Abstract
Amyotrophic lateral sclerosis (ALS) is a progressive and fatal neurodegenerative disorder resulting from selective death of motor neurons in the brain and spinal cord. In ≈25% of familial ALS cases, the disease is caused by dominantly acting point mutations in the gene encoding cytosolic Cu,Zn superoxide dismutase (SOD1). In cell culture and in rodent models of ALS, mutant SOD1 proteins exhibit dose-dependent toxicity; thus, agents that reduce mutant protein expression would be powerful therapeutic tools. A wealth of recent evidence has demonstrated that the mechanism of RNA-mediated interference (RNAi) can be exploited to achieve potent and specific gene silencing in vitro and in vivo. We have evaluated the utility of RNAi for selective silencing of mutant SOD1 expression in cultured cells and have identified small interfering RNAs capable of specifically inhibiting expression of ALS-linked mutant, but not wild-type, SOD1. We have investigated the functional effects of RNAi-mediated silencing of mutant SOD1 in cultured murine neuroblastoma cells. In this model, stable expression of mutant, but not wild-type, human SOD1 sensitizes cells to cytotoxic stimuli. We find that silencing of mutant SOD1 protects these cells against cyclosporin A-induced cell death. These results demonstrate a positive physiological effect caused by RNAi-mediated silencing of a dominant disease allele. The present study further supports the therapeutic potential of RNAi-based methods for the treatment of inherited human diseases, including ALS.
Amyotrophic lateral sclerosis (ALS) is an age-dependent, paralytic disorder resulting from selective death of motor neurons in the brain and spinal cord. ALS is characterized by progressive muscle weakness and atrophy and is usually fatal within 5 years of clinical presentation (1). Although the majority of ALS cases are sporadic, ≈10% of cases are familial (FALS) and display autosomal dominant inheritance (2). Approximately 25% of FALS cases are caused by mutations in the gene encoding the cytosolic Cu,Zn superoxide dismutase (SOD1) (3), an abundant cellular homodimeric enzyme whose normal function is the scavenging of superoxide radicals. To date, >100 FALS-linked mutations in human SOD1 have been identified (4), and the vast majority of these are missense mutations resulting in single amino acid substitutions.
Both the association of SOD1 mutations with dominantly inherited human disease and compelling data gleaned from expression of FALS-linked mutant SOD1 in experimental model systems suggest that mutant SOD1 causes ALS through an acquired toxic property (or properties) of the mutant proteins. Transgenic mice (5-7) and rats (8) that express clinically relevant forms of mutant human SOD1 quite remarkably recapitulate the major features of the disease; in contrast, mice in which endogenous SOD1 expression has been eliminated by means of targeted deletion develop normally and do not exhibit ALS-like symptoms (9). Further, in vitro studies demonstrate that forced expression of mutant, but not wild-type, SOD1 promotes apoptosis in cultured rodent primary motor neurons (10) and neural-derived cell lines (11, 12). Both in vivo and in vitro model systems have provided valuable tools for the dissection of mutant SOD1-mediated pathology; nevertheless, the molecular etiology of ALS is not yet understood.
In cell culture and in rodent models of ALS, mutant SOD1 proteins exert dose-dependent toxic effects (5, 13, 14); inhibition of mutant SOD1 expression therefore represents an attractive target for therapy. Therapeutic inhibition of mutant SOD1 could be feasible by using RNA-mediated interference (RNAi), an evolutionarily conserved mechanism for sequence-specific posttranscriptional gene silencing in which double-stranded RNAs promote selective degradation of homologous cellular mRNAs (15, 16). RNAi mediated by endogenous noncoding RNAs has been shown to function in eukaryotic development, in defense against viruses and transposons, and in maintenance of chromatin structure (17).
RNAi is mediated directly by short RNA duplexes of ≈21-23 nucleotides, termed small interfering RNAs (siRNAs), which target cognate cellular mRNAs for degradation by recruitment to a ribonuclease-containing complex (RNA-induced silencing complex, or RISC) (18-20). RNAi-mediated inhibition of specific gene expression has been achieved in cultured mammalian cells by using synthetic 21-nt siRNA duplexes (21, 22) or intracellular expression of short hairpin RNAs (shRNAs) (23). The ability of shRNAs encoded from DNA vectors to promote RNAi has greatly enhanced the therapeutic potential of this approach and has led to the development of viral vector systems that permit persistent gene silencing in cultured cells and in mice (24-26).
A critical feature of the RNAi technology is its exquisite specificity: a single base mismatch between an siRNA and its target mRNA can abolish silencing (21, 23). In recent months, several studies have reported RNAi-mediated inactivation of dominant disease alleles differing from their wild-type counterparts by a single nucleotide (27, 28), and one report demonstrated specific inhibition of mutant SOD1 alleles (29). The present study confirms and extends these observations and identifies previously uncharacterized siRNA sequences for selective silencing of FALS-linked mutant SOD1. In addition, we sought to validate the RNAi approach in a cell-based model of mutant SOD1-mediated toxicity. We report in this article that selective silencing of mutant SOD1 by RNAi rescues cells from death induced by cyclosporin A (CsA). These results demonstrate that RNAi-mediated silencing of a dominant disease allele, in this case mutant SOD1, can be advantageous to cells and underline the potential value of this approach for the treatment of human disease.
Materials and Methods
siRNA and shRNA Design. siRNA duplexes were purchased from Qiagen (Valencia, CA). shRNA expression constructs were generated by annealing complementary DNA oligonucleotides with sense strand sequence 5′-N19-T TCA AGAGA-N19antiparallel-3′, where N19 is the target sequence in SOD1. Annealed oligonucleotides were subcloned into the pSilencer expression plasmid (Ambion, Austin, TX) and sequence-verified. Two control siRNAs were purchased from Qiagen: an siRNA targeting GFP (22) and a 5′-rhodamine labeled nonsilencing siRNA.
Plasmid Construction. Plasmids containing human wild-type or G93A mutant SOD1 cDNAs have been described (30). We used PCR-based cloning methods to generate SOD1-enhanced GFP (EGFP) reporter plasmids as follows. The coding sequence of wild-type or mutant SOD1 was amplified from cDNA with the primers 5′-ATAGGTACCGTTATGGCGACGAAGGCCGTGTGCGTGCTGAAG-3′ and 5′-TTGTCTAGAGTTGGATCCTTGGGCGATCCCAATTACACCACAAGCCAAACG-3′. PCR products were cloned into a pcDNA3-based expression vector containing EGFP as a C-terminal tag (31). All constructs were sequence-verified before use.
Cell Culture and Transfections. Cells were cultured at 37°C in a humidified chamber containing 5% CO2. HeLa cells were maintained in DMEM supplemented with 10% heat-inactivated FBS, 2 mM l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Neuro-2a (N2a) murine neuroblastoma cell lines stably expressing human mutant SOD1G37R (12, 32) were maintained in DMEM supplemented with 7% FBS, 2 mM l-glutamine, and 400 μg/ml G418. DNA and siRNA transfections were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendations and published procedures (33). HeLa cells, at 60% confluence in 24-well plates, were transfected in serum-free medium (OptiMEM; Invitrogen). For general silencing experiments, cells were transfected with 0.5 μg of SOD1-EGFP reporter plasmid, 0.2 μg of siRNA, and 0.2 μg of pDsRed1-N1 (Clontech; as a control for transfection efficiency) or with 0.2 μg of SOD1-EGFP reporter plasmid, 0.6 μg of shRNA expression plasmid, and 0.1 μg of pDsRed. For allele-specific silencing experiments, we used 0.2 μg of SOD1-EGFP reporter plasmid, 0.03 μg of siRNA, and 0.1 μg of pDsRed. Silencing efficiency was monitored in live cells by fluorescence microscopy at 24, 48, and 72 h after transfection, and cells were harvested at 60-72 h. SOD1G37R N2a cells, at 2 × 104 cells per well in 96-well plates, were transfected with 0.1 μg of siRNA per well. N2a cells were transfected in growth medium (described above), including serum but without G418. Transfection efficiency of siRNA into N2a cells was estimated by using a rhodamine-labeled nonsilencing siRNA (Qiagen) and found to be >70% (data not shown).
Immunoblotting. Total cell protein extracts were prepared by lysing cells in RIPA buffer (50 mM Tris, pH 8/150 mM NaCl/1% Triton X-100/10 mg/ml NaDeoxycholate/1 mg/ml SDS) supplemented with 2 mM EGTA, 1 mM PMSF, and Complete protease inhibitor tablets (Roche Diagnostics). Proteins were quantified by using the bicinchoninic acid (BCA) protein assay kit (Pierce). For immunoblotting, 10 to 20 μg of total cell lysate was fractionated by SDS/PAGE and transferred to poly(vinylidene difluoride) membrane (Immobilon-P; Millipore). SOD1 and SOD1-EGFP fusion proteins were detected by using sheep anti-SOD1 antibody (Calbiochem) followed by horseradish peroxidase-conjugated anti-sheep IgG (Rockland, Gilbertsville, PA). For loading controls, blots were probed with mouse monoclonal anti-β-tubulin antibody followed by horseradish peroxidase-conjugated anti-mouse IgG (Sigma). Bound antibodies were detected by using the SuperSignal West Dura kit (Pierce), and blots were visualized and quantified by using a Fluor-S MultiImager and multianalyst software (Bio-Rad).
Fluorescent Imaging and Analysis. For HeLa cell experiments, silencing efficiency was monitored in live cells by visual comparison of GFP fluorescence; transfection efficiency was confirmed by comparison of DsRed fluorescence. GFP and DsRed fluorescence in transfected cells was measured by using a Victor25 multilabel plate reader (Perkin-Elmer) with excitation/emission filters at 485/510 nm and 560/590 nm, respectively (data not shown). For N2a experiments, viable cells were visualized after a 30-min exposure to 0.4 μM Calcein acetoxymethyl ester (AM) (Molecular Probes). All cells were analyzed and photographed with a Leica DM IL inverted microscope equipped for phase contrast and fluorescence and fitted with a Leica DC300 F cooled digital camera.
N2a Survival Assays. N2a cells that stably express human SOD1G37R (12, 32) were transfected with siRNA as described above. After a 24-hour incubation, to permit recovery and activation of the RNAi pathway, the growth medium was removed and replaced with serum-free medium. Forty-eight hours after transfection, CsA (Calbiochem) was added to culture medium at 0, 7, and 10 μg/ml. Cells were treated with CsA for 24 h, and visual estimation of cell viability was assessed by using fluorescence microscopy as described above. Survival was quantified by using the WST-1 viability kit (Roche Diagnostics) according to the manufacturer's recommendations. The WST-1 assay yields a colorimetric signal that directly correlates with the number of viable cells; the assay was calibrated on each experiment day to ensure that readings were within the linear range. WST-1 assay results were measured by using a Victor25 multilabel plate reader (Perkin-Elmer) with a 420-nm photometry filter. Survival was defined, within each transfection experiment, as the ratio of CsA-treated to untreated cells. Statistical analysis of survival data were performed by using ANOVA, followed by multiple pairwise comparisons by using Student-Newman-Keuls test. For immunoblot analysis of SOD1 protein levels in these experiments, untreated (no CsA) cells were harvested at the end of the experiment (72 h after transfection).
Results
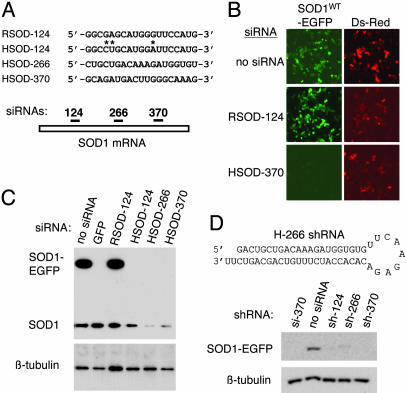
RNAi-Mediated Silencing of SOD1. We first identified siRNA sequences that permit potent and selective silencing of human SOD1 fused to EGFP (SOD1WT-EGFP). siRNAs were designed to target three distinct regions of the human SOD1 (HSOD) coding sequence (Fig. 1A): an upstream region, HSOD-124; a central region, HSOD-266; and a downstream region, HSOD-370. To control for silencing specificity, we designed an additional siRNA that targets rat SOD1 (RSOD) at the region defined by HSOD-124; we termed this siRNA RSOD-124. The sequences of HSOD-124 and RSOD-124 differ at three nucleotide positions (Fig. 1A). These four siRNAs were tested for their ability to silence the SOD1WT-EGFP fusion protein in transient transfection in HeLa cells. As shown in Fig. 1B, HSOD siRNA-transfected cells exhibited a marked reduction in SOD1WT-EGFP fluorescence (Fig. 1B Bottom Left, HSOD-370). All three siRNAs matching the human SOD1 sequence inhibited SOD1WT-EGFP expression by >90% (Fig. 1C Upper, upper band) and reduced endogenous SOD1 by >50% (Fig. 1C Upper, lower band and data not shown). Inhibition of SOD1WT-EGFP was similar to that obtained by using a validated siRNA directed against EGFP (22). In contrast, cells transfected with the rat-specific siRNA RSOD-124 showed no evidence of silencing and contained SOD1WT-EGFP levels comparable to control (Fig. 1 B Middle Left and C, lane 3).
Fig. 1.
Design and testing of siRNAs and shRNA constructs. (A Upper) Sequence of one rat and three human siRNAs; asterisks indicate mismatches between the rat and human SOD-124 siRNA sequences. For clarity, only the sense strand is shown. Numbers refer to the first nucleotide in the SOD1 target sequence, with the translation initiation codon designated as position 1. (Lower) Schematic representation of the relative positions, within the SOD1 coding region, of the three siRNA target sequences. (B) Fluorescence microscopy in live HeLa cells cotransfected with HSOD1-EGFP and siRNA directed against human (HSOD-370, Bottom) or rat (RSOD-124, Middle), along with pDsRed as a transfection control. (Left) HSOD1-EGFP fluorescence. (Right) DsRed-positive (i.e., transfected) cells in the same fields. (C) Immunoblot analysis of HeLa cells transfected with HSOD1-EGFP and the indicated siRNAs. A validated siRNA against GFP (22) was used as a positive control for silencing. (Upper) Upper band, SOD1-EGFP; lower band, endogenous SOD1. (Lower) β-Tubulin as a control for sample loading. (D Upper) Structure of the HSOD-266 RNA is shown as an example of a plasmid-encoded shRNA. (Lower) Immunoblot analysis of silencing in HeLa cells cotransfected with SOD1-EGFP and expression plasmids encoding the indicated shRNAs. HSOD-370 siRNA is used as a positive control for silencing.
Although synthetic siRNAs can act as potent inducers of RNAi-mediated gene silencing, their utility is limited to transient transfection experiments in cultured cells. This constraint has been overcome by the development of DNA vector-based systems that permit intracellular expression of shRNAs, which structurally resemble siRNAs and function in the RNAi pathway (23, 34). To assess whether the sequences we selected would function effectively as shRNAs, we designed DNA oligonucleotides based on the sequences of our SOD1 siRNAs (Fig. 1D and Materials and Methods) and cloned the annealed DNA duplexes into the pSilencer expression vector (Ambion). Silencing plasmids were cotransfected with SOD1-EGFP reporter constructs into cultured HeLa cells, and inhibition of SOD1-EGFP expression was assayed by using fluorescence microscopy (data not shown) and immunoblotting (Fig. 1D). Our results show that, like siRNAs, intracellularly expressed shRNAs efficiently silence human SOD1 in vitro.
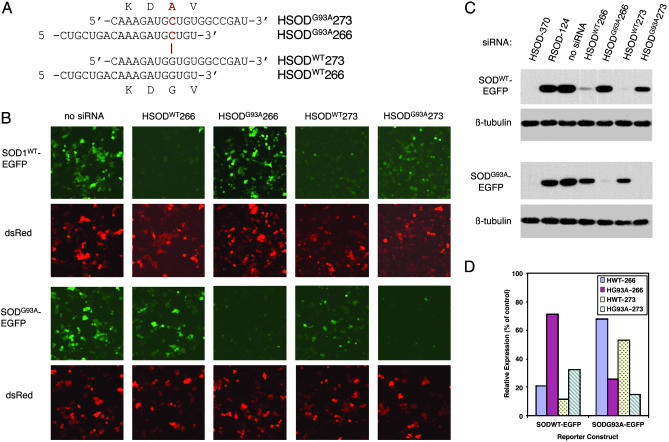
Allele-Specific Silencing of FALS-Linked SOD1G93A. We next investigated whether silencing mediated by our siRNAs could discriminate between wild-type and mutant alleles of SOD1, which differ by a single nucleotide (Fig. 2). Among the siRNAs used above, HSOD-266 (Fig. 1A) contains the site of the FALS-linked G93A mutation. This allele contains a G → C mutation at position 281, which results in a Gly → Ala transition at amino acid 93 (SOD1G93A) (3). To test the selectivity of HSOD-266, we synthesized the G93A-mutant version of this siRNA (Fig. 2A). In addition, because several reports have suggested that selectivity in RNAi-mediated silencing is sensitive to the position of the mismatched nucleotide within the siRNA (21, 27, 35), we designed a second siRNA, HSOD-273, in which the site of mutation is more centrally placed, and synthesized both the wild-type and G93A-mutant versions. The sequences of the four siRNAs used in these experiments, and the position of the G → C mismatch, are shown in Fig. 2A.
Fig. 2.
Allele-specific silencing of mutant SOD1. (A) Sequences of the G93A mutant (Upper) and wild-type (Lower) versions of H-266 and H-273 siRNAs. The site of the G → C mutation in the G93A allele is denoted in red. (B) Fluorescence microscopy of live HeLa cells transfected with either wild-type (Upper) or G93A-mutant (Lower) SOD1-EGFP, the indicated siRNA, and pDsRed. Upper shows SOD1-EGFP fluorescence. Lower shows DsRed fluorescence in same fields. (C) Immunoblots of extracts from transfected HeLa cells shown in B. (Upper) Levels of the SOD1WT-EGFP reporter. (Lower) Levels of the SOD1G93A-EGFP reporter. HSOD-370 and RSOD-124 are used as positive and negative controls, respectively, for silencing. For clarity, Upper shows only the transfected SOD1-EGFP fusion protein; the endogenous human wild-type SOD1 is omitted. (D) Quantification of SOD1-EGFP levels from immunoblots as in C from two independent experiments. SOD1-EFGP protein levels in transfected cells are expressed as percentage of control (lane 3 in C). Similar results were obtained by comparing fluorimetric measurements (see Materials and Methods) of GFP normalized to DsRed (data not shown).
As before, we monitored silencing by using reporter constructs containing either the human SOD1WT or SOD1G93A coding sequence fused to EGFP. We assessed efficiency and selectivity of silencing by fluorescence microscopy (Fig. 2B) and immunoblotting (Fig. 2C). Our results demonstrate that both HSOD-266 and HSOD-273 mediate potent silencing of SOD-EGFP and that these siRNAs exhibit selectivity for the matched allele (Fig. 2 B and C, compare Upper and Lower in each). As determined by quantitative immunoblotting, HSODWT266 reduced human SOD1WT-EGFP levels by ≈80%, but reduced SOD1G93A-EGFP by only ≈30% (Fig. 2D); conversely, HSODG93A266 silenced the mutant allele preferentially, reducing SOD1WT-EGFP levels by only ≈30% and SOD1G93A-EGFP by >70% (Fig. 2D). HSODWT273 exhibits complete (≈90%) silencing of SOD1WT-EGFP and partial (≈50%) silencing of SOD1G93A-EGFP, whereas HSODG93A273 showed the least selectivity, reducing SOD1WT-EGFP by ≈70% and SOD1G93A-EGFP by >85%.
These results confirm previous observations that not all siRNAs are equally proficient at discriminating between alleles differing by a single nucleotide, yet some siRNAs exhibit remarkable selectivity. The HeLa cells used in these experiments contain endogenous (wild-type) human SOD1 and, when transfected with SOD1G93A-EGFP, mimic the heterozygous state. Thus, in the presence of wild-type protein, HSODG93A266 potently silences the mutant allele (Fig. 2C Lower, lane 5), yet fails to silence the SOD1WT-EGFP reporter protein (Fig. 2C Upper, lane 5) or the endogenous SOD1 (data not shown). In contrast, HSODWT273 and HSODG93A273 each demonstrate considerable inhibition of expression of the noncognate allele (Fig. 2C, lanes 6 and 7). In our experiments, HSOD-266 clearly shows the best selectivity for silencing the SOD1G93A allele. We confirmed the selectivity of silencing by these siRNAs by testing the sequences shown in Fig. 2A in the plasmid-based shRNA system described above, and we obtained similar results (data not shown). These experiments show that HSODG93A-266 preferentially inhibits expression of the FALS-linked SOD1G93A allele.
Functional Analysis of RNAi-Mediated Silencing of Mutant SOD1. The above results confirm that RNAi can be used to achieve selective and potent silencing of dominantly inherited disease alleles and identify siRNA sequences for specific inactivation of the SOD1G93A allele. Ultimately, we will use these sequences to test the therapeutic utility of RNAi-mediated gene silencing in ALS model mice, which express the human SOD1G93A allele (5). In the interim, we sought to validate the RNAi approach in a cell-based model of mutant SOD1-mediated toxicity. Several studies have shown that expression of mutant SOD1 exerts toxic effects on cultured cells (11, 12). We investigated the biological effects of RNAi-mediated silencing of SOD1 in a mouse neuroblastoma (N2a) cell line that constitutively expresses high levels of FALS-linked mutant human SOD1 (SOD1G37R) (12, 32). In this system, overexpression of SOD1G37R does not affect cell viability under normal culture conditions but renders cells more sensitive to cytotoxic stimuli (12).
The SOD1G37R N2a cells have been extensively characterized and shown to exhibit enhanced sensitivity to cell death induced by CsA (P.P., L. Van Den Bosch, B. Stockwell, S. Sarang, S. Kohnstamm, S. Kaplan, E. Wilkes, V. Vleminckx, S. Gullans, W. Robberecht, and R.H.B., unpublished data). In these cells, <60% of SOD1G37R cells survive treatment with 7 μg/ml CsA, whereas SOD1WT cells show >80% survival. At higher concentrations, CsA-induced toxicity is less selective for SOD1G37R-expressing cells, with SOD1WT cells exhibiting ≈45% and SOD1G37R <30% viability after treatment with 10 μg/ml CsA (P.P., unpublished data, and data not shown).
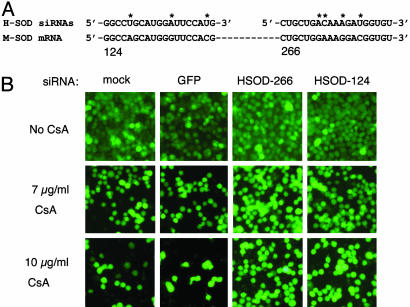
RNAi-Mediated Inhibition of Mutant SOD1 Rescues CsA-Induced Cell Death. If CsA-induced killing of SOD1G37R N2a cells is enhanced by expression of the SOD1G37R protein, we reasoned that a reduction in mutant protein levels would lessen the cells' sensitivity to this drug. We tested the effects of the siRNAs shown in Fig. 1 on CsA-induced toxicity in SOD1G37R N2a cells. To ensure selective silencing of the mutant (human) SOD1G37R, but not the endogenous wild-type (mouse) SOD1, we used the HSOD-266 and HSOD-124 siRNAs (see Fig. 1). The sequences of these siRNAs differ at 3 or more nucleotide positions from mouse SOD1 (Fig. 3A). We modified the CsA assay to include transfection of siRNAs 48 h before treatment with CsA (see Materials and Methods). In initial experiments, we used the fluorescent dye Calcein AM to stain viable cells and assessed survival by using fluorescence microscopy. Transfection with two siRNAs directed against human SOD1, HSOD-266, and HSOD-124 decreased toxicity in CsA-treated cells, whereas transfection with a control (GFP) siRNA did not (Fig. 3). Results from a typical experiment are shown in Fig. 3; we consistently observed a greater number of surviving cells after CsA treatment in HSOD1 siRNA-transfected samples (Fig. 3B, right columns) than in mock- or control siRNA-transfected samples (Fig. 3B, left columns).
Fig. 3.
siRNA directed against mutant SOD1 reduces CsA-induced death in N2a cells that express human SOD1G37R. (A) Sequences of the human SOD1 siRNAs used in these experiments (Upper) are aligned with the mouse SOD1 mRNA (Lower). The sequences of HSOD-124 and H-226 are not identical to mouse SOD1. Asterisks indicate mismatched nucleotides. (B) Transfection of SOD1G37R N2a cells with human-specific SOD1 siRNAs increases the number of viable cells after treatment with 7 μg/ml (Middle) or 10 μg/ml (Bottom) CsA treatment, as compared to mock- or control (GFP siRNA)-transfected samples. Viable cells are detected with Calcein AM.
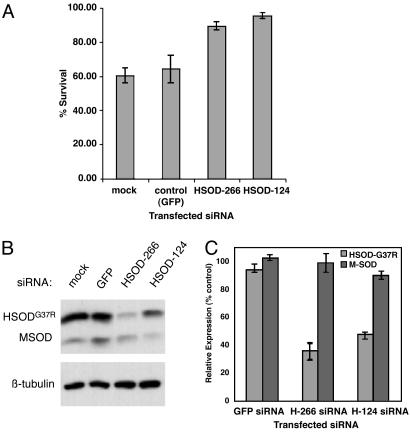
In these experiments, Calcein AM proved unsuitable for fluorimetric analyses because of the variability in staining intensity observed among samples with large differences in cell number (Fig. 3 and data not shown). We therefore used a second assay for cell viability (WST-1 assay, see Materials and Methods) to quantify the results shown in Fig. 3. The quantitative effects of RNAi-mediated silencing of SOD1G37R on cell survival are shown in Fig. 4A. After treatment with 7 μg/ml CsA, HSOD-266 or HSOD-124 siRNA-transfected SOD1G37R N2a cells exhibit 85-95% survival compared with 60-65% for mock- or control (GFP) siRNA-transfected cells (Fig. 4A). Thus, transfection of HSOD1 siRNA results in a 20-30% increase in survival in CsA-treated SOD1G37R N2a cells. One-way ANOVA of these data was significant (F = 8.61, P = 0.005); multiple comparison tests of the differences between groups (mock vs. H-124, GFP vs. H-124, mock vs. H-226, and GFP vs. H-226) were significant by the Student-Newman-Keuls test (P < 0.05). We also observed increased cell survival in HSOD1 siRNA-transfected cells at high (10 μg/ml) CsA concentration (data not shown); however, this effect was less consistent. This result is not surprising given that both SOD1WT and SOD1G37R N2a cells exhibit considerable cell death at this dose.
Fig. 4.
Selective silencing of mutant HSOD1G37R rescues CsA-induced cell death. (A) Quantitative analysis of cell survival in siRNA-transfected SOD1G37R N2a cells treated with 7 μg/ml CsA. Cells were transfected with control (GFP) siRNA or SOD1 siRNAs H-266 or H-124, or exposed to transfection reagent but no siRNA (mock). Viable cells were estimated by using the WST-1 assay (see Materials and Methods); survival is expressed as the ratio of CsA-treated to untreated cells within each transfection experiment. Data represent the survival mean ± standard deviation of three independent experiments. (B) Representative immunoblot showing relative levels of mutant human (HSOD1G37R; Upper, upper band) and endogenous wild-type mouse SOD1 (MSOD; Upper, lower band) in samples from A. Proteins were extracted from untreated (no CsA) cells at the conclusion of the survival assay. Lower shows β-tubulin as loading control. (C) Quantification of HSOD1G37R and MSOD1 levels from immunoblots as in B from two independent experiments. SOD1 protein levels are expressed as percentage of control (mock-transfected) normalized to tubulin. Error bars represent standard deviation.
Under these experimental conditions, we achieved a 50-70% reduction in SOD1G37R protein levels in cells transfected with HSOD1 siRNAs, as determined by immunoblotting. Fig. 4B shows representative immunoblots from the siRNA transfection experiments shown above, and results from two independent experiments are quantified in Fig. 4C. Importantly, introduction of HSOD-266 or HSOD-124 siRNAs did not significantly affect levels of the wild-type (mouse) SOD1 in these cells (Fig. 4C). A third siRNA (HSOD-370), which silences both human and mouse SOD1, yielded inconsistent results in this assay (data not shown). These results demonstrate that selective, RNAi-mediated reduction of mutant SOD1 protein levels can confer a significant survival advantage on cultured cells, and they further validate RNAi-based approaches to disease therapy.
Discussion
Gene silencing by RNAi has emerged as a promising candidate for treatment of dominantly inherited human disease. The potency and selectivity of this approach has recently been documented for several dominant disease genes (27, 28), including SOD1. Our findings confirm and extend the recent report of Ding et al. (29), which described RNAi-mediated, allele-specific silencing of the SOD1G85R and SOD1G93A alleles. In the present study, we have identified a previously uncharacterized siRNA that permits allele-specific silencing of the FALS-linked SOD1G93A allele. In addition, we now document an important functional corollary to the inhibition of disease gene expression by using RNAi: the rescue of N2a cell death mediated by mutant SOD1 after exposure to CsA.
RNAi-mediated gene silencing is a sequence-specific process. This specificity may be critical to effective therapy for mutant SOD-mediated FALS, because even though loss of SOD1 does not cause ALS, the wild-type protein performs important cellular functions (9, 36). Our results indicate that some siRNAs can discriminate between wild-type and mutant alleles of SOD1 but that not all siRNAs exhibit comparable selectivity. Of the siRNAs we tested for allele-specific silencing, HSODG93A266 demonstrates the greatest degree of selectivity for its cognate allele, SOD1G93A-EGFP. This finding is somewhat surprising, given that several reports have suggested that selective silencing of alleles differing by a single nucleotide is more easily achieved when the mismatched base is located near the center of the siRNA (28, 29). In our HSOD-266 siRNAs, the mismatched nucleotide resides near the 3-′ terminus of the sense strand. Nevertheless, in our experiments, this siRNA exhibits greater selectivity for silencing of the cognate allele than does HSOD-273, which closely resembles a recently described shRNA for specific inactivation of SOD1G93A (29). The list of guidelines for siRNA design is rapidly evolving; however, our results suggest that straightforward application of design rules will not always yield the best (i.e., most potent and/or selective) siRNA for every experiment. These findings underscore the importance of empirical testing of multiple siRNAs to identify the optimal sequence for a particular application.
Our data show that selectively silencing mutant SOD1 without altering wild-type protein levels has a positive effect on survival in a cell culture-based model of mutant SOD1-mediated toxicity. In our experiments, a 50% reduction in mutant SOD1 protein levels was sufficient to rescue CsA-induced cell death. This finding is consistent with previous reports establishing that mutant SOD1-mediated toxicity is dose-dependent in vitro and in vivo. Disease phenotype in mutant SOD1 transgenic animals, for instance, depends on transgene copy number, and mice with high transgene copy number display pathological symptoms at an earlier age and have more rapidly progressing disease than do mice with low transgene copy number (5, 13, 14). Taken together, these results support the hypothesis that even a partial reduction of mutant SOD1 in vivo could impart a significant therapeutic benefit.
The results of the present study provide further support for the development of RNAi-based therapeutics. The long-term, in vivo efficacy of RNAi-mediated silencing can be tested by systemic expression, in ALS model mice, of the siRNA sequences reported here. Notably, viral vectors that permit in vivo delivery of shRNA expression cassettes to the central nervous system recently have been reported (37). Ultimately, viral delivery of shRNAs in vivo will be critical to the evaluation of RNAi-based approaches for the treatment of dominantly inherited human diseases, including ALS.
Acknowledgments
We thank Deborah Russel and Mary Elizabeth Belford for technical assistance and Alexander McCampbell for helpful discussions. This work was supported by National Institute on Aging Grants 5P01 AG12992-9 (to R.H.B.) and R03 AG 22152-01 (to P.P.), National Institute of Neurological Disorders and Stroke Grant 5PO1 NS40828-02 (to R.H.B.), Angel Fund, Project ALS, ALS Association, and Pierre L. Bourgknecht ALS Foundation. M.M.M. is an Angel Fund Fellow.
Abbreviations: ALS, amyotrophic lateral sclerosis; FALS, familial ALS; SOD1, Cu,Zn superoxide dismutase; HSOD, human SOD1; RSOD, rat SOD1; RNAi, RNA-mediated interference; siRNA, small interfering RNA; shRNA, short hairpin RNA; EGFP, enhanced GFP; CsA, cyclosporin A; N2a, Neuro-2a.
References
- 1.Traynor, B. J., Codd, M. B., Corr, B., Forde, C., Frost, E. & Hardiman, O. (1999) Neurology 52, 504-509. [DOI] [PubMed] [Google Scholar]
- 2.Mulder, D. W., Kurland, L. T., Offord, K. P. & Beard, C. M. (1986) Neurology 36, 511-517. [DOI] [PubMed] [Google Scholar]
- 3.Rosen, D. R., Siddique, T., Patterson, D., Figlewicz, D. A., Sapp, P., Hentati, A., Donaldson, D., Goto, J., O'Regan, J. P., Deng, H. X., et al. (1993) Nature 362, 59-62. [DOI] [PubMed] [Google Scholar]
- 4.Andersen, P. M., Sims, K. B., Xin, W. W., Kiely, R., O'Neill, G., Ravits, J., Pioro, E., Harati, Y., Brower, R. D., Levine, J. S., et al. (2003) Amyotroph. Lateral Scler. Other Motor Neuron Disord. 4, 62-73. [DOI] [PubMed] [Google Scholar]
- 5.Gurney, M. E., Pu, H., Chiu, A. Y., Dal Canto, M. C., Polchow, C. Y., Alexander, D. D., Caliendo, J., Hentati, A., Kwon, Y. W., Deng, H. X., et al. (1994) Science 264, 1772-1775. [DOI] [PubMed] [Google Scholar]
- 6.Wong, P. C., Pardo, C. A., Borchelt, D. R., Lee, M. K., Copeland, N. G., Jenkins, N. A., Sisodia, S. S., Cleveland, D. W. & Price, D. L. (1995) Neuron 14, 1105-1116. [DOI] [PubMed] [Google Scholar]
- 7.Bruijn, L. I., Becher, M. W., Lee, M. K., Anderson, K. L., Jenkins, N. A., Copeland, N. G., Sisodia, S. S., Rothstein, J. D., Borchelt, D. R., Price, D. L. & Cleveland, D. W. (1997) Neuron 18, 327-338. [DOI] [PubMed] [Google Scholar]
- 8.Nagai, M., Aoki, M., Miyoshi, I., Kato, M., Pasinelli, P., Kasai, N., Brown, R. H., Jr., & Itoyama, Y. (2001) J. Neurosci. 21, 9246-9254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reaume, A. G., Elliott, J. L., Hoffman, E. K., Kowall, N. W., Ferrante, R. J., Siwek, D. F., Wilcox, H. M., Flood, D. G., Beal, M. F., Brown, R. H., Jr., et al. (1996) Nat. Genet. 13, 43-47. [DOI] [PubMed] [Google Scholar]
- 10.Durham, H. D., Roy, J., Dong, L. & Figlewicz, D. A. (1997) J. Neuropathol. Exp. Neurol. 56, 523-530. [DOI] [PubMed] [Google Scholar]
- 11.Rabizadeh, S., Gralla, E. B., Borchelt, D. R., Gwinn, R., Valentine, J. S., Sisodia, S., Wong, P., Lee, M., Hahn, H. & Bredesen, D. E. (1995) Proc. Natl. Acad. Sci. USA 92, 3024-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pasinelli, P., Borchelt, D. R., Houseweart, M. K., Cleveland, D. W. & Brown, R. H., Jr. (1998) Proc. Natl. Acad. Sci. USA 95, 15763-15768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiu, A. Y., Zhai, P., Dal Canto, M. C., Peters, T. M., Kwon, Y. W., Prattis, S. M. & Gurney, M. E. (1995) Mol. Cell. Neurosci. 6, 349-362. [DOI] [PubMed] [Google Scholar]
- 14.Shibata, N., Hirano, A., Yamamoto, T., Kato, Y. & Kobayashi, M. (2000) Amyotroph. Lateral Scler. Other Motor Neuron Disord. 1, 143-161. [DOI] [PubMed] [Google Scholar]
- 15.Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E. & Mello, C. C. (1998) Nature 391, 806-811. [DOI] [PubMed] [Google Scholar]
- 16.Tuschl, T., Zamore, P. D., Lehmann, R., Bartel, D. P. & Sharp, P. A. (1999) Genes Dev. 13, 3191-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denli, A. M. & Hannon, G. J. (2003) Trends Biochem. Sci. 28, 196-201. [DOI] [PubMed] [Google Scholar]
- 18.Hamilton, A. J. & Baulcombe, D. C. (1999) Science 286, 950-952. [DOI] [PubMed] [Google Scholar]
- 19.Hammond, S. M., Bernstein, E., Beach, D. & Hannon, G. J. (2000) Nature 404, 293-296. [DOI] [PubMed] [Google Scholar]
- 20.Zamore, P. D., Tuschl, T., Sharp, P. A. & Bartel, D. P. (2000) Cell 101, 25-33. [DOI] [PubMed] [Google Scholar]
- 21.Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494-498. [DOI] [PubMed] [Google Scholar]
- 22.Caplen, N. J., Parrish, S., Imani, F., Fire, A. & Morgan, R. A. (2001) Proc. Natl. Acad. Sci. USA 98, 9742-9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brummelkamp, T. R., Bernards, R. & Agami, R. (2002) Science 296, 550-553. [DOI] [PubMed] [Google Scholar]
- 24.Brummelkamp, T. R., Bernards, R. & Agami, R. (2002) Cancer Cell 2, 243-247. [DOI] [PubMed] [Google Scholar]
- 25.Abbas-Terki, T., Blanco-Bose, W., Deglon, N., Pralong, W. & Aebischer, P. (2002) Hum. Gene Ther. 13, 2197-2201. [DOI] [PubMed] [Google Scholar]
- 26.Rubinson, D. A., Dillon, C. P., Kwiatkowski, A. V., Sievers, C., Yang, L., Kopinja, J., Rooney, D. L., Ihrig, M. M., McManus, M. T., Gertler, F. B., et al. (2003) Nat. Genet. 33, 401-406. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez-Alegre, P., Miller, V. M., Davidson, B. L. & Paulson, H. L. (2003) Ann. Neurol. 53, 781-787. [DOI] [PubMed] [Google Scholar]
- 28.Miller, V. M., Xia, H., Marrs, G. L., Gouvion, C. M., Lee, G., Davidson, B. L. & Paulson, H. L. (2003) Proc. Natl. Acad. Sci. USA 100, 7195-7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding, H., Schwarz, D. S., Keene, A., Affar el, B., Fenton, L., Xia, X., Shi, Y., Zamore, P. D. & Xu, Z. (2003) Aging Cell 2, 209-217. [DOI] [PubMed] [Google Scholar]
- 30.Hayward, L. J., Rodriguez, J. A., Kim, J. W., Tiwari, A., Goto, J. J., Cabelli, D. E., Valentine, J. S. & Brown, R. H., Jr. (2002) J. Biol. Chem. 277, 15923-15931. [DOI] [PubMed] [Google Scholar]
- 31.Kazantsev, A., Preisinger, E., Dranovsky, A., Goldgaber, D. & Housman, D. (1999) Proc. Natl. Acad. Sci. USA 96, 11404-11409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borchelt, D. R., Guarnieri, M., Wong, P. C., Lee, M. K., Slunt, H. S., Xu, Z. S., Sisodia, S. S., Price, D. L. & Cleveland, D. W. (1995) J. Biol. Chem. 270, 3234-3238. [DOI] [PubMed] [Google Scholar]
- 33.Elbashir, S. M., Harborth, J., Weber, K. & Tuschl, T. (2002) Methods 26, 199-213. [DOI] [PubMed] [Google Scholar]
- 34.Sui, G., Soohoo, C., Affar el, B., Gay, F., Shi, Y. & Forrester, W. C. (2002) Proc. Natl. Acad. Sci. USA 99, 5515-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harborth, J., Elbashir, S. M., Vandenburgh, K., Manninga, H., Scaringe, S. A., Weber, K. & Tuschl, T. (2003) Antisense Nucleic Acid Drug Dev. 13, 83-105. [DOI] [PubMed] [Google Scholar]
- 36.Shefner, J. M., Reaume, A. G., Flood, D. G., Scott, R. W., Kowall, N. W., Ferrante, R. J., Siwek, D. F., Upton-Rice, M. & Brown, R. H., Jr. (1999) Neurology 53, 1239-1246. [DOI] [PubMed] [Google Scholar]
- 37.Xia, H., Mao, Q., Paulson, H. L. & Davidson, B. L. (2002) Nat. Biotechnol. 20, 1006-1010. [DOI] [PubMed] [Google Scholar]