Abstract
Rho family guanosine triphosphatase (GTPase) 3 (Rnd3), a member of the small Rho GTPase family, is involved in the regulation of cell actin cytoskeleton dynamics, cell migration, and proliferation through the Rho kinase-dependent signaling pathway. We report a role of Rnd3 in the pathogenesis of hydrocephalus disorder. Mice with Rnd3 genetic deletion developed severe obstructive hydrocephalus with enlargement of the lateral and third ventricles, but not of the fourth ventricles. The cerebral aqueducts in Rnd3-null mice were partially or completely blocked by the overgrowth of ependymal epithelia. We examined the molecular mechanism contributing to this Rnd3-deficiency–mediated hydrocephalus and found that Rnd3 is a regulator of Notch signaling. Rnd3 deficiency, through either gene deletion or siRNA knockdown, resulted in a decrease in Notch intracellular domain (NICD) protein degradation. However, there was no correlated change in mRNA change, which in turn led to an increase in NICD protein levels. Immunoprecipitation analysis demonstrated that Rnd3 and NICD physically interacted, and that down-regulation of Rnd3 attenuated NICD protein ubiquitination. This eventually enhanced Notch signaling activity and promoted aberrant growth of aqueduct ependymal cells, resulting in aqueduct stenosis and the development of congenital hydrocephalus. Inhibition of Notch activity rescued the hydrocephalus disorder in the mutant animals.
Keywords: developmental brain defect, genetic mouse model, post-translational modification
The cerebrospinal fluid (CSF) flow tract is a dynamic circulatory system that supplies the brain with essential nutrients and growth factors throughout development and into adulthood (1). Ependymal cells, specified epithelial cells that line the aqueduct and cerebral ventricles of mammalian species, make up the CSF tract and propel CSF flow (2, 3). Malformation of the ependymal epithelium can lead to failure of cerebral irrigation and a buildup of CSF in the ventricular cavities of the brain, a pathological condition known as hydrocephalus.
Hydrocephalus in humans is a severe neurologic disorder characterized by an excessive accumulation of CSF in the brain. It affects approximately 1 in every 500 children, based on estimates from the National Institute of Neurological Disorders and Stroke. The precise causes of hydrocephalus remain largely unknown. Accumulating evidence suggests that genetic factors are critical to the pathogenesis of hydrocephalus. Congenital hydrocephalus, caused by genetic abnormalities, is the most common type. Aqueductal stenosis is the major contributing factor in congenital hydrocephalus, with an prevalence of 0.1–0.3% of all live births (4). At least nine genes have been identified as closely associated with the pathogenesis of congenital hydrocephalus in animal studies, but few of these genes have been verified in humans (5–8). Continued identification of new genes/loci linked to congenital hydrocephalus will expand our knowledge of the genetic components of this disorder.
Rho family GTPase 3 (Rnd3, also known as RhoE) was originally defined as a repressor of Rho protein kinase 1 (ROCK1) (9, 10). Initial studies of Rnd3 focused mainly on its inhibitory effect on Rho kinase-mediated biological functions, including actin cytoskeleton formation, myosin light chain phosphatase phosphorylation, and apoptosis (9–11). Recent studies have identified Rnd3 as essential in mouse neuron development through its negative regulation of the Rho signaling pathway (12, 13).
In this study, we demonstrated a different function of Rnd3. Mice with a genetic deletion of Rnd3 developed aqueductal stenosis, which led to the development of obstructive hydrocephalus owing to hyperplasia of ependymal cells in the aqueducts. We also identified the molecular mechanism of Rnd3 deficiency-induced hydrocephalus and demonstrated that Rnd3 is a regulator of Notch signaling. Rnd3 deficiency in animal studies and down-regulation in cell culture experiments were associated with obvious decreases in Notch intracellular domain (NICD) degradation, which in turn led to an increase in NICD protein levels. This eventually promoted Notch signaling activity and facilitated aqueduct ependymal cell proliferation, resulting in aqueductal stenosis and the development of congenital hydrocephalus. The pathogenesis of hydrocephalus was curtailed by the inhibition of Notch activity.
Results
High Expression of Rnd3 Was Detected in Mouse Brains, Particularly in Ependymal Cells Lining Aqueducts and Ventricles.
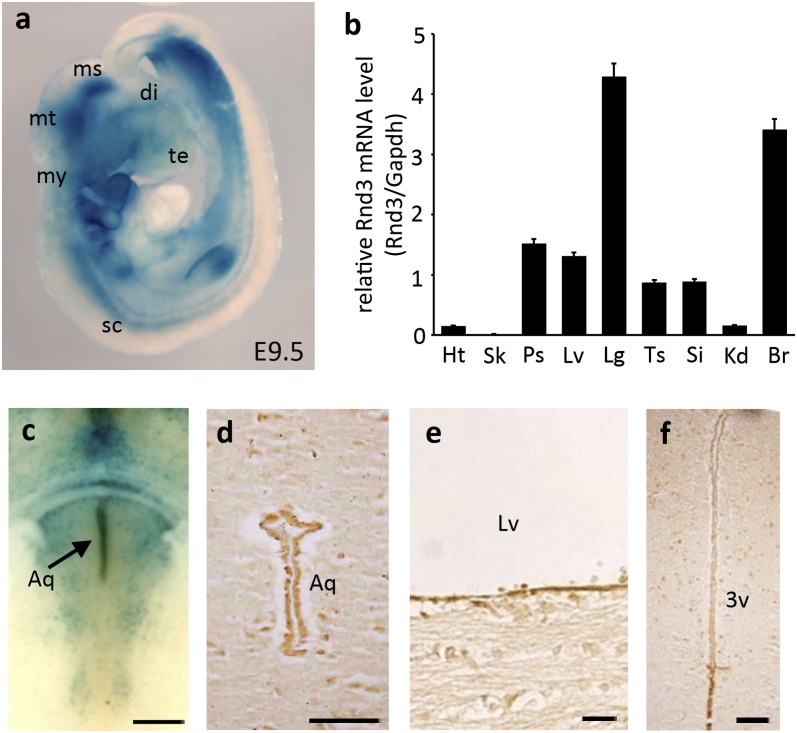
The generation of Rnd3 KO mice is described in Materials and Methods. The targeting vector carried a β-gal expression cassette with an internal ribosomal entry site. The endogenous levels of Rnd3 were detected by beta-galactosidase (lacZ) staining, showing the universal expression of Rnd3, with high expression levels in the CNS (Fig. 1A). Rnd3 transcript levels in various organs were assessed by quantitative PCR (qPCR) and found to be consistent with the lacZ staining (Fig. 1B). Immunohistochemistry analysis revealed enriched expression of Rnd3 in the ependymal cells lining the aqueductal lumina (Fig. 1 C and D) and the inner ventricle walls (Fig. 1 E and F).
Fig. 1.
Expression of Rnd3 is universal and enriched in the CNS, with the highest levels in the ependymal cells lining the aqueduct, lateral ventricle, and third ventricle. (A) LacZ staining displayed Rnd3 expression in an E9.5 mouse. (B) Transcript levels of Rnd3 were quantified by qPCR analysis. (C and D) Enriched expression of Rnd3 was detected in ependymal cells at an aqueduct (Aq) region shown by lacZ staining (C, arrow) and by immunostaining (D). (E and F) Ependymal cells at lateral ventricle (LV) (E) and third ventricle (3V) (F) also displayed high expression levels of Rnd3. qPCR data were pooled from four mice, with analyses for each organ done in triplicate. di, diencephalon; ms, mesencephalon; mt, metencephalon; my, myelencephalon; sc, spinal cord; te, telencephalon; Ht, heart; Sk, skeletal muscle; Ps, prostate; Lv, Liver; Lg, lung; Ts, testis; Si, small intestine; Kd, kidney; Br, brain. (Scale bars: 500 µm in C; 25 µm in D–F.)
Severe Hydrocephalus Was Caused by Aqueductal Stenosis Through Aberrant Growth of Ependymal Cells as a Result of Genetic Deletion of Rnd3.
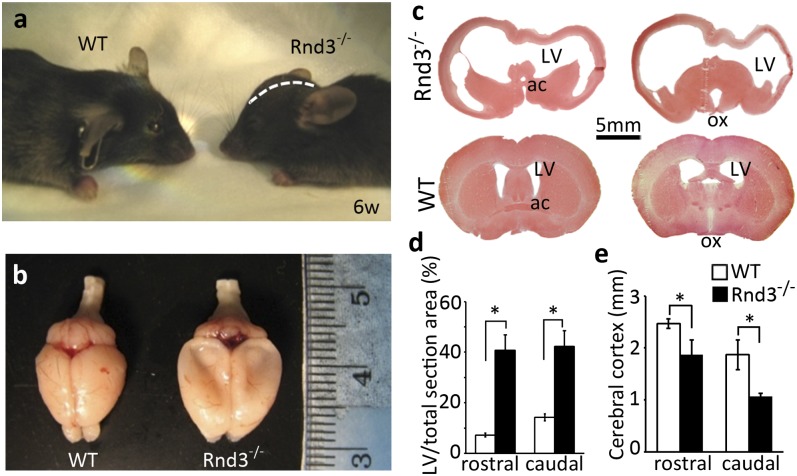
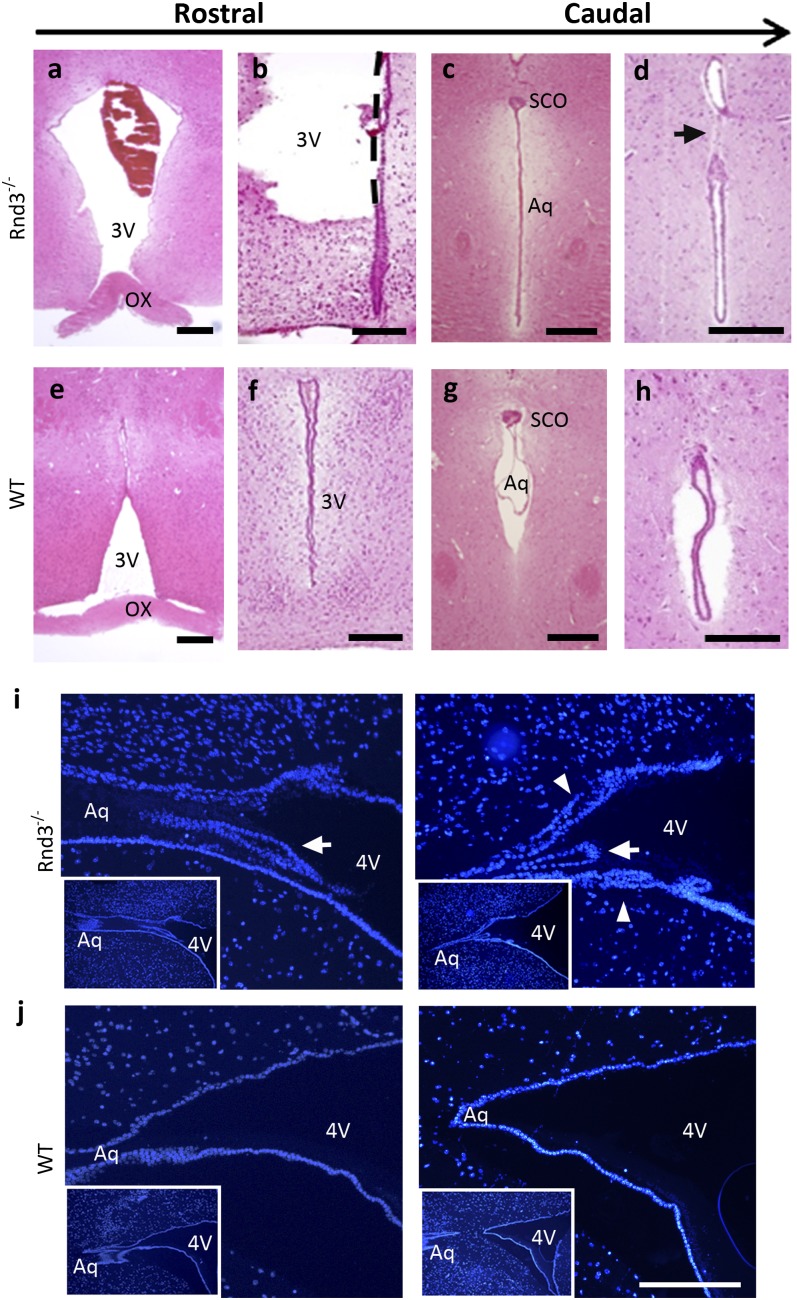
Rnd3−/− mice exhibited a dome-shaped skull (Fig. 2A). A large amount of CSF drained out of the mutant mouse brain during isolation (Fig. 2B). Significantly enlarged lateral ventricles and remarkable thinning of the occipital cortex were observed (Fig. 2C) and quantified (Fig. 2D). Further dissections revealed the extremely dilated or broken third ventricles (Fig. 3 A and B) along with the narrowed or even disconnected aqueduct (Fig. 3 C and D). No enlargement or dilation was seen in the fourth ventricles. Examination of brain sagittal revealed stenosis and multiple layers of ependymal cells at the aqueduct end in the mutant mice (Fig. 3I), compared with the open aqueduct smoothly lined with a single layer of ependymal cells in the WT mice (Fig. 3J). These findings indicate that a severe noncommunicating hydrocephalus developed in the Rnd3-null mice, and that the aqueductal stenosis contributed to the genesis of this hydrocephalus possibly through the overgrowth of ependymal cells.
Fig. 2.
Rnd3-deficient mice developed hydrocephalus. (A) The mutant mouse displayed a domed skull (white dashed line) compared with the WT mouse. (B) A large amount of CSF was drained out during brain isolation, and the global brains showed the enlarged hemispheres, compressed olfactory bulbs, and subsided cerebral cortex in the mutant mice. (C) Representative brain coronal sections across the anterior commissure (ac) (Left) and optical chiasm (ox) (Right) showed the extreme dilations of the LVs along with a thin cerebral cortex in the mutant mice (Upper) compared with the WT mice (Lower). The brain sections were stained with H&E. (D and E) The area of the LV (D) and the thickness of the cerebral cortex (E) were quantified from a total of 12 staining pictures taken from six brains from 6-wk-old mice in each group. *P < 0.05 vs. WT.
Fig. 3.
Dilated third ventricle and stenotic aqueduct in Rnd3-deficient mice. (A–H) Brain coronal sections with H&E staining displayed an enlarged (A) or damaged (B) third ventricle (3V) and stenotic (C) or completely blocked (D) aqueduct in 6-wk-old Rnd3−/− mice compared with WT control mice in the identical areas (E–H, respectively). SCO, subcommissural organ; Ox, optic chiasm; Aq, aqueduct. (Scale bars: 500 μm.) (I and J) The stenotic aqueduct was viewed in sagittal sections with DAPI staining. The stenosis (arrow) and multiple layers of ependymal cells (arrowhead) at the end of the aqueduct were detected in the mutant mice (I), but not in the WT control mice (J). The left and right sections in panels I and J represent different layers of the same brain, respectively. (Insets) Lower-magnification views of the same areas shown in I and J. (Scale bars: 50 μm.) 4V, fourth ventricle.
Rnd3 Negatively Regulated Notch Signaling Through Interaction and Degradation of NICD.
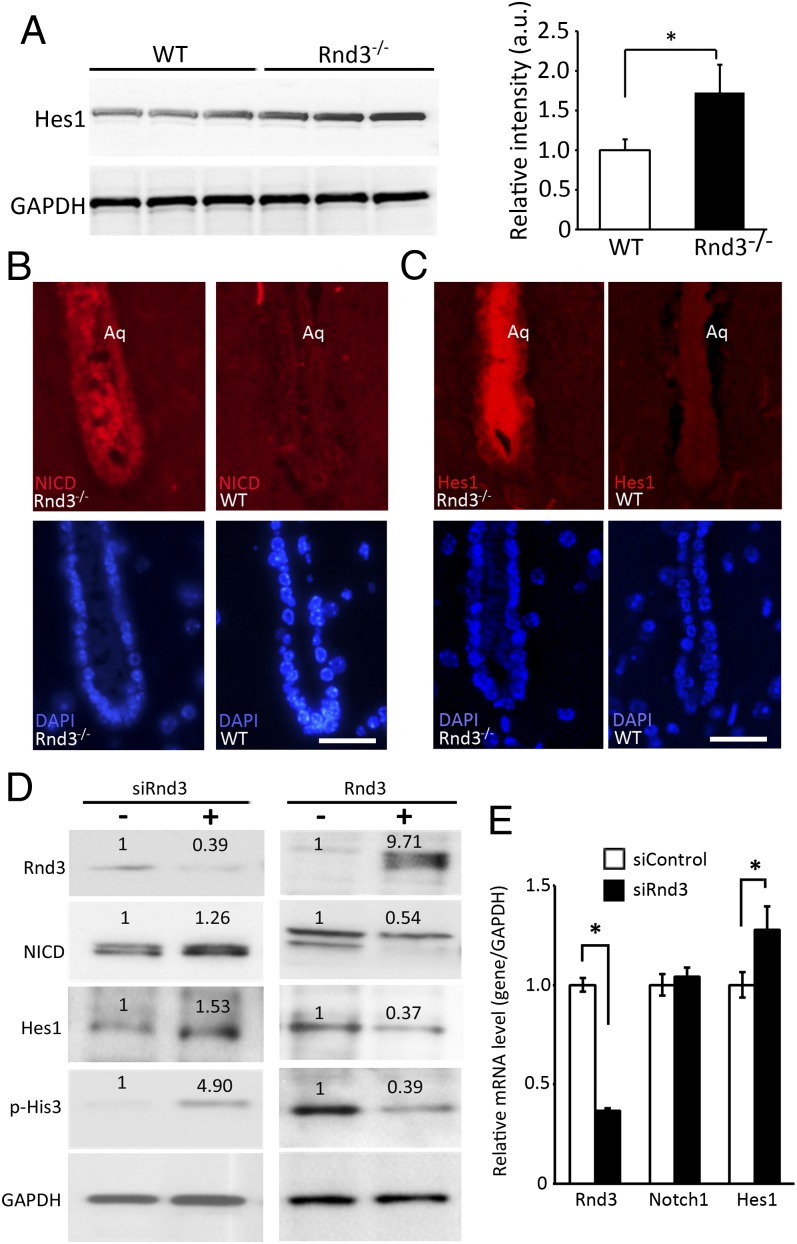
To explore the molecular mechanism of Rnd3 deficiency-mediated aqueductal ependymal cell overgrowth, we conducted a pilot microarray assessment in the mutant mice, which revealed significant up-regulation of Notch signaling (Figs. S1 and S2 and Dataset S1). Thus, we analyzed the mutant mouse brain, and found an increase in the levels of Hes1 protein (Fig. 4A). Hes1 is a Notch signaling target gene demonstrated to function as a p21 repressor to promote cell proliferation (14). Meanwhile, immunostaining specific for NICD, the active Notch isoform, revealed a strong detection in the mutant mouse aqueduct (Fig. 4B), along with strong immunostaining for Hes1 at the same location (Fig. 4C). To evaluate the relationship between Rnd3 and activation of Notch signaling, we used loss-of-function and gain-of-function approaches for in vitro studies using HEK 293T cells. Rnd3 knockdown resulted in elevations of NICD and Hes1 protein levels, along with histone 3 (His3) hyperphosphorylation (Fig. 4D, Left). The opposite results were achieved after the cells were transfected with the Rnd3 expression vector (Fig. 4D, Right). However, knockdown of Rnd3 failed to increase the NICD (Notch1) mRNA levels, even though the Hes1 mRNA levels were elevated (Fig. 4E). These data suggest that NICD is under posttranslational regulation through Rnd3.
Fig. 4.
Rnd3 negatively regulated Notch signaling at protein expression levels. (A) Protein expression level from the Notch signaling target gene, Hes1, was significantly increased in the brain tissues of Rnd3-null mice compared with the WT control mice. This up-regulation was quantified and presented in the right panel of this section. (B and C) Marked increases in NICD (B) and Hes1 (C) immunostaining were seen in the aqueduct (Aq) from a 6-wk-old Rnd3-null mouse compared with a WT control mouse. (D) Immunoblot analyses showed that the down-regulation of Rnd3 resulted in increases in the Notch1 active form NICD, Notch target gene Hes1, and p-His3 expression levels in HEK 293T cells. The opposite results were observed on Rnd3 overexpression. The number at the top of each band represents the average of densitometries from three experiments, normalized by GAPDH. (E) qPCR analysis indicated no change in Notch1, but elevated Hes1 transcript levels by Rnd3 knockdown. The data were pooled from three experiments, with analyses for each in triplicate. (Scale bar: 10 μm.) *P < 0.05 vs. control or WT.
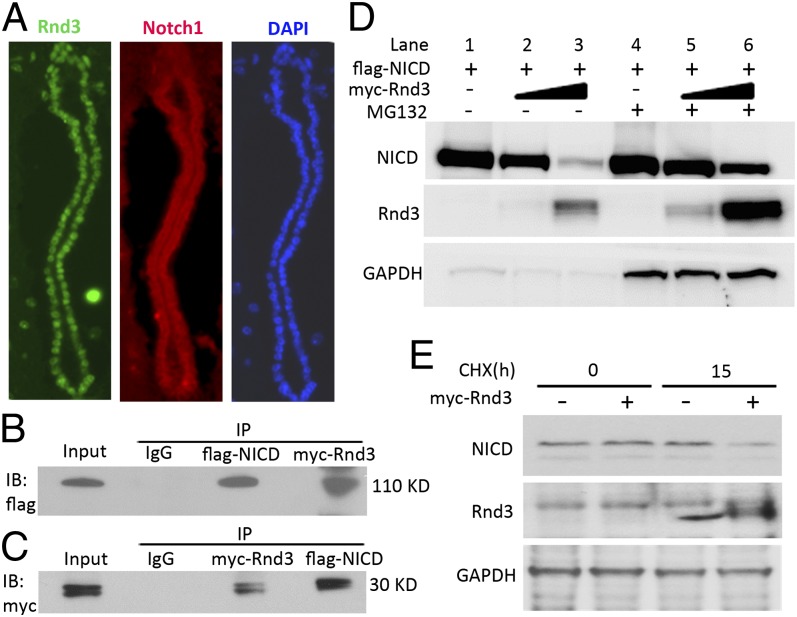
Given that the ubiquitin-proteasome system (UPS) is a critical posttranslational regulation mechanism for NICD, we investigated whether Rnd3 plays a role in UPS-mediated NICD degradation. We first examined the expression profiles of the two proteins, and found that both Rnd3 and Notch1 receptors were highly expressed in mouse aqueductal ependyma (Fig. 5A). We then explored the interaction of Rnd3 and NICD ex vivo by cotransfection of myc-Rnd3 and flag-NICD in HEK293T cells, followed by mutual coimmunoprecipitation (Fig. 5 B and C). To evaluate the biological significance of the interaction of these two proteins, we assessed the levels of NICD protein along with the increase in Rnd3 expression. Immunoblot analysis revealed decreased NICD protein levels when the cells expressed higher levels of Rnd3 (Fig. 5D, lanes 1–3). This process was significantly attenuated by treatment with a proteasome inhibitor, MG132 (Fig. 5D, lanes 4–6). The enhanced NICD degradation after the introduction of Rnd3 was verified by cycloheximide chase analysis. Lower endogenous NICD protein levels were observed in cells transfected with the Rnd3 expression vector compared with control cells (Fig. 5E). These data provide evidence that Rnd3 interacts with NICD and promotes UPS-mediated NICD degradation.
Fig. 5.
Rnd3 physically interacted with NICD and facilitated its degradation. (A) Coronal sections of a WT mouse aqueduct showed endogenous Rnd3 (green) and Notch1 (red) expression by immunofluorescent staining. (B and C) Co-immunoprecipitation (IP) pull-downs were conducted, followed by immunoblotting analyses (IB). The blots confirmed the interaction of Rnd3 and NICD proteins. (D) NICD protein levels in the entire Rnd3 expression vector transfected cell lysates were measured by immunoblot analysis. Along with the elevated Rnd3 protein levels, noticeable decreases in NICD protein levels were observed; this effect was attenuated by treatment with a proteasome inhibitor, MG132. (E) Immunoblot analysis showed that the degradation of endogenous NICD was enhanced by the introduction of Rnd3 when protein synthesis was blocked by treatment with cycloheximide (CHX). h, hours.
Inhibition of Notch Signaling Blocked Rnd3 Deficiency-Induced Aqueductal Ependymal Cell Aberrant Growth and Prevented Hydrocephalus in Rnd3-Null Mice.
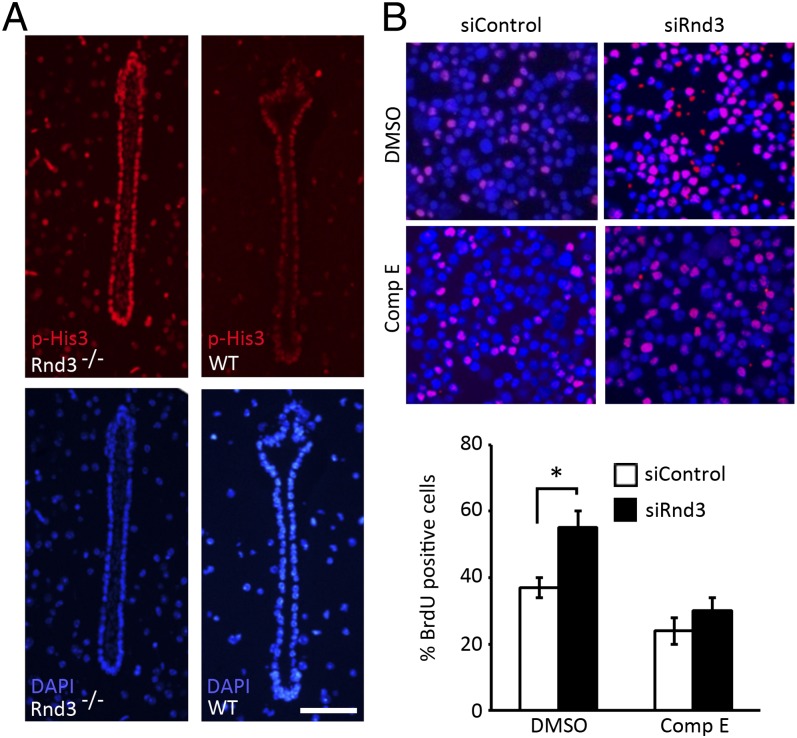
Consistent with our early findings indicating the overgrowth of aqueductal ependymal cells (Fig. 3I), we detected a more intense immunostaining of phosphorylated His3 (p-His3) in aqueducts of the mutant mice compared with WT mice (Fig. 6A). To investigate whether the abnormal ependymal cell proliferation, aqueductal stenosis, and eventual hydrocephalus observed in Rnd3-null mice could be rescued by the inhibition of Notch signaling, we conducted in vitro and in vivo experiments using compound E, a Notch signaling inhibitor. We found that the knockdown of Rnd3 in HEK 293T cells led to a significant increase in bromodeoxyuridine (BrdU)-positive cells, and that this increase was completely impeded by the compound E treatment (Fig. 6B).
Fig. 6.
Genetic deletion of Rnd3 promoted aqueductal ependymal cell proliferation, which was attenuated by treatment with compound E (Comp E). (A) p-His3 was detected in Rnd3−/− aqueduct ependymal cells. The lower panels were DAPI staining. (B) A noticeable increase in BrdU-positive cells (pink) was observed in the HEK 293T cells treated with the siRNA specific for Rnd3 (siRnd3) (upper right section of the image). This increase was attenuated by treatment with compound E (lower right section of the image). The BrdU-positive cells were quantified from nine images, taken from three slides, and presented in the lower panel. (Scale bar: 25 μm.) *P < 0.05 vs. control siRNA (siControl) group.
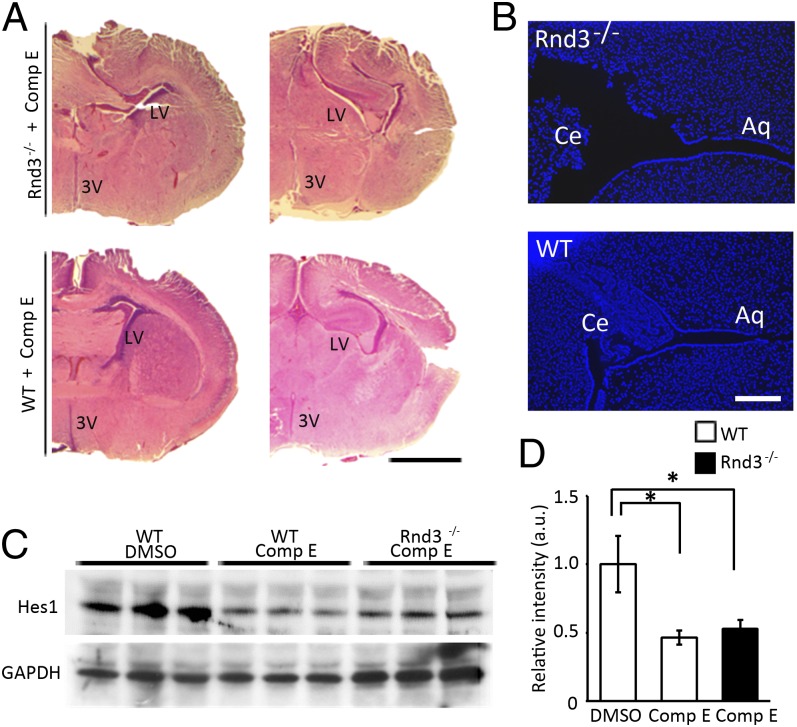
For the animal rescue experiments, because mouse aqueduct development starts at E13.5, we delivered compound E into 10 pregnant mice at E12.5 through i.p. injection until birth. We found no development of hydrocephalus in the newborn Rnd3-null mice (Fig. 7A). An open aqueduct, smoothly lined with a single layer of ependymal cells, was observed in the compound E-treated Rnd3-null mice (Fig. 7B). As expected, lower expression of Hes1 protein was detected in the mouse brain tissues after compound E treatment (Fig. 7 C and D). Thus, the inhibition of Notch signaling by compound E treatment is sufficient to abolish the development of hydrocephalus in mice caused by Rnd3 deficiency.
Fig. 7.
Inhibition of Notch signaling blocked the development of hydrocephalus. (A) The development of an enlarged lateral ventricle (LV) and third ventricle (3V) induced by Rnd3 deficiency was prevented by treatment with compound E (Comp E). (Scale bar: 1 mm.) (B) Aqueduct stenosis was not seen in the brains of newborn Rnd3−/− mice after treatment with compound E. Ce, cerebellum; Aq, aqueduct. (Scale bar: 200 μm.) (C) Expression levels of Hes1 protein in the brain tissues of WT and Rnd3−/− mice were decreased by treatment with compound E. (D) Quantified results from C. *P < 0.05 compared with DMSO-treated WT mice.
Discussion
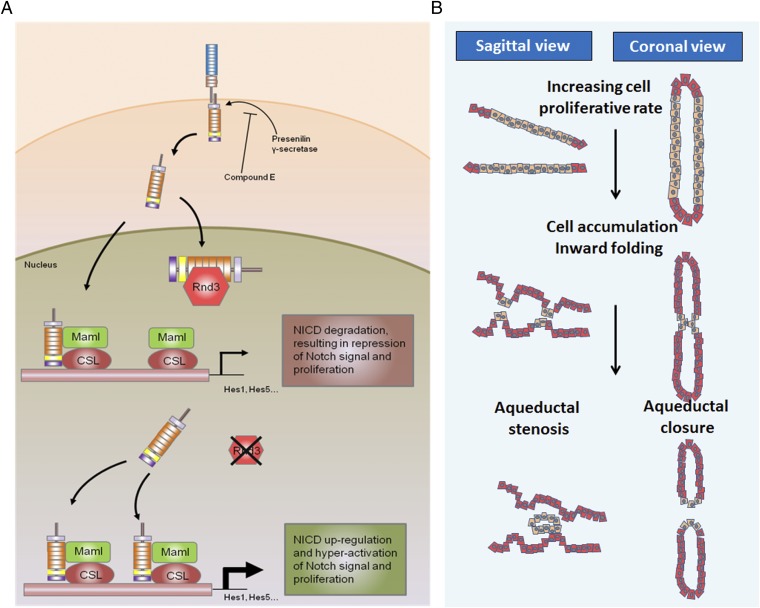
Notch receptor is a single-pass transmembrane protein involved in numerous biological processes, particularly cell-to-cell signaling communication. Notch activation consists of multiple cascade processes. In brief, the binding of Notch receptor to its ligand triggers two proteolytic cleavages. The initial cleavage removes the extracellular domain of Notch by a metalloprotease α-secretase, and the second cleavage is catalyzed by a presenilin/γ-secretase complex within the transmembrane domain of Notch. The proteolysis releases an active Notch isoform, NICD, which translocates to the nucleus. In the nucleus, NICD forms a transcriptional complex with other factors, including mastermind-like protein (Maml) and CSL (CBF1, suppressor of hairless, Lag-1), to regulate downstream target genes, including hairy and enhancer of split-1 (Hes1) and Hes5 (Fig. 8). Dysregulation of Notch signaling results in severe neurogenesis, including hydrocephalus during development (15, 16).
Fig. 8.
Proposed model outlining the molecular mechanism of the Rnd3 deficiency-mediated development of hydrocephalus. (A) On activation, Notch receptors on ependymal cell membranes are cleaved into the intracellular isoform NICD, which is translocated into the nucleus. In the nucleus, Rnd3 controls the accessibility of NICD protein to Maml and CSL by physically interacting with NICD. In the absence of Rnd3, Notch signaling is significantly enhanced owing to the extra amount of NICD available for Maml and CSL to form transcriptomes, which then facilitates ependymal cell proliferation, resulting in aqueduct stenosis and hydrocephalus. (B) The process of aqueduct stenosis formation. Depletion of Rnd3 promotes ependymal cell proliferation through the enhanced Notch signaling mechanism. The overgrowth of ependymal cells leads to the formation of multiple ependymal cell layers, inward cellular folding, and eventually luminal stenosis or closure.
An aqueduct is the narrowest part in CSF circulation tunnels. Aqueductal stenosis is considered the major cause of congenital hydrocephalus (4). Because of this tight anatomic structure, aqueducts are susceptible to many pathological conditions that result in aqueductal stenosis, such as the overgrowth of ependymal cells. Ependymal cells are a single layer of neuroepithelial cells lining the wall of the aqueductal lumen, as well as the ventricles. During mouse embryonic development, the ependymal epithelium is derived from the neuroepithelial cells lining the lumen of the dorsal-caudal aspect of the diencephalon. Proliferation and differentiation of primitive neuroepithelial progenitors are driven toward a specialized ependymal cell fate under the control of numerous signal pathways, including Notch signaling.
Several previous studies have demonstrated that the normal function of Notch signaling is crucial for neuroepithelial cell growth. Lethal giant larvae 1 (Lgl1) is a negative Notch regulator. Neuroepithelial cells with Lgl1 deficiency were hyperproliferative with high Notch activity, and the Lgl1-null mice even developed severe hydrocephalus at the embryonic stage (15). Genetic deletion of another Notch antagonist protein, numb homolog gene Numb or Numblike (Numbl), also led to obstructive hydrocephalus owing to the multilayered stratified epithelium developed in the CSF circulation tunnels (16). Numb promoted Notch receptor ubiquitination, which in turn facilitated Notch protein degradation (17). Interestingly, Lgl1 and Numb/Numbl act in concert to regulate the Notch signaling pathway. A lack of either one perturbs Notch signaling, resulting in hydrocephalus, which suggests the importance of Notch signaling integrity in the pathogenesis of hydrocephalus.
This study provides evidence that Rnd3 is a Notch signaling antagonist. Rnd3 regulates Notch activity through posttranslational regulation of NICD. It physically interacts with NICD and facilitates NICD protein degradation. We propose a working model in which Rnd3 functions as a NICD cofactor (Fig. 8), binding to NICD and preventing NICD from forming a functional complex with Maml and CSL in the cell nucleus. This process is dynamically regulated. The down-regulation of Rnd3 releases more NICD, which is then available for the formation of the regulatory complex that hyperactivates Notch signaling. The latter promotes ependymal cell overgrowth, leading to aqueductal stenosis or closure.
Although previous studies of the biological function of Rnd3 focused mostly on its inhibitory effect on the Rho kinase signaling pathway, recent emerging evidence indicates that Rnd3 also participates in the regulation of cell proliferation, consistent with our results through different mechanisms. In fibroblasts, forced expression of Rnd3 blocked cell cycle progression at the G1 phase through down-regulation of cyclin D1 (18). In prostate cancer cells, Rnd3 expression induced the arrest of cells in the G2/M phase by repressing cyclin B1 and cdc2 expression (19). Although we cannot rule out the possibility that these cell-cycling regulatory mechanisms by Rnd3 may apply to ependymal cells, this Rnd3 deficiency-mediated proproliferative effect is in line with our findings. Moreover, the up-regulated Hes1 expression observed in our Rnd3 knockdown studies also support Rnd3 as a factor involved in cell proliferation, because Hes1 can inhibit p21 to promote cell proliferation (14).
In summary, we have demonstrated that Rnd3 is highly expressed in ependymal cells of the CNS. Genetic deletion of Rnd3 results in hyperplasia of the ependymal cells, causing aqueductal stenosis or closure, in turn leading to the development of obstructive hydrocephalus. Rnd3 is a regulator of Notch. Genetic or siRNA knockdown of Rnd3 attenuates NICD protein degradation, resulting in increased Notch signaling activity, which then promotes cell proliferation and contributes to the pathogenesis of hydrocephalus. Inhibition of Notch activity is sufficient to rescue the Rnd3-mediated hydrocephalus disorder in the mutant animals. The finding of the effect of Rnd3 on NICD adds a regulatory layer of Notch signaling. Given the fundamental role of the Notch pathway in cell signaling and cell-to-cell communication, our findings indicate a potential target for pharmacologic manipulations.
Materials and Methods
Generation of Rnd3 KO Mouse Lines and Assessment of Rnd3 Expression and Hydrocephalus Disorder in Mice.
We generated Rnd3 KO mice derived from a Texas Institute for Genomic Medicine gene trap ES cell line. The targeting vector containing a lacZ-expressing cassette with an internal ribosomal entry site was inserted into intron 2 of Rnd3. All animal experiments were approved by the Institutional Animal Care and Use Committee of the Texas A&M Health Science Center-Houston.
The Rnd3 expression profile was assessed by lacZ staining in E9.5 heterozygous embryos, qPCR analysis, and immunostaining in WT adult (8-wk-old) littermates. Hydrocephalus disorder was evaluated in 6- to 8-wk-old mice. For the compound E rescue experiment, 10 pregnant Rnd3+/− mice at E12.5 were given compound E (10 nmol/g/day i.p.; Adipogen) until birth, and elimination of hydrocephalus disorder was assessed in postnatal day 3 (P3) mice. The same amount of DMSO was administered to control groups. The cell culture experiments used 10 µM compound E.
qPCR Analysis.
Transcripts were quantified by qPCR analysis (StepOnePlus; Life Technologies) using the SYBR Green method with a MasterMix buffer system containing Taq polymerase, as described previously (20). Total RNA was prepared by TRIzol extraction. The forward and reverse PCR primers were as follows (5′ to 3′): Rnd3: CTATGACCAGGGGGCAAATA/TCTTCGCTTTGTCCTTTCGT; Notch1: GCAGTTGTGCTCCTGAAGAA/CGGGCGGCCAGAAAC; Hes1: CGGACATTCTGGAAATGACA/CATTGATCTGGGTCATGCAG; GAPDH: GGTGAAGGTCGGTGTGAACGGATTT/GCAGAAGGGGCGGAGATGATGA. GAPDH expression levels were used for qPCR normalization. Expression levels were determined by the 2-∆∆Ct threshold cycle method.
Immunostaining, Immunoblotting, and Immunoprecipitation.
The following antibodies were used for immunoanalyses: anti-Rnd3 (Cocalico Biologicals), anti-Notch1 (Abcam; ab27526), anti-cleaved Notch1 (Valv 1744) (Cell Signaling; 2421), anti-Hes1 (Abcam; ab71559), anti-pHis3 (Santa Cruz Biotechnology; sc-8656), anti-c-Myc (9E10, Santa Cruz Biotechnology; sc-40), APC-conjugated anti-BrdU antibody (BD; 552598). Protein loading for immunoblotting analysis was verified by the intensity of the GAPDH blot (Santa Cruz Biotechnology; sc-20357).
siRNA specific for Rnd3 was purchased from Thermo Scientific (l-007794-00-0005). The myc-Rnd3 expression construct was generated with a pCMV-Myc backbone (Clontech). The Flag-NICD construct was kindly provided by Chundong Yu (Xiamen University, China).
β-Gal (lacZ) Staining.
Brain tissues or mouse embryos at E9.5 were incubated in the fixing solution consisting of 4% paraformaldehyde and 0.2% glutaraldehyde, and then stained by the lacZ staining solution containing 1 mg/mL X-gal, 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, and 2 mM MgCl2 in PBS.
Cell Culture, Gene Transient Transfection, and BrdU Staining Analysis.
Human embryonic kidney A293T (HEK 293T) cells were cultured in DMEM with 10% FBS. All of the gene transient transfections were performed using the NEON transfection system (MPK5000; Life Technologies). The final concentration of 50 µg/mL cycloheximide was applied in the cycloheximide chase experiment. For BrdU staining analysis, cells were incubated with BrdU for 45 min before being harvested. Images were acquired by fluorescence microscopy.
Microarray, Data Analysis, and Signaling Pathway Analysis.
Total RNA was isolated from the brains of 8-wk-old mice, and 0.5 µg was used for cDNA preparation. The assessment was performed using the Qiagen RT2 Profile PCR Array. Data analyses were performed with R (www.r-project.org), a publicly available statistical tool for data analysis. Signaling pathway analysis was performed with Ingenuity (www.ingenuity.com).
Statistical Analysis.
Data are expressed as mean ± SEM. Multiple comparisons were done using one-way ANOVA, followed by the Holm–Sidak method. Two-group comparisons were done using the unpaired two-tailed Student t test (SigmaPlot, version 11.0). A P value < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Dr. Vladimir N. Potaman and Kelsey Andrade for editorial assistance. This work was supported by the National Institutes of Health-National Heart, Lung, and Blood Institute Grants R01 HL102314, R21 HL094844 and K02 HL098956 (to J.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219995110/-/DCSupplemental.
References
- 1.Huh MS, Todd MA, Picketts DJ. SCO-ping out the mechanisms underlying the etiology of hydrocephalus. Physiology (Bethesda) 2009;24:117–126. doi: 10.1152/physiol.00039.2008. [DOI] [PubMed] [Google Scholar]
- 2.Yamadori T, Nara K. The directions of ciliary beat on the wall of the lateral ventricle and the currents of the cerebrospinal fluid in the brain ventricles. Scan Electron Microsc. 1979;(3):335–340. [PubMed] [Google Scholar]
- 3.Lehtinen MK, Walsh CA. Neurogenesis at the brain-cerebrospinal fluid interface. Annu Rev Cell Dev Biol. 2011;27:653–679. doi: 10.1146/annurev-cellbio-092910-154026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pérez-Fígares JM, Jimenez AJ, Rodríguez EM. Subcommissural organ, cerebrospinal fluid circulation, and hydrocephalus. Microsc Res Tech. 2001;52(5):591–607. doi: 10.1002/1097-0029(20010301)52:5<591::AID-JEMT1043>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 5.Zhang J, Williams MA, Rigamonti D. Genetics of human hydrocephalus. J Neurol. 2006;253(10):1255–1266. doi: 10.1007/s00415-006-0245-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin S, Liu M, Niu W, Zhang CL. Dysregulation of Kruppel-like factor 4 during brain development leads to hydrocephalus in mice. Proc Natl Acad Sci USA. 2011;108(52):21117–21121. doi: 10.1073/pnas.1112351109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krebs DL, et al. Development of hydrocephalus in mice lacking SOCS7. Proc Natl Acad Sci USA. 2004;101(43):15446–15451. doi: 10.1073/pnas.0406870101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Town T, et al. The stumpy gene is required for mammalian ciliogenesis. Proc Natl Acad Sci USA. 2008;105(8):2853–2858. doi: 10.1073/pnas.0712385105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riento K, Guasch RM, Garg R, Jin B, Ridley AJ. RhoE binds to ROCK I and inhibits downstream signaling. Mol Cell Biol. 2003;23(12):4219–4229. doi: 10.1128/MCB.23.12.4219-4229.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ongusaha PP, et al. RhoE is a pro-survival p53 target gene that inhibits ROCK I-mediated apoptosis in response to genotoxic stress. Curr Biol. 2006;16(24):2466–2472. doi: 10.1016/j.cub.2006.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Riento K, Ridley AJ. ROCKs: Multifunctional kinases in cell behaviour. Nat Rev Mol Cell Biol. 2003;4(6):446–456. doi: 10.1038/nrm1128. [DOI] [PubMed] [Google Scholar]
- 12.Pacary E, et al. Proneural transcription factors regulate different steps of cortical neuron migration through Rnd-mediated inhibition of RhoA signaling. Neuron. 2011;69(6):1069–1084. doi: 10.1016/j.neuron.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mocholí E, et al. RhoE deficiency produces postnatal lethality, profound motor deficits and neurodevelopmental delay in mice. PLoS ONE. 2011;6(4):e19236. doi: 10.1371/journal.pone.0019236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kabos P, Kabosova A, Neuman T. Blocking HES1 expression initiates GABAergic differentiation and induces the expression of p21(CIP1/WAF1) in human neural stem cells. J Biol Chem. 2002;277(11):8763–8766. doi: 10.1074/jbc.C100758200. [DOI] [PubMed] [Google Scholar]
- 15.Klezovitch O, Fernandez TE, Tapscott SJ, Vasioukhin V. Loss of cell polarity causes severe brain dysplasia in Lgl1 knockout mice. Genes Dev. 2004;18(5):559–571. doi: 10.1101/gad.1178004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuo CT, et al. Postnatal deletion of Numb/Numblike reveals repair and remodeling capacity in the subventricular neurogenic niche. Cell. 2006;127(6):1253–1264. doi: 10.1016/j.cell.2006.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGill MA, McGlade CJ. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J Biol Chem. 2003;278(25):23196–23203. doi: 10.1074/jbc.M302827200. [DOI] [PubMed] [Google Scholar]
- 18.Villalonga P, Guasch RM, Riento K, Ridley AJ. RhoE inhibits cell cycle progression and Ras-induced transformation. Mol Cell Biol. 2004;24(18):7829–7840. doi: 10.1128/MCB.24.18.7829-7840.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bektic J, et al. Small G-protein RhoE is underexpressed in prostate cancer and induces cell cycle arrest and apoptosis. Prostate. 2005;64(4):332–340. doi: 10.1002/pros.20243. [DOI] [PubMed] [Google Scholar]
- 20.Lin X, et al. Protein tyrosine phosphatase-like A regulates myoblast proliferation and differentiation through MyoG and the cell cycling signaling pathway. Mol Cell Biol. 2012;32(2):297–308. doi: 10.1128/MCB.05484-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.