila melanogaster11, both NAD synthase domain activity and the presence of other domains are needed in vertebrates for the neuroprotective function of NMNAT1 (ref. 12). In addition, little is known about the endogenous function of NMNAT1 in neuronal cells in vertebrates. Nmnat1 mutation resulting in complete loss of function in mice results in embryonic lethality, whereas heterozygous loss of Nmnat1 does not cause acceleration of neurodegeneration after injury13. To date, no mutations in NMNAT1 have been associated with any human disease. Here, we report that biallelic NMNAT1 mutations cause LCA, a severe neurodegenerative condition of the retina, which is the most active metabolic tissue of the human body14.
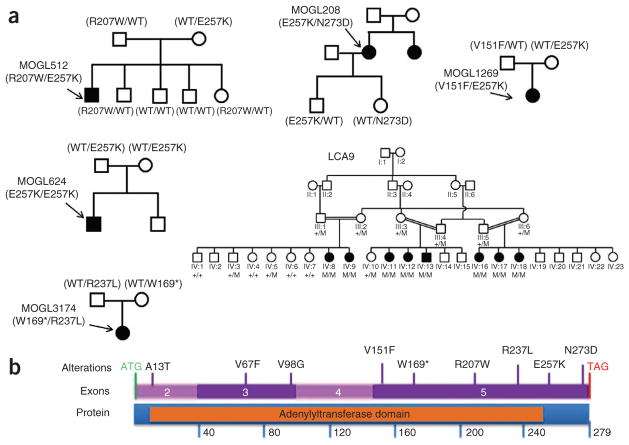
Early-onset neurodegeneration in the human retina can lead to LCA (MIM 204000), the most severe human form of inherited photoreceptor-neuron degeneration resulting in congenital blindness, with an incidence of ~1 in 80,000 (ref. 15). LCA is genetically and clinically heterogeneous, with mutations in 16 genes implicated so far in the disease, explaining 70% of cases. The proteins encoded by these genes are involved in a range of functions, including ciliary transport, phototransduction, retinoid cycling and photoreceptor development15–17. To identify new LCA-associated genes underlying the remaining 30% of unresolved LCA cases, we performed whole-exome sequencing experiments. We first screened individuals with LCA for mutations in all known LCA-associated genes using APEX technology (Asper Ophthalmics) and Sanger sequencing. We then captured the exomes of the first 50 individuals with LCA who lacked mutations in known LCA-causing genes as described18. Next- generation sequencing of these samples identified three unrelated individuals with LCA (MOGL208, MOGL512 and MOGL1269) with compound heterozygous variants in NMNAT1 (NM_022787). All three of these individuals have the same p.Glu257Lys missense variant (Table 1). Additionally, subject MOGL208 carries the missense variant p.Asn273Asp, subject MOGL512 carries the missense variant p.Arg207Trp and subject MOGL1269 carries the missense variant p.Val151Phe. Cosegregation of the alleles encoding these variants in the pedigrees (Fig. 1) and the absence of these mutations in 200 normal controls indicate that NMNAT1 is likely to be a new LCA-causing gene. To confirm this finding, we sequenced the coding region of NMNAT1 in 150 additional individuals with LCA. Another four affected individuals were found to carry either compound heterozygous or homozygous mutations in NMNAT1 (Fig. 1 and Table 1). Notably, NMNAT1 maps to 1p36 within the LCA9 locus that was first identified through linkage analysis in a large consanguineous Pakistani family with LCA, making it an excellent candidate gene. Indeed, in a parallel sequencing experiment, we identified a homozygous NMNAT1 mutation (encoding p.*280Gln) that segregated perfectly with the disease in the original family in which LCA9 was identified19 (Fig. 1). Thus, in total, we identified ten mutant alleles of NMNAT1 in eight families with LCA, including a nonsense mutation (encoding p.Trp169*), a read-through mutation (encoding p.*280Gln) and eight missense mutations that are likely to cause partial loss of function (Supplementary Fig. 1a, b and Supplementary Table 1). We validated all mutant alleles by direct PCR Sanger sequencing (Supplementary Fig. 2).
Table 1.
NMNAT1 mutations and clinical phenotypes in individuals with LCA
| MOGL number | Age (years) | Allele 1 | Allele 2 | Clinical information | Nationality |
|---|---|---|---|---|---|
| 208 | 56 | c.769G>A (p.Glu257Lys) | c.817A>G (p.Asn273Asp) | NLP and CF vision, hyperopia, nystagmus, pseudophakia | French Canadian |
| 512 | 13 | c.619C>T (p.Arg207Trp) | c.769G>A (p.Glu257Lys) | CF vision, hyperopia, nystagmus | French Canadian |
| 1269 | N/A | c.451G>A (p.Val151Phe) | c.769G>A (p.Glu257Lys) | N/A | European |
| 624 | 10 | c.769G>A (p.Glu257Lys) | c.769G>A (p.Glu257Lys) | NLP vision, hyperopia, nystagmus | Saudi Arabian |
| 3174 | 33 | c.507G>A (p.Trp169*) | c.710G>T (p.Arg237Leu) | 20/2000 vision, hyperopia, nystagmus | Irish |
| 3294 | 41 | c.199G>T (p.Val67Phe) | c.769G>A (p.Glu257Lys) | CF vision, hyperopia, nystagmus | Russian |
| 3698 | 10 | c.37G>A (p.Ala13Thr) | c.293T>G (p.Val98Gly) | CF vision, hyperopia, nystagmus | Haitian (African descent) |
| LCA9 | 2–30 | c.838T>C (p.*280Gln) | c.838T>C (p.*280Gln) | LP vision, congenital visual loss, nystagmus, nyctalopia, all (n = 8) have acquired macular colobomas, cataracts (PSCC), white retinal dots, retinal pigment, one has keratoconus | Pakistani |
All patients have macular colobomas. NLP, no light perception; LP, light perception; CF, counting fingers; N/A, not available.
Figure 1.
NMNAT1 variants identified in individuals with LCA. (a) NMNAT1 variants and the cosegregation of the alleles encoding these variants in pedigrees. Filled symbols indicate individuals with LCA. Arrows indicate affected individuals who were genotyped in this study. WT or +, wild type; M, mutant. (b) Gene and protein structures of NMNAT1. All variants found in individuals with LCA are indicated. The nicotinic acid mononucleotide adenylyltransferase (NMNAT) protein domain is shown in orange.
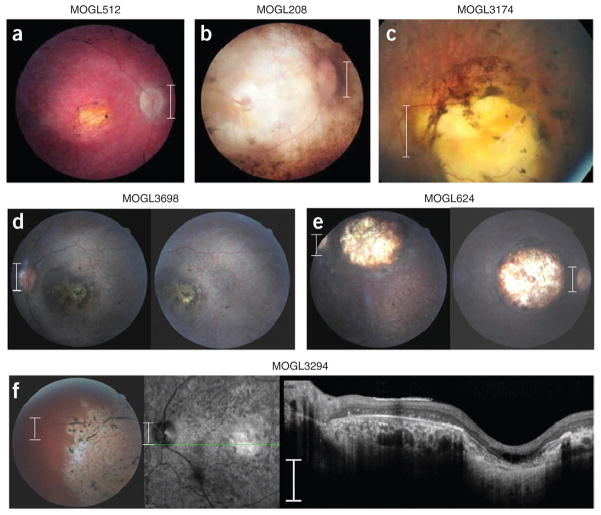
Because NMNAT1 is ubiquitously expressed, we re-examined the phenotype of all individuals with NMNAT1 mutations and interviewed children and parents about their general health. All individuals with biallelic NMNAT1 mutations had severe LCA but normal physical and mental health. Notably, in addition to the typical LCA phenotype of nystagmus, severe loss of vision and abnormal electroretinogram (ERG), all individuals with biallelic NMNAT1 mutations were found to have a peculiar, prominent retinal feature termed ‘macular coloboma’, which consists of an atrophic lesion in the central retina with a pigmented border, signifying complete loss of neural tissue in the fovea, including of photoreceptors, bipolar cells and ganglion cells (Fig. 2). This suggests that NMNAT1 mutations are associated with severe and rapid foveal degeneration. The rest of the retina was abnormal as well, with pigmentary changes, attenuated retinal blood vessels and optic disc pallor. In addition, other layers of the retina, such as the ganglion cell layer, were also severely affected (Fig. 2). Some infants with macular colobomas are misdiagnosed with parasitic Toxoplasma retinal infection, which has a similar presentation but a very different etiology and prognosis.
Figure 2.
Retinal phenotypes of six individuals with NMNAT1 mutations. All individuals in our study with NMNAT1 mutations were found to have prominent macular colobomas. Presented is the phenotypic spectrum of the colobomas found in our cohort. (a) The fundus of a 13-year-old individual with LCA from Quebec (MOGL512) harboring the p.Glu257Lys and p.Arg207Trp variants. (b) The fundus of a 56-year-old individual with LCA (MOGL208) with a very similar macular coloboma who harbors the p.Glu257Lys and p.Asn273Asp variants. (c) The fundus of a 32-year-old individual with LCA (MOGL3174) with a severe macular coloboma, misdiagnosed as a toxoplasmosis scar, harboring the p.Trp169* and p.Arg237Leu variants. (d) The macula of a 9-year-old girl from Haiti (MOGL3698) with macular colobomas harboring the p.Val98Gly and p.Ala13Thr variants. (e) The macula of a 10-year-old boy from Saudi Arabia (MOGL624) homozygous for the mutation encoding the p.Glu257Lys variant with deep colobomas. (f) The macula (left), optical coherence tomography (OCT) (center) and retinal periphery (right) of a 41-year-old Russian individual with LCA (MOGL3294) harboring the p.Val67Phe and p.Glu257Lys variants showing prominent macular colobomas, extensive retinal remodeling with abnormalities of all retinal layers and a well-defined boundary between normal and abnormal retina, as seen in several of the affected individuals. Scale bars in a–e and f (left, center), 1.5 mm; scale bar in f (right), 300 μm.
On the basis of our observation that five families carried the p.Glu257Lys variant, we tested whether the allele encoding this variant represents a founder mutation. Genotyping of surrounding SNPs and haplotype analysis confirmed that all individuals of European descent carrying the mutation encoding p.Glu257Lys share the same haplotype, strongly suggesting that this is a founder mutation (Supplementary Fig. 3). On the basis of this finding, we developed an amplification-refractory mutation system (ARMS) primer set that easily distinguishes between the wild-type and mutant alleles in one PCR reaction to facilitate rapid and inexpensive molecular diagnosis for newborns with LCA carrying NMNAT1 mutations and those with macular colobomas (Supplementary Fig. 1c).
Close examination of the position of altered residues in the NMNAT1 crystal structure suggests that its structure and function are likely to be affected by the mutations (Supplementary Fig. 1b). For example, two charged residues, Arg207 and Glu257, are located at a protein-interaction domain interface that has an important role during hexamerization of the enzyme. Substituting a hydrophobic tryptophan for a positively charged arginine (p.Arg207Trp) or a positively charged lysine for a negatively charged glutamic acid (p.Glu257Lys) is likely to directly interfere with hexamer formation. In addition, the Asn273 residue is in close proximity to the pyridine-binding site and is thought to be involved in the coordination of an active site water molecule and to have a role in substrate recognition (Fig. 1b and Supplementary Fig. 1b)20. Therefore, these missense mutations may negatively affect the function of the protein rather than altering NMNAT1 expression. Indeed, mutant alleles of NMNAT1 are expressed in subjects MOGL208 and MOGL512 (Supplementary Fig. 4).
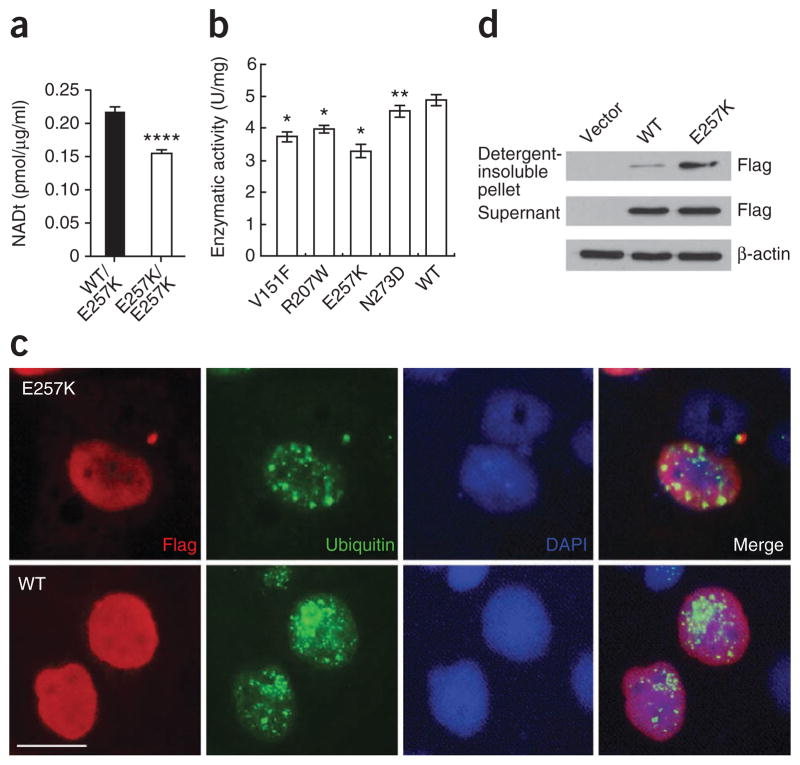
On the basis of these in silico predictions, we performed both in vivo and in vitro functional assays. We first assessed whether enzymatic function in NAD biosynthesis was negatively affected in vivo in red blood cells (RBCs) from an individual with LCA and his heterozygous mother. We obtained fresh RBCs from the MOGL624 family and measured the level of NAD production. We observed significantly lower concentrations of NAD in the affected individual, who is homozygous for the mutation encoding the p.Glu257Lys variant, compared to his heterozygous mother, suggesting reduced enzymatic function of the mutant NMNAT1 protein (Fig. 3a). To further examine the impact of these mutations on enzymatic activity, we performed in vitro assays using affinity-purified wild-type and mutant NMNAT1 proteins. Consistent with the in vivo findings, NMNAT1 proteins with p.Val151Phe, p.Arg207Trp, p. Glu257Lys and p.Asn273Asp alterations also had significantly reduced enzymatic activity compared to the wild-type protein in the in vitro assays (Fig. 3b). In parallel, we engineered the most common mutation found in our study (encoding p.Glu257Lys) into NMNAT1 cDNA. We transfected HeLa cells with the constructs encoding either wild-type or mutant protein and then examined protein expression by immunohistochemistry. Consistent with previous studies, we observed strong nuclear staining of NMNAT1 in cells transfected with the wild-type construct (Fig. 3c). Conversely, we observed strong staining of NMNAT1 p.Glu257Lys outside the cell nucleus in the cytoplasm (Fig. 3c). The mutant NMNAT1 was likely aggregated in the cytoplasm, as it was insoluble, and was positive for ubiquitin staining, indicating that the p.Glu257Lys substitution likely affects proper protein folding (Fig. 3c, d).
Figure 3.
Functional assays of mutant NMNAT1 proteins. (a) Total NAD levels in fresh RBCs from an affected individual homozygous for the mutation encoding p.Glu257Lys and his heterozygous mother. NAD is present at significantly lower levels in the affected proband compared with his carrier mother (****P < 0.0001). (b) HeLa cells were transfected with constructs encoding wild-type or mutant NMNAT1 fused with a Flag tag. Compared with wild-type (WT) NMNAT1, the p.Val151Phe, p.Arg207Trp, p.Glu257Lys and p.Asn273Asp variants had lower enzymatic activity. *P ≤ 0.0001, **P < 0.05. Results in a, b are given as mean ± s.d. (c) Cells transfected with constructs encoding Flag-tagged wild-type or p.Glu257Lys Nmnat1. Immunofluorescence staining using antibody to Flag and to ubiquitin showed that Nmnat1 p.Glu257Lys could form ubiquitin-positive protein aggregates in the cell. The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Scale bar, 20 μm. (d) Nmnat1 p.Glu257Lys forms protein aggregates in cells. Cells were transfected with constructs encoding Flag-tagged wild-type or p.Glu257Lys Nmnat1 or with vector alone. Protein blot results showed increased detergent-insoluble Nmnat1 protein in the cells transfected with the vector encoding Flag-tagged Nmnat1 p.Glu257Lys construct. β-actin was used as a loading control.
In summary, we have identified NMNAT1 as the disease-causing gene at the LCA9 locus and have provided the first link between a human nervous system disease and endogenous NMNAT1 function. As has been shown in Drosophila, human NMNAT1 seems to be an essential protein for photoreceptor neuron function and survival; however, the underlying mechanisms may be distinct6,11,12,21–23. Consistent with the idea that the protective effects of NMNAT1 are correlated with NAD production in the vertebrate model, our results suggest that LCA-causing NMNAT1 mutations reduce enzyme activity and lower NAD levels in affected individuals. However, as the molecular mechanisms of NMNAT1 in neuroprotection remain controversial and the intrinsic differences between the peripheral and central nervous systems are not yet fully understood, it is possible that mechanisms other than alterations in NAD production account for the retinal phenotypes observed in individuals with NMNAT1 mutations. Finally, it is worth noting that, as NAD supplementation has been shown to significantly delay axonal degeneration in the mouse after injury10, our work raises the possibility that increasing NAD levels in the retinas of individuals with LCA with NMNAT1 mutations may be a potential therapeutic option.
URLs
Exome sequencing data, https://www.hgsc.bcm.edu/content/retinaldisease; Primer3, http://frodo.wi.mit.edu/primer3/.
ONLINE METHODS
Study subjects
LCA samples were collected at the Montreal Children’s Hospital at the McGill University Health Centre (MUHC) and at the University of Leeds after obtaining informed consent. This study was approved by the McGill University Health Center Research Institute Research Ethics Board and by the Leeds Teaching Hospitals Trust Research Ethics Committee and adhered to the tenets of the declaration of Helsinki. Blood samples were collected from all available family members, and DNA was extracted with the Qiagen blood genomic DNA extraction kit following the manufacturer’s protocol.
Whole-exome capture sequencing and validation
We generated Illumina paired-end libraries according to the manufacturer’s protocol. Briefly, 1 μg of genomic DNA was sheared into fragments of approximately 300–500 bp in length. Fragments were end repaired, and an extra A base was added to the 3′ end. Illumina index adaptors were ligated to the fragments, and ten cycles of PCR amplification were applied to each sample after ligation. DNA was quantified by PicoGreen assay (Invitrogen). We used 3 μg of pre-capture library for each capture reaction. NimbleGen SeqCap EZ Hybridization and Wash kits were used for whole-exome capture following the standard manufacturer’s protocol. Captured libraries were quantified and sequenced on the Illumina HiSeq 2000 as 100-bp paired-end reads following the manufacturer’s protocols. Illumina sequencing was performed at the BCM-FGI core and at the McGill University and Genome Quebec Innovation Centre.
Sequence data were analyzed using the data analysis pipeline established at the BCM-HGSC as described18. Briefly, the fastq read files were aligned to the hg19 human reference genome using bwa24. Aligned reads were recalibrated and realigned using GATK25. SNPs were called using Atlas-SNP2 (ref. 26). The minimum posterior probability cutoff was set as 0.9, and the minimum number of variant reads was set as 3. Indels were called using Atlas-Indel developed at BCM-HGSC27. For each subject, all SNPs and indels were filtered against dbSNP130, 1000 Genomes Project and BCM-HGSC internal databases. Mutations were first given cDNA positions with nucleotide changes and were then described as amino-acid changes at residues in the protein. Genomic evolutionary rate profiling (GERP) scores were calculated with the GERP++ package in the UCSC Genome Browser hg19 version. Scores equal to or greater than 2 indicate high confidence of the constraint intensity. I-Mutant 3.0.6 and Mupro 1.1 were used to calculate the difference in the free energy changes (ΔΔG) from unfolding the wild-type versus the mutant protein. Negative ΔΔG (kcal/mol) indicates that the mutation can destabilize the protein.
To confirm mutations in affected individuals and their family members, we used a direct PCR sequencing approach. Primers were designed using Primer3 (see URLs). To ensure completeness and quality of the sequences, primers were designed from intronic sequences such that the entire exon and at least 50 bp of intronic DNA on each side of the exon were amplified for each exon.
NAD production experiments in fresh RBCs
We performed a NAD+/NADH assay in affected individuals and heterozygous family members with NMNAT1 mutations. Intracellular NADt (NAD+ and NADH) concentrations in RBCs were measured by colorimetric NAD(H) assays using the commercially available kit from BioVision (K337-100) according to the manufacturer’s instructions. Briefly, approximately 4 ml of fresh venous blood was collected from each of the subjects in lavender tubes containing EDTA as an anti-coagulant. RBC pellets (and white blood cell fractions) were obtained by centrifugation at 4 °C after which the supernatant was discarded. Pellets were washed twice with cold PBS, and cells were lysed in the recommended NAD/NADH extraction buffer (Biovision). Cell lysates were filtered through a filter with a 10-kDa molecular weight cutoff (BioVision, 1997–25). Samples (100 μl per time-point per biological replicate) were processed following the instructions, and all samples were processed in a single run to enhance quantification and comparability. Colorimetric measurements were made at 25 °C using optical density (OD)450 measurements on an Epoch microplate spectrophotometer (Biotek). We performed two technical replicates for each sample, which was collected twice. At each time point, OD450 measurements were converted to picomoles per microgram of protein per milliliter using a standard curve. Protein concentrations of cell lysates were determined using the Bio-Rad protein assay kit. For data analyses, we used the last six time points, as these represent times at which there are stable amounts of NAD. The mean value between two groups was compared by Student’s t test. Differences were considered to be statistically significant at P < 0.05.
Site-directed mutagenesis
Full-length human NMNAT1 cDNA was amplified from placenta RNA and cloned into PCR-blunt vector (Invitrogen). Site-directed mutagenesis was carried out on this plasmid using the QuikChange kit (Agilent). The resulting DNA was sequenced to confirm the nucleotide changes.
RT-PCR
RNA was extracted from the blood of affected individuals MOGL208 and MOGL512 using the PAXgene blood RNA kit. Reverse transcription was carried out using oligo(dT). NMNAT1-specific PCR was performed (Supplementary Table 2), and PCR products were sequenced using Sanger sequencing.
Immunofluorescence staining
Constructs encoding wild-type and mutant NMNAT1 were transfected into HeLa cells using Lipofectamine 2000 (Invitrogen). After 24–48 h, cells were fixed with a 1:1 methanol:acetone mixture at −20 °C for 10 min. After washing and blocking, cells were incubated with primary antibody overnight at 4 °C and were then incubated with fluorescent dye–conjugated secondary antibody at a 1:1,000 dilution for 1 h at room temperature. The primary antibodies used were an anti-Flag M2 antibody (1:200 dilution; F1804, Sigma), a rabbit antibody to Flag (1:200 dilution; F7425, Sigma) and an antibody to ubiquitin (P4D1) (1:200 dilution; sc-8017, Santa Cruz Biotechnology). DAPI counterstaining was performed to stain the nuclei. Vectashield mounting medium for fluorescence (Vector Laboratories) was used to mount the slides for observation.
In vitro enzymatic activity assays
The enzymatic activity of NMNAT1 was measured using a continuous coupled assay as previously described11,28. Briefly, the assay was performed at 37 °C in 100 μl of reaction solution containing 30 mM HEPES pH 7.4, 12 mM MgCl2, 16 mM Semicarbacide pH 7.4, 1.5 mM ATP, 0.625% ethanol, 15 U alcohol dehydrogenase, 0.625 mM NMN (Sigma) and affinity-purified NMNAT1 proteins. The production of NADH, represented by an increase in the absorbance at 340 nm, was monitored using a Synergy 2 reader (Biotek).
Differential detergent extraction assays
The procedure of fractioning cellular proteins on the basis of their solubility was performed as described with minor modification29. After transfection, cells were lysed with a lysis buffer containing 10 mM Tris, 1 mM EDTA pH 8.0, 150 mM NaCl, 1% NP-40, 0.1% DOC and proteinase inhibitors. Detergent-soluble (supernatant) and detergent-insoluble (pellet) fractions were collected and analyzed by protein blot.
Supplementary Material
Acknowledgments
We thank all of the individuals with LCA and their parents who were involved in this study. We thank R. Sifers for scientific discussion and J.E. Zaneveld for critical reading of the manuscript. R.K.K. is supported by the Foundation Fighting Blindness Canada, the Canadian Institutes for Health Research, the US National Institutes of Health (NIH), Reseau Vision, the Fonds de la Recherche en Santé du Québec (FRSQ) and FORGE Canada. We acknowledge the FORGE Canada Consortium. We thank R. Pigeon for coordinating all the individuals with LCA. G.A.F. acknowledges the Pangere Corporation, Grousbeck Foundation and the Wynn-Gund Foundation for financial support. We sincerely acknowledge The Royal Society (C.T. is a university research fellow), Yorkshire Eye Research and The Sir Jules Thorn Charitable Trust (grant 09/JTA). Exome sequencing was performed at the BCM–Functional Genome Initiative (BCM-FGI) core facility, which is supported by NIH shared instrument grant 1S10RR026550 to R.C. H.W. was supported by postdoctoral fellowship F32EY19430. J.C. was supported by the Graduate Innovation Foundation of Hunan Province (CX2011B388). This work is supported by grants from the Retinal Research Foundation and the National Eye Institute (R01EY018571) to G.M. and R.C.
The FORGE Steering Committee comprises K. Boycott (leader; University of Ottawa, Ottawa, Ontario, Canada), J. Friedman (co-lead; University of British Columbia, Vancouver, British Columbia, Canada), J. Michaud (co-lead; Université de Montréal, Montreal, Quebec, Canada), F. Bernier (University of Calgary, Calgary, Alberta, Canada), M. Brudno (University of Toronto, Toronto, Ontario, Canada), B. Fernandez (Memorial University, St. John’s, Newfoundland, Canada), B. Knoppers (McGill University, Montreal, Quebec, Canada), M. Samuels (Université de Montréal, Montreal, Quebec, Canada) and S. Scherer (University of Toronto, Toronto, Ontario, Canada). This work was funded in part by the Government of Canada through Genome Canada, the Canadian Institutes of Health Research and the Ontario Genomics Institute (OGI-049). Additional funding was provided by Genome Quebec and Genome British Columbia. We thank J. Marcadier (Clinical Coordinator) and C. Beaulieu (Project Manager) for their contribution to the infrastructure of the FORGE Canada Consortium.
Footnotes
Accession codes. Exome sequencing data have been deposited in the secure ftp server hosted at the Baylor College of Medicine–Human Genome Sequencing Center (BCM-HGSC) and can be accessed through the HGSC website (see URLs).
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
R.K.K., C.F.I., C.T. and R.C. conceived the study. Y.C., Y.L. and H.X. performed the biochemistry and cell culture studies, H.W., I.L., H.R., N.S. and J.C. performed the sequencing, J.M., I.L., G.A.F., M.G., J.S., E.I.T., C.V.L., M.M., B.E.H., D.A.P., C.A.J., M.N., J.A.P., M.D.M., H.J., Y.R., G.R.T., V.K. and Q.F. carried out subject recruitment, and H.W. and I.L. performed pedigree analyses. R.K.K., H.W., J.M., X.W., C.F.I., C.T. and R.C. analyzed the data. R.K.K., H.W., X.W., G.M., C.F.I., C.T. and R.C. wrote and edited the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Patel VP, Chu CT. Nuclear transport, oxidative stress, and neurodegeneration. Int J Clin Exp Pathol. 2011;4:215–229. [PMC free article] [PubMed] [Google Scholar]
- 2.Lassmann H. Mechanisms of neurodegeneration shared between multiple sclerosis and Alzheimer’s disease. J Neural Transm. 2011;118:747–752. doi: 10.1007/s00702-011-0607-8. [DOI] [PubMed] [Google Scholar]
- 3.Wishart TM, et al. Design of a novel quantitative PCR (QPCR)-based protocol for genotyping mice carrying the neuroprotective Wallerian degeneration slow (Wlds) gene. Mol Neurodegener. 2007;2:21. doi: 10.1186/1750-1326-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ludwin SK, Bisby MA. Delayed wallerian degeneration in the central nervous system of Ola mice: an ultrastructural study. J Neurol Sci. 1992;109:140–147. doi: 10.1016/0022-510x(92)90160-m. [DOI] [PubMed] [Google Scholar]
- 5.Hoopfer ED, et al. Wlds protection distinguishes axon degeneration following injury from naturally occurring developmental pruning. Neuron. 2006;50:883–895. doi: 10.1016/j.neuron.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 6.Feng Y, Yan T, He Z, Zhai Q. WldS, Nmnats and axon degeneration—progress in the past two decades. Protein Cell. 2010;1:237–245. doi: 10.1007/s13238-010-0021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lunn ER, Perry VH, Brown MC, Rosen H, Gordon S. Absence of Wallerian degeneration does not hinder regeneration in peripheral nerve. Eur J Neurosci. 1989;1:27–33. doi: 10.1111/j.1460-9568.1989.tb00771.x. [DOI] [PubMed] [Google Scholar]
- 8.Gillingwater TH, Ribchester RR. Compartmental neurodegeneration and synaptic plasticity in the Wlds mutant mouse. J Physiol (Lond) 2001;534:627–639. doi: 10.1111/j.1469-7793.2001.00627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mack TG, et al. Wallerian degeneration of injured axons and synapses is delayed by a Ube4b/Nmnat chimeric gene. Nat Neurosci. 2001;4:1199–1206. doi: 10.1038/nn770. [DOI] [PubMed] [Google Scholar]
- 10.Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- 11.Zhai RG, et al. Drosophila NMNAT maintains neural integrity independent of its NAD synthesis activity. PLoS Biol. 2006;4:e416. doi: 10.1371/journal.pbio.0040416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Avery MA, Sheehan AE, Kerr KS, Wang J, Freeman MR. WldS requires Nmnat1 enzymatic activity and N16-VCP interactions to suppress Wallerian degeneration. J Cell Biol. 2009;184:501–513. doi: 10.1083/jcb.200808042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conforti L, et al. Reducing expression of NAD+ synthesizing enzyme NMNAT1 does not affect the rate of Wallerian degeneration. FEBS J. 2011;278:2666–2679. doi: 10.1111/j.1742-4658.2011.08193.x. [DOI] [PubMed] [Google Scholar]
- 14.Niven JE, Laughlin SB. Energy limitation as a selective pressure on the evolution of sensory systems. J Exp Biol. 2008;211:1792–1804. doi: 10.1242/jeb.017574. [DOI] [PubMed] [Google Scholar]
- 15.den Hollander AI, Roepman R, Koenekoop RK, Cremers FP. Leber congenital amaurosis: genes, proteins and disease mechanisms. Prog Retin Eye Res. 2008;27:391–419. doi: 10.1016/j.preteyeres.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Wang H, et al. Mutations in SPATA7 cause Leber congenital amaurosis and juvenile retinitis pigmentosa. Am J Hum Genet. 2009;84:380–387. doi: 10.1016/j.ajhg.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Estrada-Cuzcano A, et al. IQCB1 mutations in patients with Leber congenital amaurosis. Invest Ophthalmol Vis Sci. 2011;52:834–839. doi: 10.1167/iovs.10-5221. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, et al. Whole-exome sequencing identifies ALMS1, IQCB1, CNGA3 and MYO7A mutations in patients with Leber congenital amaurosis. Hum Mutat. 2011;32:1450–1459. doi: 10.1002/humu.21587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keen TJ, et al. Identification of a locus (LCA9) for Leber’s congenital amaurosis on chromosome 1p36. Eur J Hum Genet. 2003;11:420–423. doi: 10.1038/sj.ejhg.5200981. [DOI] [PubMed] [Google Scholar]
- 20.Zhou T, et al. Structure of human nicotinamide/nicotinic acid mononucleotide adenylyltransferase. Basis for the dual substrate specificity and activation of the oncolytic agent tiazofurin. J Biol Chem. 2002;277:13148–13154. doi: 10.1074/jbc.M111469200. [DOI] [PubMed] [Google Scholar]
- 21.Coleman MP, Freeman MR. Wallerian degeneration, wlds, and nmnat. Annu Rev Neurosci. 2010;33:245–267. doi: 10.1146/annurev-neuro-060909-153248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasaki Y, Milbrandt J. Axonal degeneration is blocked by nicotinamide mononucleotide adenylyltransferase (Nmnat) protein transduction into transected axons. J Biol Chem. 2010;285:41211–41215. doi: 10.1074/jbc.C110.193904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conforti L, et al. NAD+ and axon degeneration revisited: Nmnat1 cannot substitute for WldS to delay Wallerian degeneration. Cell Death Differ. 2007;14:116–127. doi: 10.1038/sj.cdd.4401944. [DOI] [PubMed] [Google Scholar]
- 24.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen Y, et al. A SNP discovery method to assess variant allele probability from next-generation resequencing data. Genome Res. 2010;20:273–280. doi: 10.1101/gr.096388.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Challis D, et al. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics. 2012;13:8. doi: 10.1186/1471-2105-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balducci E, et al. Assay methods for nicotinamide mononucleotide adenylyltransferase of wide applicability. Anal Biochem. 1995;228:64–68. doi: 10.1006/abio.1995.1315. [DOI] [PubMed] [Google Scholar]
- 29.Prudencio M, Durazo A, Whitelegge JP, Borchelt DR. An examination of wild-type SOD1 in modulating the toxicity and aggregation of ALS-associated mutant SOD1. Hum Mol Genet. 2010;19:4774–4789. doi: 10.1093/hmg/ddq408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.