Abstract
Edema factor (EF), a key virulence factor in anthrax pathogenesis, has calmodulin (CaM)-activated adenylyl cyclase activity. We have found that adefovir dipivoxil, a drug approved to treat chronic infection of hepatitis B virus, effectively inhibits EF-induced cAMP accumulation and changes in cytokine production in mouse primary macrophages. Adefovir diphosphate (PMEApp), the active cellular metabolite of adefovir dipivoxil, inhibits the adenylyl cyclase activity of EF in vitro with high affinity (Ki = 27 nM). A crystal structure of EF-CaM-PMEApp reveals that the catalytic site of EF forms better van der Waals contacts and more hydrogen bonds with PMEApp than with its endogenous substrate, ATP, providing an explanation for the ≈10,000-fold higher affinity EF-CaM has for PMEApp versus ATP. Adefovir dipivoxil is a clinically approved drug that can block the action of an anthrax toxin. It can be used to address the role of EF in anthrax pathogenesis.
The spore-forming Bacillus anthracis secretes three major toxins: edema factor (EF), protective antigen (PA), and lethal factor (LF) (1, 2). As an adenylyl cyclase, EF raises the concentration of a second messenger, cyclic AMP (cAMP), inside host cells to supraphysiological levels (3, 4). An inactivating mutation in EF results in reduced survival of germinated anthrax spores in macrophages, indicating an active role for EF at early stages of anthrax infection (5). EF also modulates the profile of cytokines such as tumor necrosis factor α (TNF-α) and interleukin 6 (IL-6) produced by human monocytes, which could impair cellular antimicrobial responses (6). Consequently, a strain of anthrax with a defective EF gene has 100-fold reduced lethality in mice (7).
EF enters host cells via a complex with PA, which is a pH-dependent protein transporter (8). LF, a zinc metalloprotease that inactivates mitogen-activated protein kinase kinase, also enters into host cells by its association with PA (9, 10). LF works coordinately with EF to facilitate bacterial survival in macrophages and to impair host innate immunity (5-7, 11, 12). The combination of toxemia caused by anthrax toxins and bacteremia due to the rapid growth of anthrax bacteria in vital organs can result in sepsis, pulmonary edema, and/or meningitis within days, making inhalational anthrax a deadly disease.
Natural isolates of B. anthracis are sensitive to a broad spectrum of antibiotics; thus antibiotics have been the primary recourse for therapy (13). However, antibiotics are ineffective against either toxemia or antibiotic-resistant strains of anthrax. The antibiotic treatment used for victims of the 2001 bioterrorism-related anthrax attack in the United States resulted in a survival rate of slightly better than 50% for cases of inhalational anthrax. Some survivors have experienced illness with symptoms such as fatigue, shortness of breath, chest pain, and memory loss. However the limited patient sample size does not allow an accurate assessment as to whether such symptoms are anthrax sequelae or not. This situation highlights an urgent need for a more effective treatment to improve the survival rate and quality of life of patients suffering from inhalational anthrax due to future acts of bioterrorism (14).
Clinically approved drugs represent the chemical space that has the favorable pharmacological properties necessary to provide patients with therapeutic benefits (15). To take advantage of this chemical space, we examined a series of nucleotide analogues that mimic ATP, the natural substrate of EF. Here we report that a clinically approved viral drug, adefovir dipivoxil {9-[2-[[bis[(pivaloyloxy)methoxy]phosphinyl]methoxy]ethyl]adenine; bis-POM-PMEA}, can effectively block the pathological effects of anthrax EF on mammalian cells, including EF-induced cAMP accumulation and altered cytokine production by primary macrophages. The cellular metabolite of this drug, adefovir diphosphate {9-[2-(phosphonomethoxy)ethyl]adenine diphosphate; PMEApp}, is a potent and specific inhibitor of the adenylyl cyclase activity of EF in vitro. The crystal structure of PMEApp in complex with EF and its activator, calmodulin (CaM), elucidates the molecular basis of the 10,000-fold higher affinity EF has for PMEApp compared with its endogenous substrate, ATP.
Methods
Materials. bis-POM-PMEA, adefovir {9-[2-(phosphonomethoxy)ethyl]adenine, PMEA}, PMEApp, PMPApp, PMEDAPpp, and PMPDAPpp were synthesized and supplied by Gilead Sciences. (For structures, see Fig. 1 A and B.)
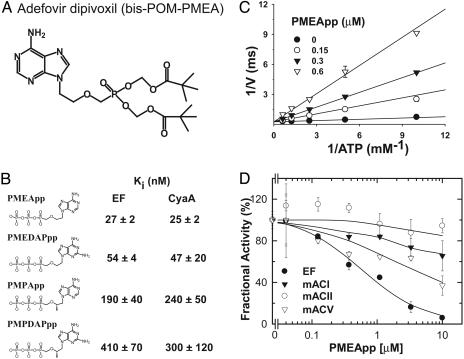
Fig. 1.
Effect of acyclic nucleotide phosphonates (ANPs) on the enzymatic activity of anthrax EF and host adenylyl cyclase. (A) The chemical structure of adefovir dipivoxil (bis-POM-PMEA). (B) Chemical structures of four ANPs and their Ki values for EF and CyaA. (C) Lineweaver-Burk plot for the inhibition of EF by PMEApp. (D) Inhibition of EF and three mammalian adenylyl cyclase isoforms, bovine type I (mACI), rat type II (mACII), and mouse type V (mACV), by PMEApp. Adenylyl cyclase assays were performed with 17 pM EF3, 1 μM CaM, and 1 μM free Ca2+ (B and C) and with 20 μg of Sf9 cell membrane, 0.5 μM Gsα, 100 μM forskolin, 30 μM AlF3, 5 mM MgCl2, and 10 mM NaF (D). Mean ± SE is representative of at least two experiments, and the representative data for B are shown in Figs. 5 and 6, which are published as supporting information on the PNAS web site.
In Vitro Adenylyl Cyclase Assay. The plasmid for the expression of the catalytic domains of EF and adenylyl cyclase toxin (EF3 and CyaA-N) as well as EF3 mutants were constructed and the recombinant proteins were purified from Escherichia coli as described (16). Sf9 insect cells were infected with recombinant baculoviruses for the expression of type I, type II, and type V adenylyl cyclase, and membranes of Sf9 cells containing the overexpressed cyclases were prepared as described (17). Recombinant α-subunit of GS protein (Gsα) was purified from E. coli by using Ni-NTA and Q-Sepharose columns (18). Adenylylcyclase activity of EF-3 and CyaA-N was measured at 30°C for 10 min in the presence of 20 mM Hepes (pH 7.2), 10 mM MgCl2, 1 mM EDTA, 1 μM free Ca2+ (added as CaCl2), and ≈10 nM [32P]ATP with either a fixed concentration of ATP (5 mM) or variable ATP concentrations as indicated (16). ATP and cAMP were separated by a two-column method (Dowex and alumina) and adenylyl cyclase activities were calculated. The adenylyl cyclase activity of 20 μg of Sf9 cell membrane, stimulated by 500 nM Gsα and 100 μM forskolin, was measured at 30°C for 20 min in the presence of 50 μM AlCl3, 10 mM MgCl2, and 10 mM NaF as described (17).
Tissue Culture. Cells were maintained in DMEM/F12 supplemented with 1% l-glutamine and 1% penicillin/1% streptomycin. For Chinese hamster ovary (CHO) cells, 10% calf serum was added; for adrenocortical Y1 cells, 2.5% FBS and 12.5% horse serum were added. Mouse bone marrow (BM) cells were collected by flushing femurs and tibias of C57BL/6 mice with Hanks' balanced salt solution. BM-derived macrophages (BMMΦ) were propagated from BM progenitors in RPMI medium 1640 supplemented with 10% FCS (RPMI-10) and 30% L929-conditioned medium. After 5 days, nonadherent cells were removed and adherent ones were recovered by incubation with trypsin/EDTA at 37°C for 5 min and plated for experiments.
cAMP Accumulation in CHO and BMMΦ Cells and Morphology Change in Adrenocortical Y1 Cells. Recombinant EF and PA were expressed and purified as described (19). To measure cAMP accumulation inside cells, the cells were seeded in a 24-well plate either at 7.5 × 104 cells per well (CHO) or at 5.0 × 105 cells per well (BMMΦ) and allowed to grow for ≈16 h before addition of PMEA. To ensure the accumulation and conversion of bis-POM-PMEA to PMEApp inside cells, bis-POM-PMEA was added 5-8 h before treatment of the cells with ET, and fresh drug was added again 1 h before addition of ET. ET or forskolin was added and cells were incubated for an additional 1-2 h. The levels of cAMP were determined by enzyme immunoassay (Biotrak EIA, Amersham Pharmacia). Morphological changes in adrenocortical Y1 cells were examined as described (19).
Cytokine Production. To study the effect of ET on TNF-α production, BMMΦ (5 × 105 cells per well) were treated with various concentrations of ET and cultured in the presence (for TNF-α) or absence (for IL-6) of E. coli O111:B4 lipopolysaccharide (LPS; 5 μg/ml, Sigma) in a final volume of 500 μl of RPMI-10. To examine the effect of bis-POM-PMEA on cytokine production, BMMΦ were pretreated with various concentrations for 5 h. Cells were then stimulated with ET plus LPS (for TNF-α) or ET alone, and received a second drug treatment. Twenty hours later, the culture supernatants were harvested, and the levels of TNF-α and IL-6 were quantified by sandwich ELISA (Pharmingen).
Structure Determination. Crystals of the EF3-CH6-CaM complex were grown to a size greater than 0.4 mm × 0.4 mm × 0.4 mm. The isolated single crystals were soaked with 2 mM PMEApp overnight during cryoprotection and flash-frozen in liquid nitrogen as described (20). Data were collected at 100 K at the Advanced Photon Source Structural Biology Center Beamline 19-ID, Argonne National Laboratory, and processed by using hkl2000 (21). The initial solutions were obtained by difference Fourier method using cns and a model of the EF3-CaM complex (22). The final model was refined by cns, o, and turbo-frodo (Table 1, which is published as supporting information on the PNAS web site). The coordinates for EF3-CaM-PMEApp are available from the Protein Data Bank (ID code 1PK0).
Results
Characterization of PMEApp as a Potent and Specific Inhibitor of EF. Acyclic nucleoside phosphonates represent a class of nucleotide analogues approved for a variety of viral infections, which include cidofovir [(cytomegalovirus (CMV)], tenofovir [human immunodeficiency virus (HIV)], and adefovir [hepatitis B virus (HBV)] (23). bis-POM-PMEA (Hepsera, Gilead Sciences) (Fig. 1A) is an orally bioavailable prodrug of adefovir (PMEA), an analogue of adenosine monophosphate (AMP) approved for the treatment of chronic HBV infection (23, 24). Inside the cells, PMEA is converted by cellular kinases to the diphosphate derivative PMEApp, a noncyclizable ATP analogue (Fig. 1B). PMEApp is a competitive inhibitor and chain terminator of HBV DNA replication by HBV DNA polymerase (Ki = 100 nM) (25). In an in vitro adenylyl cyclase assay, we found that PMEApp has almost 10,000-fold higher affinity for the EF-CaM complex (Ki = 27 nM, Fig. 1B) than the endogenous substrate, ATP (Km = 168-194 μM; Fig. 5) and 4-fold higher affinity than the affinity for its original therapeutic target, HBV DNA polymerase (Figs. 1B and 5) (25). Kinetic analysis revealed that PMEApp inhibits the catalytic activity of EF by competing with the binding of ATP (Fig. 1C). Quantitative structure-activity relationship (QSAR) analysis (Figs. 1B and 5) revealed that the addition of a methyl group at the 2′ position of PMEApp to create 9-[2-(phosphonomethoxy)propyl)]adenine diphosphate (PMPApp) resulted in 7-fold lower affinity, whereas the addition of an amino group at the C2 position of the adenine base to create 9-[2-(phosphonomethoxy)ethyl]-2,6-diaminopurine diphosphate (PMEDAPpp) generated ≈2-fold lower affinity. Furthermore, when these two substitutions were combined in 9-[2-(phosphonomethoxy)propyl]-2,6-diaminopurine diphosphate (PMPDAPpp), the effects were additive and a 15-fold reduction in affinity was observed.
We also tested the effect of PMEApp and its analogues on the catalytic activity of CyaA, an adenylyl cyclase toxin with 35% amino acid sequence identify to EF. CyaA is secreted by Bordetella pertussis, the causative agent of whooping cough, and it is vital for bacterial colonization in vivo (26). We found that PMEApp also competitively inhibits the activity of CyaA with a Ki of 25 nM (Figs. 1B and 6). The QSAR profile of PMEApp analogues against CyaA was parallel to the profile for EF (Fig. 1B). Because it was the most potent inhibitor of EF, we focused our subsequent efforts on PMEApp.
We examined the ability of PMEApp to inhibit host mammalian adenylyl cyclases (mACs). At least nine isoforms of membrane-bound mAC as well as one testis-specific soluble mAC have been characterized (27, 28). We examined the effect of PMEApp on the Gsα- and forskolin-stimulated activity of mAC types I, II, and V, representing the three major families of mAC. We found that PMEApp had 10- to 20-fold lower affinity for mouse type V and bovine type I mAC than for EF (Fig. 1D), similar to the reported affinity of PMEApp for rat brain adenylyl cyclase (29). Even at a concentration of 10 μM, PMEApp exhibited little inhibition of type II mAC (Fig. 1D). Such selectivity for EF and CyaA over mAC suggests that PMEApp may be used at concentrations that neutralize the bacterial toxins effectively without deleterious side effects through interference with host mACs.
Effectiveness of the Antiviral Drug Bis-POM-PMEA in Blocking EF-Induced cAMP Accumulation and Altering Cytokine Production in Primary Macrophages. Encouraged by the selectivity demonstrated in enzymatic assays, we tested whether bis-POM-PMEA, the prodrug of PMEApp (Fig. 1 A), could block the biological effects of edema toxin (ET, the combination of EF and PA) on cells. We found that bis-POM-PMEA could effectively inhibit ET-induced cAMP accumulation in CHO cells with an IC50 value around 0.1 μM (Fig. 2A). Bis-POM-PME was 20-fold less effective in inhibiting the cAMP accumulation stimulated by forskolin, a diterpene that stimulates most mACs (Fig. 2 A) (27), consistent with the selectivity for EF over mACs observed in the enzymatic assays. We also found that bis-POM-PMEA could prevent ET-induced morphological changes in Y1 cells, a mouse adrenocortical cell line previously used to monitor ET effects (Fig. 7, which is published as supporting information on the PNAS web site) (19).
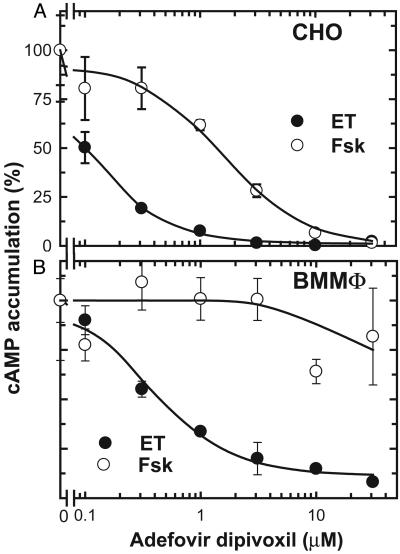
Fig. 2.
Effects of bis-POM-PMEA on ET-induced cAMP accumulation in CHO cells (A) and mouse BMMΦ (B). Cells were pretreated with bis-POM-PMEA for 5 h, then fresh bis-POM-PMEA was reapplied together with ET (5 ng/ml EF and 25 ng/ml PA for CHO and 30 ng/ml EF and 300 ng/ml PA for BMMΦ). For comparison, forskolin (Fsk; 100 μM for CHO cells and 10 μM for BMMΦ) was used to elevate intracellular cAMP levels. Mean ± SE is representative of at least two experiments.
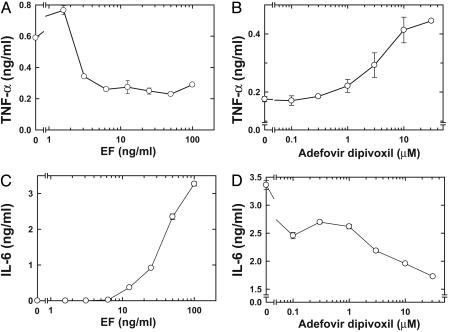
Macrophages play key roles in anthrax pathogenesis (5, 6, 30). In the early stages of infection, spores germinate in and are disseminated by macrophages. However, in the later stage of infection, anthrax toxins modify the functions of or kill macrophages. We thus examined the effect of bis-POM-PMEA on BMMΦ. We found that ET could raise intracellular cAMP levels in BMMΦ in a dose-dependent manner that could be blocked by bis-POM-PMEA. Bis-POM-PMEA was significantly less effective in blocking cAMP accumulation induced by forskolin (Fig. 2B). ET has been shown to down-regulate the production of TNF-α in human mononuclear phagocytes after stimulation with LPS (6). Similarly, we found that ET significantly reduced LPS-induced TNF-α production by BMMΦ (Fig. 3A). Preincubation of BMMΦ with bis-POM-PMEA effectively reversed the EF-mediated down-regulation of TNF-α production (Fig. 3B). ET has also been shown to induce IL-6 production by human monocytes; we found a similar effect in BMMΦ (Fig. 3C) (6). The addition of bis-POM-PMEA significantly reduced ET's effect on IL-6 production in BMMΦ (Fig. 3D). Taken together, our data indicate that bis-POM-PMEA can effectively inhibit ET-induced cAMP accumulation and the alteration of cytokine production in macrophages.
Fig. 3.
Effect of bis-POM-PMEA on cytokine production by BMMΦ. (A) Effect of ET on LPS-induced TNF-α production. (B) Bis-POM-PMEA reversed the suppressive effect of ET on LPS-induced TNF-α production. (C) Effect of ET on IL-6 production. (D) Bis-POM-PMEA blocked ET-induced IL-6 production. To study the effect of ET on cytokine production, BMMΦ were treated with indicated amounts of EF and a 10-fold higher concentration of PA in the presence (A) or absence (C) of LPS. To examine the effect of PMEA (B and D), BMMΦ were pretreated with various concentrations of bis-POM-PMEA as described for Fig. 2. Cells were then stimulated with ET (12 ng/ml EF and 120 ng/ml PA) and 5 μg/ml LPS (B) or 30 ng/ml EF and 300 ng/ml PA (D) and received a second drug treatment. Culture supernatants were harvested 20 h later, and the amount of TNF-α (A and B) or IL-6 (C and D) was measured by using ELISA. Mean ± SE is representative of at least two experiments.
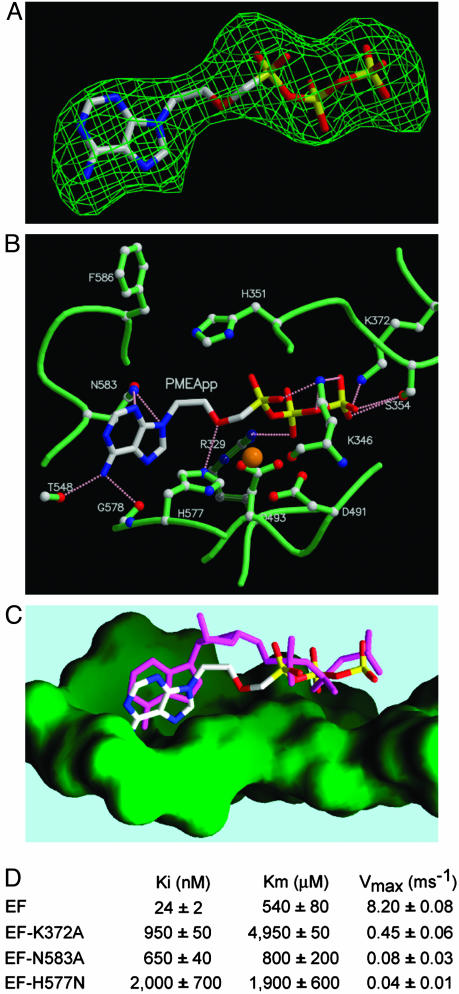
Molecular Basis for the High Affinity Between EF-CaM and PMEApp. PMEApp displays an affinity for the EF catalytic site that is four orders of magnitude higher than the affinity of ATP for EF. To determine the molecular basis for this higher affinity, we determined the molecular structure of EF-CaM in complex with PMEApp at a resolution of 3.3 Å (Table 1). The electron density of PMEApp is clearly visible at the catalytic site (Fig. 4A). PMEApp forms a network of salt bridges and hydrogen bonds with the amino acids at the catalytic site of EF (Fig. 4B). The adenine base and triphosphate moieties of PMEApp make contacts with EF similar to those of 3′-deoxy-ATP (3′dATP, a noncyclizable ATP analogue) (Fig. 4C) (4). Consistent with our structures, in which Lys-372 of EF forms a salt bridge with the γ-phosphate of both substrates, a mutation replacing Lys-372 with Ala (K372A) resulted in a 40-fold increase in the Ki for PMEApp as well as a 10-fold increase in the Km for ATP (Figs. 4D and 8). Although PMEApp and 3′dATP use similar modes of binding to EF, PMEApp has significantly more van der Waals interactions with the ventral cleft of the catalytic site of EF than does 3′dATP (Fig. 4C). Furthermore, PMEApp forms additional hydrogen bonds with EF, including the interaction of adenine N3 and N9 with Asn-583 and that of the methoxy group with His-577 (Fig. 4C). The importance of these hydrogen bonds in the PMEApp-EF-CaM complex is supported by the finding that the replacement of Asn-583 with Ala (N593A) and His-577 with Asn (H577N) significantly increased the Ki values for PMEApp (30- and 80-fold reduction, respectively) but had only a minor effect on the Km for ATP (Figs. 4D and 8). In addition, PMEApp has more negative charges in the nonbridging oxygens of the α-phosphate, which mimic the pentavalent transition state for the cyclization reaction of EF (31). Because enzymes have evolved to have substantially better bonding in the transition state than in the ground state, mimicking the transition state could significantly contribute to the affinity of PMEApp for EF (32). Taken together, better van der Waals contacts, additional hydrogen bonds, and transition state mimicry likely explain why PMEApp has 10,000-fold higher affinity for EF than does ATP.
Fig. 4.
Structural analyses of the interactions of PMEApp with EF. (A) Omit map of PMEApp in the catalytic site of EF. The model of PMEApp is shown with the 3σ cutoff electron density map. The color of PMEApp is kept the same throughout, in which carbon, oxygen, nitrogen, and phosphorus are white, red, blue, and yellow, respectively. (B) The interactions of the EF-CaM complex with PMEApp. The structure of EF-CaM in the presence of PMEApp is in green, and the atoms of carbon, oxygen, and nitrogen are white, red, and blue, respectively. A catalytic metal, ytterbium, is orange, and hydrogen bonds are represented in pink. (C) Comparison of PMEApp with 3′-deoxy-ATP. The molecular surface of the ventral side of the active site is green. 3′-Deoxy-ATP is in magenta. (D) Kinetic analysis of wild-type and mutant forms of EF, EF3-K372A, EF3-N583A, and EF3-H577N. Representative kinetic data for D are supplied in Fig. 8, which is published as supporting information on the PNAS web site.
Discussion
The fatalities and long term disabilities resulting from the bioterrorism-related anthrax in 2001 have highlighted the need to effectively prevent and treat the symptoms and sequelae of inhalational anthrax. The pathogenesis of anthrax is mediated by the combination of toxemia and bacteremia. Therefore, effective treatment must block both the actions of anthrax toxins and the growth of anthrax bacteria. Several potential antitoxins have been developed recently (19, 33-36). However, extensive modifications and testing will be required to realize their therapeutic potential. Bis-POM-PMEA is the first clinically approved drug that can block the action of an anthrax toxin in vitro. Our studies suggest that bis-POM-PMEA may be a promising adjunctive therapy against anthrax and other human diseases caused by pathogenic bacteria that secrete adenylyl cyclase toxins, such as Bordetella pertussis (whooping cough), Pseudomonas aeruginosa (nosocomial infections), and Yersinia pestis (plague) (26, 37, 38).
The active metabolite of bis-POM-PMEA, PMEApp, is a slightly more potent inhibitor of the adenylyl cyclase activity of EF (Ki = 27 nM) than of its approved therapeutic target, HBV DNA polymerase (Ki = 100 nM) (25). Correspondingly, the effective concentrations of bis-POM-PMEA required for inhibition of the cellular effects induced by ET (IC50 = 0.1-0.5 μM) are comparable to those reported for inhibition of HBV DNA replication by bis-POM-PMEA in hepatoblastoma cells in vitro (IC50 = 0.7-1.2 μM) (39). Because bis-POM-PMEA possesses favorable pharmacological properties that permit once-daily oral administration of a relatively low dose (10 mg) for the treatment of chronic hepatitis B, further studies are warranted to determine whether bis-POM-PMEA can exert antitoxin activity in vivo. In addition, the molecular structures of EF-CaM in complex with PMEApp as well as the molecular structure of the catalytic domain of mACs are available to provide a structural blueprint to further improve the affinity and selectivity of PMEA for EF for the next generation of drugs (40). Further studies are required to determine whether bis-POM-PMEA (or a related member of this class of acyclic nucleotide analogues) can be used experimentally to assess the role of adenylyl cyclase toxins in bacterial pathogenesis as well as clinically to provide protection against several deadly human diseases.
Supplementary Material
Acknowledgments
We are grateful to Xiaojing Yang at Renz Research, Inc., for the maintenance of computers for the laboratory of W.-J.T., the beamline personnel Younchang Kim and Andrzej Joachimiak at the Advanced Photon Source Structural Biology Center for their assistance in data collection and processing, and Michael D. Miller at Gilead Sciences and Jan Florián at Loyola University for critical review of the manuscript. This work was supported by National Institutes of Health Grants GM-62548 and GM-53459 (to W.-J.T.) and AI-40301 (to C.-R.W.). Use of the Argonne National Laboratory Structural Biology Center and BioCARS beamlines at the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Energy Research, under Contract W-31-109-ENG-38.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: EF, edema factor; PA, protective antigen; ET, edema toxin; TNF-α, tumor necrosis factor α; CaM, calmodulin; PMEA or adefovir, 9-[2-(phosphonomethoxy)ethyl]adenine; bis-POM-PMEA or adefovir dipivoxil, 9-[2-[[bis[(pivaloyloxy)methoxy]phosphinyl]methoxy]ethyl]adenine; PMEApp or adefovir diphosphate, PMEA diphosphate; PMPApp, 9-[2-(phosphonomethoxy)propyl)]adenine diphosphate; PMEDAPpp, 9-[2-(phosphonomethoxy)ethyl]-2,6-diaminopurine diphosphate; PMPDAPpp, 9-[2-(phosphonomethoxy)propyl]-2,6-diaminopurine diphosphate; CyaA, adenylyl cyclase toxin; BMMΦ, bone marrow-derived macrophage(s); LPS, lipopolysaccharide; CHO, Chinese hamster ovary; HBV, hepatitis B virus; mAC, mammalian adenylyl cyclase.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1PK0).
References
- 1.Mock, M. & Fouet, A. (2001) Annu. Rev. Microbiol. 55, 647-671. [DOI] [PubMed] [Google Scholar]
- 2.Mourez, M., Lacy, D. B., Cunningham, K., Legmann, R., Sellman, B. R., Mogridge, J. & Collier, R. J. (2002) Trends Microbiol. 10, 287-293. [DOI] [PubMed] [Google Scholar]
- 3.Leppla, S. H. (1982) Proc. Natl. Acad. Sci. USA 79, 3162-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drum, C. L., Yan, S. Z., Bard, J., Shen, Y., Lu, D., Soelaiman, S., Grabarek, Z., Bohm, A. & Tang, W.-J. (2002) Nature 415, 396-402. [DOI] [PubMed] [Google Scholar]
- 5.Guidi-Rontani, C., Levy, M., Ohayon, H. & Mock, M. (2001) Mol. Microbiol. 42, 931-938. [DOI] [PubMed] [Google Scholar]
- 6.Hoover, D. L., Friedlander, A. M., Rogers, L. C., Yoon, I. K., Warren, R. L. & Cross, A. S. (1994) Infect. Immun. 62, 4432-4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brossier, F., Weber-Levy, M., Mock, M. & Sirard, J. C. (2000) Infect. Immun. 68, 1781-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petosa, C., Collier, R. J., Klimpel, K. R., Leppla, S. H. & Liddington, R. C. (1997) Nature 385, 833-838. [DOI] [PubMed] [Google Scholar]
- 9.Duesbery, N. S., Webb, C. P., Leppla, S. H., Gordon, V. M., Klimpel, K. R., Copeland, T. D., Ahn, N. G., Oskarsson, M. K., Fukasawa, K., Paull, K. D. & Vande Woude, G. F. (1998) Science 280, 734-737. [DOI] [PubMed] [Google Scholar]
- 10.Vitale, G., Bernardi, L., Napolitani, G., Mock, M. & Montecucco, C. (2000) Biochem. J. 352, 739-745. [PMC free article] [PubMed] [Google Scholar]
- 11.Agrawal, A., Lingappa, J., Leppla, S. H., Agrawal, S., Jabbar, A., Quinn, C. & Pulendran, B. (2003) Nature 424, 329-334. [DOI] [PubMed] [Google Scholar]
- 12.Pellizzari, R., Guidi-Rontani, C., Vitale, G., Mock, M. & Montecucco, C. (1999) FEBS Lett. 462, 199-204. [DOI] [PubMed] [Google Scholar]
- 13.Dixon, T. C., Meselson, M., Guillemin, J. & Hanna, P. C. (1999) N. Engl. J. Med. 341, 815-826. [DOI] [PubMed] [Google Scholar]
- 14.Atlas, R. M. (2002) Annu. Rev. Microbiol. 56, 167-185. [DOI] [PubMed] [Google Scholar]
- 15.Lipinski, C. A., Lombardo, F., Dominy, B. W. & Feeney, P. J. (2001) Adv. Drug Delivery Rev. 46, 3-26. [DOI] [PubMed] [Google Scholar]
- 16.Shen, Y., Lee, Y. S., Soelaiman, S., Bergson, P., Lu, D., Chen, A., Beckingham, K., Grabarek, Z., Mrksich, M. & Tang, W. J. (2002) EMBO J. 21, 6721-6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang, W. J., Krupinski, J. & Gilman, A. G. (1991) J. Biol. Chem. 266, 8595-8603. [PubMed] [Google Scholar]
- 18.Yan, S. Z. & Tang, W. J. (2002) Methods Enzymol. 344, 171-175. [DOI] [PubMed] [Google Scholar]
- 19.Soelaiman, S., Wei, B. Q., Bergson, P., Lee, Y. S., Shen, Y., Mrksich, M., Shoichet, B. K. & Tang, W. J. (2003) J. Biol. Chem. 278, 25990-25997. [DOI] [PubMed] [Google Scholar]
- 20.Drum, C. L., Shen, Y., Rice, P. A., Bohm, A. & Tang, W. J. (2001) Acta Crystallogr. D 57, 1881-1884. [DOI] [PubMed] [Google Scholar]
- 21.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 22.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 23.Marcellin, P., Chang, T. T., Lim, S. G., Tong, M. J., Sievert, W., Shiffman, M. L., Jeffers, L., Goodman, Z., Wulfsohn, M. S., Xiong, S., et al. (2003) N. Engl. J. Med. 348, 808-816. [DOI] [PubMed] [Google Scholar]
- 24.Hadziyannis, S. J., Tassopoulos, N. C., Heathcote, E. J., Chang, T. T., Kitis, G., Rizzetto, M., Marcellin, P., Lim, S. G., Goodman, Z., Wulfsohn, M. S., et al. (2003) N. Engl. J. Med. 348, 800-807. [DOI] [PubMed] [Google Scholar]
- 25.Xiong, X., Flores, C., Yang, H., Toole, J. J. & Gibbs, C. S. (1998) Hepatology 28, 1669-1673. [DOI] [PubMed] [Google Scholar]
- 26.Goodwin, M. S. & Weiss, A. A. (1990) Infect. Immun. 58, 3445-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang, W.-J. & Hurley, J. H. (1998) Mol. Pharmacol. 54, 231-240. [DOI] [PubMed] [Google Scholar]
- 28.Chen, Y., Cann, M. J., Litvin, T. N., Iourgenko, V., Sinclair, M. L., Levin, L. R. & Buck, J. (2000) Science 289, 625-628. [DOI] [PubMed] [Google Scholar]
- 29.Shoshani, I., Laux, W. H., Perigaud, C., Gosselin, G. & Johnson, R. A. (1999) J. Biol. Chem. 274, 34742-34744. [DOI] [PubMed] [Google Scholar]
- 30.Park, J. M., Greten, F. R., Li, Z. W. & Karin, M. (2002) Science 297, 2048-2051. [DOI] [PubMed] [Google Scholar]
- 31.Florián, J., Strajbl, M. & Warshel, A. (1998) J. Am. Chem. Soc. 120, 7959-7966. [Google Scholar]
- 32.Warshel, A. & Florián, J. (1998) Proc. Natl. Acad. Sci. USA 95, 5950-5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bradley, K. A., Mogridge, J., Mourez, M., Collier, R. J. & Young, J. A. (2001) Nature 414, 225-229. [DOI] [PubMed] [Google Scholar]
- 34.Mourez, M., Kane, R. S., Mogridge, J., Metallo, S., Deschatelets, P., Sellman, B. R., Whitesides, G. M. & Collier, R. J. (2001) Nat. Biotechnol. 19, 958-961. [DOI] [PubMed] [Google Scholar]
- 35.Sellman, B. R., Mourez, M. & Collier, R. J. (2001) Science 292, 695-697. [DOI] [PubMed] [Google Scholar]
- 36.Tonello, F., Seveso, M., Marin, O., Mock, M. & Montecucco, C. (2002) Nature 418, 386. [DOI] [PubMed] [Google Scholar]
- 37.Parkhill, J., Wren, B. W., Thomson, N. R., Titball, R. W., Holden, M. T., Prentice, M. B., Sebaihia, M., James, K. D., Churcher, C., Mungall, K. L., et al. (2001) Nature 413, 523-527. [DOI] [PubMed] [Google Scholar]
- 38.Yahr, T. L., Vallis, A. J., Hancock, M. K., Barbieri, J. T. & Frank, D. W. (1998) Proc. Natl. Acad. Sci. USA 95, 13899-13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heijtink, R. A., De Wilde, G. A., Kruining, J., Berk, L., Balzarini, J., De Clercq, E., Holy, A. & Schalm, S. W. (1993) Antiviral Res. 21, 141-153. [DOI] [PubMed] [Google Scholar]
- 40.Tesmer, J. J., Sunahara, R. K., Gilman, A. G. & Sprang, S. R. (1997) Science 278, 1907-1916. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.