Abstract
African pygmies are at the lower extreme of human variation in adult stature and many evolutionary hypotheses have been proposed to explain this phenotype. We showed in a recent study that the difference in average stature of about 10 cm observed between contemporary pygmies and neighboring non-pygmies has a genetic component. Nevertheless, the genetic basis of African pygmies' short stature remains unknown. Using a candidate-gene approach, we show that intronic polymorphisms in GH receptor (GHR) and insulin-like growth factor 1 (IGF1) genes present outlying values of the genetic distance between Baka pygmies and their non-pygmy Nzimé neighbors. We further show that GHR and IGF1 genes have experienced divergent natural selection pressures between pygmies and non-pygmies throughout evolution. In addition, these SNPs are associated with stature in a sample composed of 60 pygmies and 30 non-pygmies and this association remains significant when correcting for population structure for the GHR locus. We conclude that the GHR and IGF1 genes may have a role in African pygmies' short stature. The use of phenotypically contrasted populations is a promising strategy to identify new variants associated with complex traits in humans.
Keywords: pygmies, height, human evolution
Introduction
There is great variation in stature among human populations, and African pygmies are known as the shortest human populations: the mean male stature varies from 141.7 (Mbuti from the Democratic Republic of Congo) to 160.9 cm (Twa from the Democratic Republic of Congo) across pygmy populations, compared to 166.8 cm on average across sub-Saharan non-pygmy populations.1 Previous studies have shown that the divergence between ancestral pygmy populations and ancestral non-pygmy populations is quite ancient and occurred around 60 000 years ago.2, 3 Many evolutionary hypotheses have been proposed to explain this phenotype,4 which could be due to an adaptation to food scarcity,5 to the hot and humid climate of the rainforest,6 to the density of this environment where mobility is uneasy,7 to a life-history tradeoff in a context of high mortality8, 9 or to particular mate choices.10 However, none of these hypotheses has proven to be true4, 11 and the evolution of the short stature of African pygmies remains largely unknown. In a recent paper, we showed that phenotypic plasticity alone cannot explain the differential phenotype between pygmies and neighboring non-pygmies and that unknown genetic factors are probably involved in the determination of pygmies' stature.12
In the search for the molecular mechanism underlying the pygmies' short stature, endocrinologists have shown that this phenotype is characterized by a normal serum level of the growth hormone (GH), but a lower level of insulin-like growth factor 1 (IGF1) and of GH binding protein (GHBP) as compared to controls.13, 14, 15, 16, 17 Furthermore, a recent study showed that the expression of the GH receptor gene (GHR, which is the same gene as GHBP) is 8 times lower in pygmy subjects, as compared with neighboring non-pygmies.18 Small regions of the GHR and IGF1 genes have been previously tested for association with the stature of African pygmies with negative results.18, 19, 20, 21 A recent genome-wide study has identified regions in the genome that might be involved in the difference in stature observed between pygmies from Cameroon and their non-pygmy neighbors, although this signal was not significant.22 Some of these regions contain genes linked to the GH–IGF1 signaling pathway.
The aim of this study is to test for the implication of four candidate genes (GHR, IGF1, and the signal transducers and activators of transcription STAT5A and STAT5B) in the short stature of African pygmies, using a population genetics approach and phenotypic data. The genes under study have been chosen in view of the studies mentioned above and all the four participate in the GH–IGF1 signaling pathway (see Materials and methods).
Materials and methods
Study design and choice of the candidate genes
Several genes are known to be involved in height variation in humans, among which the genes of the GH response cascade have a major role in prenatal and postnatal growth.23, 24 According to endocrinological assays, pygmy individuals have normal levels of GH but lower serum levels of IGF1, and exhibit an eightfold underexpression of the GHR gene as compared with the controls.16, 18, 19 Therefore, the GHR and IGF1 genes represent good candidates to explain pygmies' short stature. We develop here a candidate-gene approach focusing on these two genes and on the signal transducers and activators of transcription STAT5A and STAT5B. The latter genes code for two proteins mediating the signal from the activated GHR to the nucleus and inducing the synthesis of various proteins including IGF1. The four candidate genes are rather long (from 24.4 kb for STAT5A to 297.9 kb for GHR) and we had limited DNA quantities at our disposal for pygmies and non-pygmies. We therefore chose to re-sequence only portions of each one of these genes. Taking into account the fine genetic map of the genes (see Supplementary Figure S1), these portions were chosen in order to cover most of the genes' variability through linkage disequilibrium (Supplementary Methods and Supplementary Table S1).
Sampling populations
The re-sequencing was performed for 90 unrelated individuals (Table 1) of known adult height from two pygmy populations: the Baka from South-Eastern Cameroon (16 men and 14 women; mean height in the population: 155.6 cm for men and 146.5 cm for women), the taller and more admixed Bongo from South-Eastern Gabon2, 12 (19 men and 11 women; 159.7 cm and 151.2 cm) and from the non-pygmy Nzimé (6 men and 24 women; 168.5 cm and 155.8 cm), who are the immediate neighbors of the Baka.
Table 1. Sample sizes and mean stature (cm) by sex in the present sample and in a larger samplea and their standard deviations.
| Men | Women | |||||||
|---|---|---|---|---|---|---|---|---|
|
Present sample |
Larger samplea |
Present sample |
Larger samplea |
|||||
|
Population |
Nb |
Sample height; SD |
Nb |
Pop height; SD |
Nb |
Sample height; SD |
Nb |
Pop height; SD |
| Bongo Gabonb | 19 | 159.6; 7.1 | 25 | 159.7; 7.6 | 11 | 152.1; 4.9 | 19 | 151.2; 4.8 |
| Baka Cameroonc | 16 | 151.4; 4.9 | 182 | 155.6; 5.7 | 14 | 145.7; 4.5 | 184 | 146.5; 4.9 |
| Nzimé Cameroonc | 6 | 169.8; 7.0 | 42 | 168.5; 6.5 | 24 | 155.3; 6.6 | 58 | 155.8; 6.8 |
Mean stature estimated on larger samples of the same populations by Becker et al.12
Anthropometric data and DNA samples gathered by:
P Verdu, 2006–2007.
A Froment, 2002–2007.
Blood (Baka and Nzimé) or saliva (Bongo) was sampled from each participant and stature was measured with a height gauge according to standard procedures.25 Mean height in the populations and in our samples are presented in Table 1.
Oral and video-recorded informed consent was obtained from each donor and the sampling program was accepted by the Cameroonian and Gabonese ethics committees, as required by the French laws of bioethics.
Amplification and analysis of the sequences
Two to three sequences in each candidate gene were chosen using information on the linkage disequilibrium in the region and on previously published sequences (see Supplementary Methods). Most of the primers were designed using Primer 326 and others were adapted from previous studies.21, 27, 28 Primer sequences can be found in Supplementary Table S1. Amplifications were performed using standard PCR conditions (available upon request). PCR products were analyzed in an Applied Biosystems (Carlsbad, CA, USA) 3100 or 3130 XL automated sequencer. The sequences obtained were aligned with Genalys 3.3 software.29 All sequences are available upon request.
Population genetics analyses
Haplotypes were reconstructed from unphased data using PHASE 2.1 software.30, 31
Summary statistics were calculated using DNAsp 50.1 software32 and Arlequin 3.11 software33 (nucleotide diversity π34 can be found in Supplementary Table S3). No significant departures from Hardy–Weinberg equilibrium were detected for any of the SNPs discovered in any of the three populations under study.
Genetic distances between pairs of populations (unilocus and multilocus FST35 in Supplementary Table S2) were estimated using Arlequin 3.11 software.33 Pairwise FST P-values were estimated using a standard permutation procedure considering 10 000 haplotype permutations between individuals across populations.
To evaluate whether the FST values observed in our sequences between the two extreme populations (Baka pygmies and Nzimé non-pygmies) were significantly different from neutral expectations, we performed a test proposed by Beaumont and Nichols.36 This test compares the FST values, conditional on heterozygosity (He estimated as the average pairwise difference between all possible pairs of genes between each sample36), obtained for each polymorphism, with a distribution of values obtained on data sets simulated under a neutral model. The test was performed using a version of the Dfdist software modified for bi-allelic co-dominant markers.37 Following Segurel et al,37 we performed 500 000 coalescent simulations with a two-deme island model, the maximum frequency of the most common allele set to 0.99 and θ=2nNμ=0.01, where n=2 is the number of demes of size N and μ is the mutation rate. This particular θ value corresponds to N=125 and μ=2 × 10−5. Different values of θ (0.1 and 0.001) were also tested and gave very similar results (Table 2). These neutral data sets had to be simulated with a mean FST value equal to that estimated on neutral sequences. No neutral sequence-based FST estimates were available between the Baka and the non-pygmy Nzimé here under study, but FST values between the Baka pygmies and two different non-pygmy populations from Western Central Africa have been published elsewhere25 (FST=0.033 with the Gabonese Akele and 0.038 with the Cameroonian Ngumba). Moreover, all non-pygmy populations of this region, including the Nzimé, exhibit reduced pairwise genetic distances.2, 3 The mean FST value was thus set to 0.033, corresponding to the genetic distance between the Baka pygmies and the non-pygmy Gabonese Akele.3 The same simulations were also performed using a mean FST value of 0.038 (corresponding to the distance between the Baka pygmies and the Cameroonian Ngumba) and gave similar results (Table 2). The P-value for the joint (FST, He) estimate (Table 2) at each SNP was calculated as the proportion of the bivariate probability distribution of the full set of simulations smaller than the (FST, He) estimate obtained for the observed polymorphism.37
Table 2. Heterozygozities and genetic distances between Baka pygmies and Nzimé non-pygmies estimated for two SNPs from GHR and IGF1 genes, associated P-values for the test of Beaumont and Nichols with different parameter valuesa and Spearman's estimates for the correlation between genotypes and height.
| Gene | Sequence | SNP | He | FST | P-valueb | P-valuec | P-valued | P-valuee | rho | P-value | rho with adf | P-value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GHR | Intron 1b | rs2972419 | 0.255 | 0.232 | 0.011 | 0.011 | 0.011 | 0.018 | 0.35 | <0.001 | 0.28 | 0.016 |
| IGF1 | Intron 2 | rs11831436 | 0.427 | 0.238 | 0.020 | 0.019 | 0.018 | 0.026 | −0.34 | <0.001 | −0.17 | 0.15 |
FST values were compared to the distribution of FST values calculated on 500 000 simulations using a modified version of the DFDIST software36, 37 (see Materials and methods for details) with the following parameter values:
Mean FST set to 0.033 and θ=0.01.
Mean FST set to 0.033 and θ=0.1.
Mean FST set to 0.033 and θ=0.001.
Mean FST set to 0.038 and θ=0.01.
Partial correlation with admixture as a covariate (see Materials and methods).
Intra- and inter-specific tests for detecting natural selection (Tajima's D,38 Fu's F's,39 Fu and Li's D, D*, F and F*40 and Fay and Wu's H41) were performed on each sequence using DNAsp 50.1 software32 and Arlequin 3.11 software33 (Supplementary Table S3). For inter-specific tests, re-sequencing with the same primers and PCR conditions was performed on chimpanzee or bonobo samples. These sequences were also used to determine the ancestral state of each polymorphism.
Phenotype–genotype correlations
To detect associations between stature and alleles at SNPs showing an outlying FST value, we performed Spearman's correlation tests using R 2.7.0 software42 (Table 2). As the numbers of men and women sampled in the various populations differed and sample sizes were small, we chose to perform the correlations with individuals from both sexes grouped together and therefore transformed a woman's height into the height of a man of the same population who has the same Z-score.12 Z-scores were calculated using the mean and variance of each population by sex (Table 1). Spearman's correlations were calculated, on data from the three populations pooled together, between individual stature and number of copies of the derived allele for each locus.
Spurious significant correlations could however be obtained because of population structure.43 As pygmy populations have been shown to present a substantial level of admixture with the neighboring non-pygmy populations,2 we also performed partial Spearman's correlations (Table 2) adding the individual level of admixture with the non-pygmy population as a covariate. For individual estimates of the admixture level, we used previously published3 levels of genetic clustering in the non-pygmy cluster estimated using k=2 clusters with Structure v. 2.1.44, 45 In this previous publication, the genetic clustering was performed using data on 26 microsatellite polymorphisms from 604 pygmy and non-pygmy individuals as well as from 119 African individuals from the HGDP-CEPH panel.2, 12, 46 We determined the individual levels of genotype membership proportions in the non-pygmy (blue) cluster for the 73 individuals of our study overlapping with the sample studied by Becker et al.12 For each pygmy or non-pygmy individual, the admixture estimate was thus the proportion of genotypes assigned to the non-pygmy cluster. This value ranged from 0.23 to 0.73% for Baka pygmies, from 0.50 to 0.94% for Bongo pygmies and from 0.59 to 0.93% for Nzimé non-pygmies.
Results
In total, 44 SNPs were found in the 10 studied sequences (population frequencies in Supplementary Table S2), in addition to the GHR deletion of exon 3, which has previously been shown to be associated with a higher response to GH in GH-deficient children.47, 48 This latter polymorphism was not found to be significantly more frequent in pygmies than in non-pygmies.
For each polymorphism, we estimated the genetic distance between the Baka pygmies and their direct non-pygmy neighbors, the Nzimé (FST35 in Supplementary Table S2). We found significant FST values between the Baka and the Nzimé for three SNPs located in the two sequences of the first intron of the GHR gene (FST=0.232, FDR-corrected P-value <0.001 for two SNPs of Intron 1a sequence and FST=0.232, FDR-corrected P-value=0.006 for one SNP of Intron 1b sequence) and for one SNP in the IGF1 intron 2 (FST=0.238, FDR-corrected P-value=0.004). Multilocus FST35 estimated on the GHR intron 1 sequences and on the IGF1 intron 2 sequence were also found to be significant between Baka and Nzimé (results not shown). No significant genetic distances were found on the STAT5A and STAT5B genes.
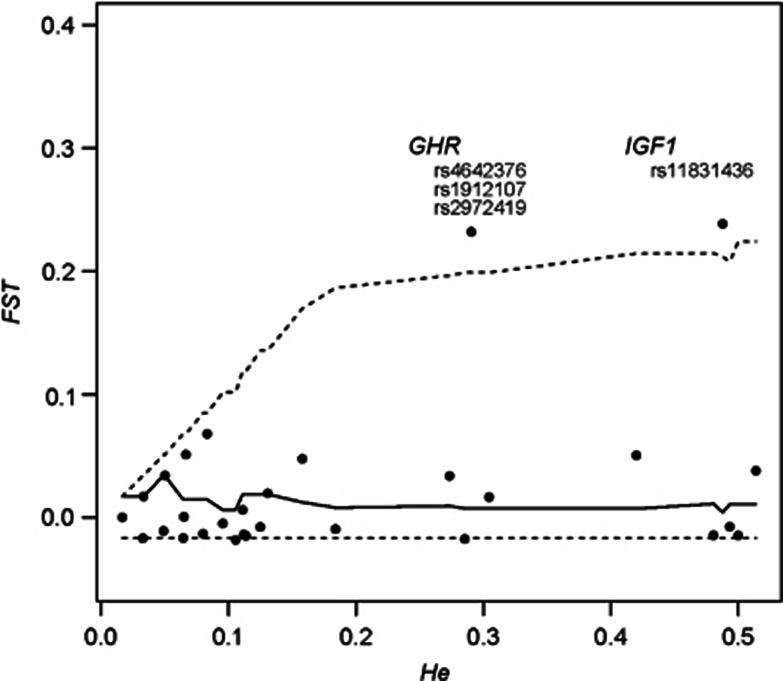
FST values can be influenced by demographic processes such as migrations and genetic drift, as well as natural selection forces.49, 50 To disentangle the respective effects of demography and natural selection on the genetic distances between the Baka pygmies and the Nzimé non-pygmies, we compared the FST estimates for each SNP with values obtained on 500 000 simulated data sets (see Materials and methods). These simulations were performed with a mean FST value equal to that obtained previously using neutral sequences between Baka and non-pygmy populations from Western Central Africa (0.033 with the Gabonese Akele and 0.038 with the Cameroonian Ngumba3). Two regions had significantly higher FST values as compared to the simulated data sets (Table 2, Figure 1). One of these regions was located on the first intron of the GHR gene and contained three SNPs in complete linkage disequilibrium (rs4642376, rs1912107 and rs2972419). For the rest of the analyses only SNP rs2972419 was kept as a representative of the GHR region (we chose this SNP because it presented no missing data). The other region was located on the IGF1 intron 2 and contained one significant SNP: rs11831436. No regions in the two other candidate genes showed a significant difference in FST values between pygmies and non-pygmies.
Figure 1.
Genetic distance (FST) as a function of heterozygosity (He) for the 42 SNPs present in the Baka and Nzimé samples. The lines represent the median value (solid line) and the 95% intervals (dashed lines) for 500 000 simulations performed under neutrality with a mean FST set to 0.033 and θ value to 0.01.
We further searched for evidence of natural selection at the intra-population level by performing seven tests that detect natural selection (see Materials and methods) at the sequence level. No sequence exhibited two significant test results for the same population (Supplementary Table S3).
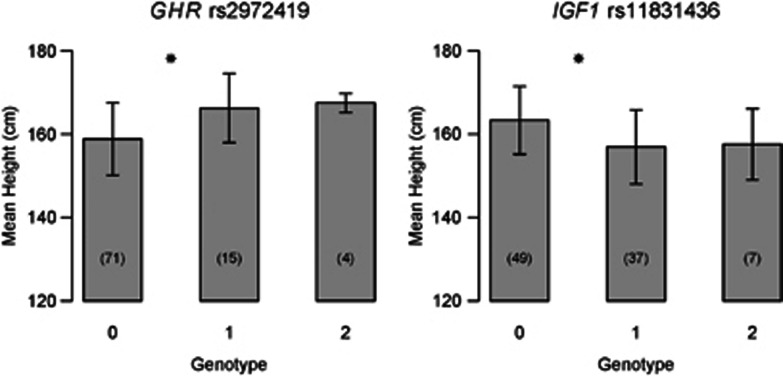
As two regions, located respectively on GHR intron 1 and IGF1 intron 2, showed a significantly higher genetic distance between Baka pygmies and Nzimé non-pygmies compared to neutral expectations, we searched for an association with individual stature for these two regions. Both showed a significant Spearman's correlation (Table 2) between the number of copies of the derived allele and individual unisex height equivalent (see Materials and methods). For the GHR SNP rs2972419 the ancestral allele was associated with a shorter stature, whereas for the IGF1 SNP the ancestral allele was associated with a taller stature (Figure 2). Population structure can have a strong confounding effect in association studies43 and pygmy populations have been shown to present variable levels of admixture with the neighboring non-pygmy populations.2 Furthermore, these variable levels of individual admixture have been shown to be positively correlated with adult height in pygmy populations.12 In order to avoid false positives (ie, the Chinese chopstick effect51, 52), we tested the correlation of the two previously identified regions, including a correction for individual admixture level (see Materials and methods). This partial correlation was significant for the GHR region only (rho estimate=0.278, P-value=0.016) (Table 2).
Figure 2.
Mean height by genotype (number of derived alleles) for the two loci showing an association with stature. Sample sizes for each genotype are given between parentheses. Female height was transformed into male equivalents (see Materials and methods). Genotype: number of copies of the derived allele identified by comparison with the chimpanzee or bonobo sequences. Bars represent SD. *Tukey's post hoc test significant at 95% between these two genotypes.
The Bongo pygmies are taller and have a higher genetic similarity with the non-pygmy populations than the Baka pygmies.2, 12 Assuming an additive effect of height-associated SNPs, we expect that the Bongo would either have an intermediate frequency between the Baka and the Nzimé at the height-associated SNPs or share alleles with the Baka for some of these polymorphisms and with the Nzimé for others. The present data match this second hypothesis. Indeed, we found high but non-significant FST values between the Bongo and the Nzimé at the GHR region (FST=0.11, FDR-corrected P-value=0.125 for rs2972419) and significant FST values with the Baka at the IGF1 region (FST=0.17, FDR-corrected P-value=0.032 for rs11831436). The Bongo were closer to the other pygmy group for the GHR gene but closer to the non-pygmy group for the IGF1 gene, thus consistent with their intermediate height between the Baka and the Nzimé.
Discussion
In this study, we have found one region composed by three linked polymorphisms: rs4642376, rs1912107 and rs2972419 in the GHR intron 1 showing outlying genetic differentiation between the Baka pygmies and the Nzimé non-pygmies as compared to the neutral expectation. For this region, pygmies have a higher proportion of the ancestral allele compared to non-pygmies, and its derived alleles are associated with a taller stature. The association with stature remained significant even after correction for admixture levels, hence showing that the effect of these SNPs on stature may not be only due to population structure. An eightfold under-expression of the GHR gene was previously found in pygmies, but no variation in the exons of the gene was found in 13 pygmies as compared to eight non-pygmies.18 The three polymorphisms found in our study are located in the first intron of the gene, which had not been re-sequenced in this previous study. Our three height-associated SNPs may have a role in the expression of the gene or may be linked to an exonic functional polymorphism that was not identified in the smaller sample used in the previous study.7 Using the CEPH-HGDP genome-wide genotyping data for almost one million SNPs,53 we calculated the r2 linkage disequilibrium value54 of all SNPs within 1 Mb of rs2972419. Among the Biaka pygmies who are the closest to our Baka sample, there are 16 SNPs that showed a r2>0.2. None of them are located in an exon, whereas two are in a GHR intron. However, functional polymorphisms absent in the HGDP-CEPH database could still be linked to the SNPs identified here.
As in previous studies,20, 21, 55 we could find no polymorphism in the 5′ region of the IGF1 gene associated with stature among Central African pygmies and non-pygmies. As a recombination hotspot separates most of the IGF1 gene from its 5′ region (see the recombination rate estimated from HapMap Phase 256 in Supplementary Figure S1), we re-sequenced other portions of this gene and found that a SNP (rs11831436) in the second intron presented an outlying value of the genetic distance between the Baka pygmies and the Nzimé non-pygmies. This SNP had a higher frequency of the derived allele in the Baka pygmies as compared to non-pygmies and we observed a suggestive association with height, although it was not significant after admixture correction. As this gene is known to be involved in growth, our results suggest that further study should be conducted to investigate the potential linkage between rs11831436 and yet undiscovered functional or regulatory polymorphisms in this gene.
In the two STAT5 genes, we found common polymorphisms that all showed similar allele frequencies and no significant genetic distances between pygmies and non-pygmies. As these genes exhibit a low recombination rate (Supplementary Figure S1) we think that they can be considered as non-associated with the genetic determination of African pygmies' short stature.
Altogether, these results show that the GH–IGF1 axis is probably involved in the difference in stature observed between African pygmies and their non-pygmy neighbors. This confirms the suggestive associations found by a recent genome-wide study.22 Interestingly, the SNPs identified here in the GHR and IGF1 genes have not been shown to be associated with height in large-scale genome-wide association studies.57 Indeed, their frequency is low in the European population (CEU) from HapMap56 (GHR rs4642376: 10.17%, rs1912107: 10.0%, rs2972419: 11.67% and IGF1 rs11831436: 5.83%). We can thus hypothesize that the genetic determination of the difference in stature between pygmies and non-pygmies is at least partly different from the genetic determination of stature variation in other populations worldwide. Height is known to be a highly multigenic trait;57 it is therefore plausible that the genetic factors involved in the determination of height may differ across human populations, having experienced different evolutionary histories; variable natural selection pressures affecting human variation in stature could have triggered the evolution of different genes or mutations across populations. Functional assays of the polymorphisms identified in this study would be of considerable interest and could contribute to a better understanding of the genetic variation in human stature.
The polymorphisms reported here in the GHR and IGF1 genes show a higher FST level as compared to neutral expectations, suggesting that divergent selection probably occurred during the evolution of pygmies and non-pygmies for these genes. No other test of detection of natural selection was found significant on these sequences. The selection event may have occurred a long time ago and may thus not be detected by such tests. As our study was focused on short sequences, no LD-based tests to detect selection such as iHS values could be performed. The HGDP-CEPH53 panel iHS values were available for the GHR SNP rs2972419 (the two others, GHR SNP and the IGF1 SNP, are not in the HGDP-CEPH SNP database). The iHS value for this SNP was small in the Mbuti pygmies (1.356) and in the South African Bantu population (1.143) and was null for the other sub-Saharian populations (Biaka pygmies, Mandeka and Yoruba). No other CEPH population reached a value higher than 1, thus further supporting that this genetic region has not been strongly influenced by recent positive selection events.
Various evolutionary hypotheses have been proposed to explain African pygmies' short stature (see Perry and Dominy4 for a review). As we show here, divergent selection may have occurred during the evolution of pygmies and non-pygmies at two genes related to growth. This supports an evolutionary hypothesis of direct selection on stature because of environmental pressures, for example, due to the climate of the equatorial forest (eg, hypotheses based on Bergman's rule that predicts the size of animals in relation to climate58, 59, 60). However, the SNPs identified here differ with respect to the effect of the ancestral allele, which is associated with shorter stature in GHR but with taller stature in IGF1. According to population genetic studies,2, 3 the common ancestral population of pygmies and non-pygmies lived about 50–70 000 years ago. Our results concerning the effect of the ancestral alleles do not allow us to make inferences on the stature of this ancestral population, reemphasizing the fundamental question of whether pygmies evolved to shorter stature or non-pygmies evolved to taller stature. Thus, the divergent selection revealed here may also have resulted in the evolution of non-pygmies towards taller stature. However, little is known about the ancestral environment of either pygmy or non-pygmy Bantu populations that live in forest areas nowadays.61 Our results open the way to further genetic studies of the GHR and IGF1 genes in other populations of short stature, in other rainforest environments, that may help understand the evolution of stature.
Acknowledgments
We warmly thank all individuals who participated in the study, as well as Frédéric Austerlitz, Serge Bahuchet, Sylvie Le Bomin and Etienne Patin for useful comments and advice, Renaud Vitalis for help with the modified version of the Dfdist software and Friso Palstra for proofreading. This work was supported by Assistance Publique Hôpitaux de Paris, ATM-MNHN: ‘Les relations Sociétés-Natures dans le long terme' 2009–2012, the Fondation pour la Recherche Médicale and the ANR-Blanc program ‘GrowingAP'.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Froment A. Biological variation and adaptation in humans: the case of African pygmies. Bull Mém Soc Anthropol Paris. 1993;5:417–448. [Google Scholar]
- Verdu P, Austerlitz F, Estoup A, et al. Origins and genetic diversity of pygmy hunter-gatherers from Western Central Africa. Curr Biol. 2009;19:312–318. doi: 10.1016/j.cub.2008.12.049. [DOI] [PubMed] [Google Scholar]
- Patin E, Laval G, Barreiro LB, et al. Inferring the demographic history of African farmers and pygmy hunter-gatherers using a multilocus resequencing data set. PLoS Genet. 2009;5:e1000448. doi: 10.1371/journal.pgen.1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry GH, Dominy NJ. Evolution of the human pygmy phenotype. Trends Ecol Evol. 2009;24:218–225. doi: 10.1016/j.tree.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Bailey RC, Head G, Tenike M, et al. Hunting and gathering in tropical rainforest: is it possible. Am Anthropol. 1989;91:59–82. [Google Scholar]
- Cavalli-Sforza LL. African Pygmies. London: Academic Press; 1986. [Google Scholar]
- Diamond JM. Why are pygmies small. Nature. 1991;354:111–112. doi: 10.1038/354111a0. [DOI] [PubMed] [Google Scholar]
- Walker R, Gurven M, Hill K, et al. Growth rates and life histories in twenty-two small-scale societies. Am J Hum Biol. 2006;18:295–311. doi: 10.1002/ajhb.20510. [DOI] [PubMed] [Google Scholar]
- Migliano AB, Vinicius L, Lahr MM. Life history trade-offs explain the evolution of human pygmies. Proc Natl Acad Sci USA. 2007;104:216–219. doi: 10.1073/pnas.0708024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker NSA, Touraille P, Froment A, et al. Short stature in African pygmies is not explained by sexual selection Evol Hum Behav 2012. advanced publication online.
- Becker NSA, Verdu P, Hewlett B. Can life history trade-offs explain the evolution of short stature in human pygmies? A response to Migliano et al. (2007) Hum Biol. 2010;82:17–27. doi: 10.3378/027.082.0101. [DOI] [PubMed] [Google Scholar]
- Becker NS, Verdu P, Froment A, et al. Indirect evidence for the genetic determination of short stature in African pygmies. Am J Phys Anthropol. 2011;145:390–401. doi: 10.1002/ajpa.21512. [DOI] [PubMed] [Google Scholar]
- Rimoin DL, Merimee TJ, Rabinowitz D, et al. Peripheral subresponsiveness to human growth hormone in the African pygmies. N Engl J Med. 1969;281:1383–1388. doi: 10.1056/NEJM196912182812502. [DOI] [PubMed] [Google Scholar]
- Merimee TJ, Rimoin DL, Cavalli-Sforza LL. Metabolic studies in the African pygmy. J Clin Invest. 1972;51:395–401. doi: 10.1172/JCI106825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merimee TJ, Zapf J, Froesch ER. Dwarfism in the pygmy. An isolated deficiency of insulin-like growth factor I. N Engl J Med. 1981;305:965–968. doi: 10.1056/NEJM198110223051701. [DOI] [PubMed] [Google Scholar]
- Merimee TJ, Zapf J, Hewlett B, et al. Insulin-like growth factors in pygmies. The role of puberty in determining final stature. N Engl J Med. 1987;316:906–911. doi: 10.1056/NEJM198704093161503. [DOI] [PubMed] [Google Scholar]
- Baumann G, Shaw MA, Merimee TJ. Low levels of highaffinity growth hormone-binding protein in African pygmies. N Engl J Med. 1989;320:1705–1709. doi: 10.1056/NEJM198906293202601. [DOI] [PubMed] [Google Scholar]
- Bozzola M, Travaglino P, Marziliano N, et al. The shortness of pygmies is associated with severe under-expression of the growth hormone receptor. Mol Genet Metab. 2009;98:310–313. doi: 10.1016/j.ymgme.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Hardouin S, Gourmelen M, Noguiez P, et al. Molecular forms of serum insulin-like growth factor (IGF)-binding proteins in man: relationships with growth hormone and IGFs and physiological significance. J Clin Endocrinol Metab. 1989;69:1291–1301. doi: 10.1210/jcem-69-6-1291. [DOI] [PubMed] [Google Scholar]
- Bowcock A, Sartorelli V. Polymorphism and mapping of the IGF1 gene, and absence of association with stature among African pygmies. Hum Genet. 1990;85:349–354. doi: 10.1007/BF00206760. [DOI] [PubMed] [Google Scholar]
- Schneid H, Le Bouc Y, Seurin D, et al. Insulin-like growth factor-I gene analysis in subjects with constitutionally variant stature. Pediatr Res. 1990;27:488–491. doi: 10.1203/00006450-199005000-00014. [DOI] [PubMed] [Google Scholar]
- Jarvis JP, Scheinfeldt LB, Soi S, et al. Patterns of ancestry, signatures of natural selection, and genetic association with stature in Western African pygmies. PLoS Genet. 2012;8:e1002641. doi: 10.1371/journal.pgen.1002641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bermejo A, Buckway CK, Rosenfeld RG. Genetic defects of the growth hormone-insulin-like growth factor axis. Trends Endocrinol Metab. 2000;11:39–49. doi: 10.1016/s1043-2760(99)00226-x. [DOI] [PubMed] [Google Scholar]
- Walenkamp MJ, Wit JM. Genetic disorders in the growth hormone – insulin-like growth factor-I axis. Horm Res. 2006;66:221–230. doi: 10.1159/000095161. [DOI] [PubMed] [Google Scholar]
- Weiner JS, Lourie JA.Practical Human Biology London; New York: Academic Press; 1981,. p439 [Google Scholar]
- Rozen S, Skaletsky H.Primer3 on the WWW for general users and for biologist programmersin Krawetz S, Misener S, (eds): Bioinformatics Methods and Protocols: Methods in Molecular Biology Totowa: Humana Press; 2000365–386. [DOI] [PubMed] [Google Scholar]
- Amselem S, Duquesnoy P, Attree O, et al. Laron dwarfism and mutations of the growth hormone-receptor gene. N Engl J Med. 1989;321:989–995. doi: 10.1056/NEJM198910123211501. [DOI] [PubMed] [Google Scholar]
- Pantel J, Machinis K, Sobrier ML, Duquesnoy P, Goossens M, Amselem S. Species-specific alternative splice mimicry at the growth hormone receptor locus revealed by the lineage of retroelements during primate evolution. J Biol Chem. 2000;275:18664–18669. doi: 10.1074/jbc.M001615200. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Matsuda F, Margetic N, Lathrop M. Automated identification of single nucleotide polymorphism from sequencing data. J Bioinform Comput Biol. 2003;1:253–265. doi: 10.1142/s021972000300006x. [DOI] [PubMed] [Google Scholar]
- Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005;76:449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Beaumont MA, Nichols RA. Evaluating loci for use in the genetic analysis of population structure. Proc R Soc Lon. 1996;263:1619–1626. [Google Scholar]
- Segurel L, Lafosse S, Heyer E, Vitalis R. Frequency of the AGT Pro11Leu polymorphism in humans: does diet matter. Ann Hum Genet. 2010;74:57–64. doi: 10.1111/j.1469-1809.2009.00549.x. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay J, Wu C. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- Thomas DC, Witte JS. Point: population stratification: a problem for case-control studies of candidate-gene associations. Cancer Epidemiol Biomarkers Prev. 2002;11:505–512. [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, et al. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Dos Santos C, Essioux L, Teinturier C, Tauber M, Goffin V, Bougneres P. A common polymorphism of the growth hormone receptor is associated with increased responsiveness to growth hormone. Nat Genet. 2004;36:720–724. doi: 10.1038/ng1379. [DOI] [PubMed] [Google Scholar]
- Wassenaar MJ, Dekkers OM, Pereira AM, et al. Impact of the exon 3-deleted growth hormone (GH) receptor polymorphism on baseline height and the growth response to recombinant human GH therapy in GH-deficient (GHD) and non-GHD children with short stature: a systematic review and meta-analysis. J Clin Endocrinol Metab. 2009;94:3721–3730. doi: 10.1210/jc.2009-0425. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza L. Population structure and human evolution. Proc R Soc Lond B Biol Sci. 1966;164:362–379. doi: 10.1098/rspb.1966.0038. [DOI] [PubMed] [Google Scholar]
- Nielsen R. Molecular signatures of natural selection. Annu Rev Genet. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet. 2003;361:598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- Hamer D, Sirota L. Beware the chopsticks gene. Mol Psychiatry. 2000;5:11–13. doi: 10.1038/sj.mp.4000662. [DOI] [PubMed] [Google Scholar]
- Cann HM, de Toma C, Cazes L, et al. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- Hill WG, Robertson A. Linkage disequilibrium in finite populations. Theor Appl Genet. 1968;38:226–231. doi: 10.1007/BF01245622. [DOI] [PubMed] [Google Scholar]
- Schneid H, Seurin D, Le Bouc Y. Pathology of the growth and abnormalities of IGF genes. Ann Endocrinol-Paris. 1991;52:479–482. [PubMed] [Google Scholar]
- Frazer K, Ballinger D, Cox D, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lango Allen H, Estrada K, Lettre G, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiernaux J.Long-term biological effects of human migration from the African savanna to the equatorial forest: a case study of human adaptation to a hot and wet climatein Harrisson GA, (ed.): Population Structure and Human Variation Cambridge: Cambridge University Press; 1977 [Google Scholar]
- Ruff CB. Climatic adaptation and hominid evolution: the thermoregulatory imperative. Evol Anthropol. 1993;2:53–60. [Google Scholar]
- Schreider E. Morphological variations and climatic differences. J Hum Evol. 1975;4:529–539. [Google Scholar]
- Eggert MKH.The Bantu problem and African archaeologyin Stahl AB, (ed.): African Archaeology Oxford: Blackwell; 2005301–326. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.