Abstract
Numerous nucleotide sugars are needed in plants to synthesize cell wall polymers and glycoproteins. The de novo synthesis of nucleotide sugars is of major importance. During growth, however, some polymers are broken down to monosaccharides. Reactivation of these sugars into nucleotide sugars occurs in two steps: first, by a substrate-specific sugar-1-kinase and, second, by UDP-sugar-pyrophosphorylase (USP), which has broad substrate specificity. A knock-out of the USP gene results in non-fertile pollen. By using various genetic complementation approaches we obtained a strong (>95%) knock-down line in USP that allowed us to investigate the physiological role of the enzyme during the life cycle. Mutant plants show an arabinose reduction in the cell wall, and accumulate mainly two sugars, arabinose and xylose, in the cytoplasm. The arabinogalactanproteins in usp mutants show no significant reduction in size. USP is also part of the myo-inositol oxygenation pathway to UDP-glucuronic acid; however, free glucuronic acid does not accumulate in cells, suggesting alternative conversion pathways of this monosaccharide. The knock-down plants are mostly sterile because of the improper formation of anthers and pollen sacks.
Keywords: nucleotide sugar, salvage pathway, cell wall precursor, UDP-arabinose, metabolism
Introduction
Nucleotide sugars provide the building blocks for plant cell wall polymers and glycoproteins. These glycosyl donors are generated initially through de novo pathways, which provide the different UDP or GDP sugars by interconversion from UDP-Glc or GDP-Man (Seifert, 2004; Reiter, 2008; Bar-Peled and O'Neill, 2011). During plant growth some of the cell wall polymers or sugar chains from glycoproteins are turned over and degraded to monomers. To be able to reuse these sugars, a parallel salvage pathway network is present in plants (Feingold and Avigad, 1980). Sugars are phosphorylated at the C1 position by specific kinases, and are subsequently converted into the corresponding UDP sugars: for example, by UDP-sugar-pyrophosphorylase (USP). This enzyme was purified from Pisum sativum (pea) sprouts, and peptide sequence data led to the isolation of the corresponding gene (Kotake et al., 2004). USP is highly conserved in plants, but was recently also identified in the genome of human parasites like Trypanosoma (Yang and Bar-Peled, 2010) and other protozoans. The USP enzyme from pea converts several sugar-1-phosphates into the corresponding UDP sugars, a property that is retained in the enzymes from Arabidopsis (Litterer et al., 2006; Kotake et al., 2007) or Trypanosoma (Yang and Bar-Peled, 2010). Because of the broad substrate specificity, USP is sometimes also referred to as Sloppy. The typical substrates of USP are shown in Figure 1. A T–DNA insertion disrupting the USP gene of Arabidopsis was shown to result in improper pollen development (Schnurr et al., 2006). A homozygous T-DNA mutant could not be obtained, as the male gametophyte does not transmit the defect usp allele. Heterozygous usp/USP plants showed no visible phenotypes, but had a 50% lower USP enzyme activity compared with the wild type. The function of USP for the metabolism of plants is unknown. USP has the highest affinity to UDP-glucuronic acid-1-phosphate (UDP-GlcA-1-P), a substrate of the myo-inositol oxygenase (MIOX) pathway to UDP-GlcA (Figure 1). This may suggest that the major function of USP is a role as terminal enzyme of the MIOX pathway to UDP-GlcA; however, a strong knock-down in a quadruple mutant of the four MIOX genes shows no visible differences compared with the wild type, and is fertile (Endres and Tenhaken, 2011). Based on microarray data and the promoter::GUS fusion (Kotake et al., 2007), USP is widely expressed during the plant life cycle, with a slight preference for flowers. This argues against a pollen-specific role. The Arabidopsis mur4 mutant has a defect in UDP-xylose-4-epimerase, which provides UDP-arabinose for polymer synthases in the Golgi apparatus (Burget and Reiter, 1999). One phenotype of mur4 is a reduction in length of the glycosylation of arabinogalactan proteins. Feeding arabinose to mur4 mutants restores the wild-type glycosylation pattern of arabinogalactan proteins, a pathway involving arabinokinase (Dolezal and Cobbett, 1991) and USP.
Figure 1.
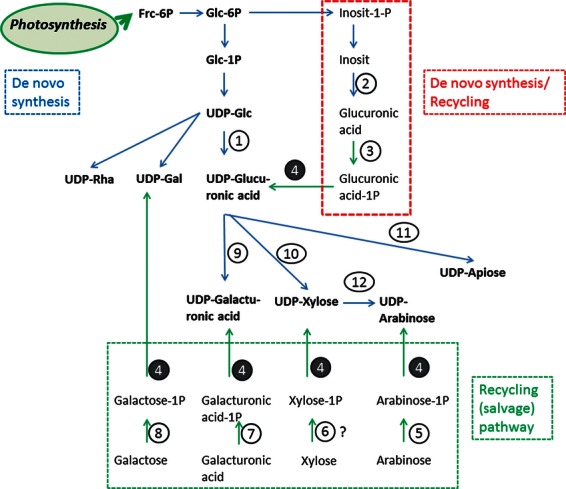
Partial overview of the nucleotide sugar pathway in plants, showing the main reaction in which USP is involved. The de novo pathway uses photosynthesis products to make UDP-GlcA, an important precursor for other UDP sugars of pectins and hemicelluloses. In a side pathway, inositol is oxygenated to GlcA, and thereafter reactivated to UDP-GlcA, involving enzymes of the sugar production and recycling pathways. The lower part of the scheme shows the classical recycling enzymes, which usually consist of a substrate-specific sugar-1-kinase and a broad range substrate using USP: 1, UDP-glucose dehydrogenase; 2, myo-inositol oxygenase; 3, glucuronokinase; 4, UDP-sugar pyrophosphorylase (USP); 5, arabinokinase; 6, putative xylokinase; 7, galacturonokinase; 8, galactokinase; 9, UDP-glucuronic acid-4-epimerase; 10, UDP-xylose synthase; 11, UDP-apiose xylose synthase; 12, UDP-xylose-epimerase.
We were interested to learn more about the role of USP for plant metabolism, and performed a series of genetic complementation experiments of heterozygous usp/USP, and searched for homozygous mutants in transformed complementing lines. This allowed us to test the importance of different possible pathways of USP.
Results
We self-fertilized two independent heterozygous T-DNA insertion lines in the USP gene and genotyped the descendants in order to obtain a homozygous knock-out line for usp. Consistent with the report from Schnurr et al. (2006), we were unable to find any homozygous usp mutants among the more than 100 plants tested. We performed scanning electron microscopy of anthers and pollen tetrads to analyse the defect in usp mutants (Figure 2). Whereas all of the pollen in wild-type plants develops normally and shows filled pollen grains, the tetrads in usp always show two filled and two collapsed pollen grains. The Mendelian inheritance during meiosis is consistent with a single gene defect in usp. In order to find out why the usp knock-out mutation is lethal, we tried to complement heterozygous usp plants with different DNA constructs, and searched in the transformed lines for homozygous usp descendants. USP uses many sugar-1-phosphates (glucose, galactose, glucuronic acid, xylose, arabinose and galacturonic acid) as substrates to convert them into the corresponding UDP sugars, as shown in Figure 1 (Litterer et al., 2006; Kotake et al., 2007). The approach with different complementation constructs allowed us to test several hypotheses about the function of USP, in particular which of the substrates of USP is likely to be responsible for the lethal pollen phenotype. The different constructs used for this approach are summarized in Table 1. We first tested whether UDP-GlcA biosynthesis is limited in usp plants causing the abortion of pollen development. During most of the life cycle, plants synthesize most of their UDP-GlcA by the oxidation of UDP-Glc by the enzyme UDP-glucose dehydrogenase (Sharples and Fry, 2007; Endres and Tenhaken, 2011; Reboul et al., 2011). In pollen however, an alternative pathway is more important, in which myo-inositol is first oxygenated to GlcA, which is subsequently phosphorylated by glucuronokinase to GlcA-1-P and finally converted into UDP-GlcA by USP (Figure 1). It has been shown previously by (Kroh and Loewus, 1968) that this pathway provides most of the precursors for pectins in Lilium pollen tubes. The enzyme UDP-glucose dehydrogenase was expressed in heterozygous kd-usp mutants either under the control of the ubiquitin10 promoter or in a dexamethasone inducible vector system to increase the pool of UDP-GlcA. We thereby tested, whether too low concentrations of UDP-GlcA are responsible for the lethal pollen phenotype in usp-mutants. We genotyped ∼100 descendants from two independent transformants for each construct, but failed to identify a homozygous usp mutant among them.
Figure 2.
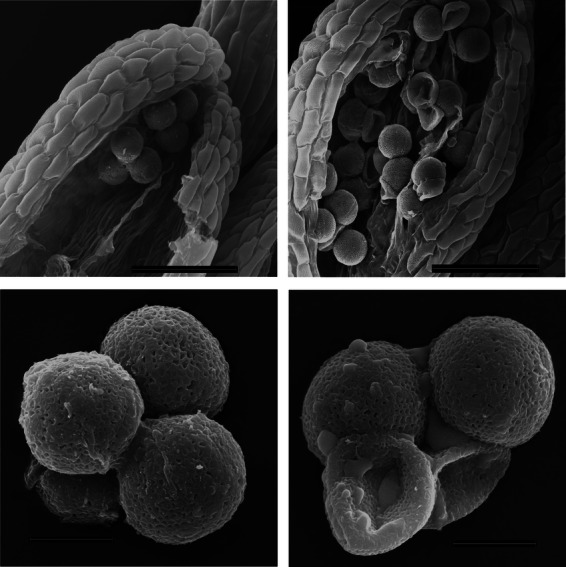
Anthers from the wild type (left) and heterozygous usp mutants (right) were analyzed by scanning electron microscopy. Scale bars: 50 μm for the anthers (upper row); 10 μm for the single pollen tetrad (lower row).
Table 1.
Overview of the complementation constructs used to transform heterozygous usp mutants
| Promotor | Structural gene | Viable homozygous T-DNA insertion lines obtained |
|---|---|---|
| pMiox4 | AtUSP::GFP | No |
| pLat52 | AtUSP::GFP | No |
| pUbq10 | AtUSP::GFP | Yes |
| Dex-inducible promotor | GmUDP-Glc DH | No |
| pMiox4 | GmUDP-Glc DH::GFP | No |
| pLat52 | GmUDP-Glc DH::GFP | No |
| pUbq10 | GmUDP-Glc DH::GFP | No |
| pCaMV35S | GMUDP-Glc DH::GFP | No |
| pUbq10 | AtUDP-Glc-pyrophosphorylase | No |
| pUbq10 | AtUDP-Glc-pyrophosphorylase::GFP | No |
Siblings of primary transformants were screened by PCR for a homozygous usp mutation. The promotors used in these experiments were either the anther/pollen-specific pMIOX4 or pLat52 promotors or the constitutive pUbq10 or CaMV35S promotors. The structural genes were: Arabidopsis USP (AtUSP) fused to GFP, the UDP-glucose dehydrogenase from soybean (GmUDP-Glc DH) or the UDP-glucose pyrophosphorylase from Arabidopsis. Typically approximately 100 descendants from at least two independent transformants were genotyped. No indicates that no homozygous usp-mutants was obtained among the 100 tested descendants.
In a second approach we tested the hypothesis that the recycling of glucose and the formation of UDP-glucose is critical during pollen maturation. The rationale behind this experiment refers to studies by Eschrich (1966), in which a massive callose breakdown was described during pollen maturation. We therefore expressed UDP-glucose pyrophosphorylase (Kleczkowski et al., 2004) under the control of the pUbq10 promoter in heterozygous usp lines. This approach also failed to obtain homozygous usp knock-out lines among the siblings.
Next we tried to complement the heterozygous usp lines with a USP::GFP fusion construct under the control of different promoters. The transgenic lines using the anther- and pollen-specific MIOX4 promoter (Kanter et al., 2005) failed to produce homozygous usp siblings. Finally, we used the ubiquitin10 promoter to drive the USP::GFP fusion. Transformed plants did not show a clearly visible GFP fluorescence, suggesting a low protein abundance of the USP::GFP fusion. Nevertheless, we obtained homozygous usp mutants from several independent primary transformants, but at a low frequency (in a ratio of roughly 1:20, instead of the theoretical ratio of 1:3). These homozygous usp mutants are comparable with knock-down plants, as they express USP::GFP fusion protein at a low level. The enzyme activity of USP was therefore measured using GlcA-1-P as a substrate. This substrate was chosen because USP has the highest affinity to it, no other side reactions are known and because the product of the reaction UDP-GlcA can easily be detected on HPLC. As shown in Figure 3, heterozygous usp lines have ∼50% activity compared with wild-type USP. The complemented usp/usp USP::GFP plants have a very low but detectable USP activity, which is ∼3% of the wild-type level, confirming the status as a strong knock-down mutant (kd-usp).
Figure 3.
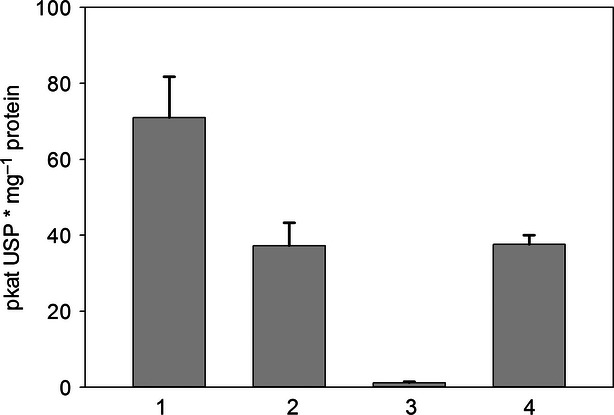
USP enzyme activity. The enzyme activity of USP was measured from leaf extracts using glucuronic acid-1-phosphate as a substrate. Products were separated and quantified by HPLC: 1, wild type; 2, heterozygous usp/USP; 3, kd-usp; 4, homozygous usp mutant, but complemented with pUSP::USP.
The phenotype of kd-usp plants is shown in Figure 4. Rosette leaves of kd-usp remain smaller compared with wild-type plants. The mutants develop normal organ structures, except that the flowers fail to produce (viable) seeds. Seeds were obtained with a very low frequency, below 0.1% of the wild-type yield, making it almost impossible to collect seeds for extensive experiments. The flowers of kd-usp have shorter anthers than the wild type and remain wet and closed, with the pollen sticking together. We also tried to manually fertilize flowers of kd-usp by streaking anthers with pollen sacs across the stigma in order to overcome any problem that might be caused by the shorter anthers. This approach was not successful, most likely because the pollen sacs did not release the pollen properly.
Figure 4.

Phenotype of the kd-usp mutant: (a) rosette of a 4-week old wild-type plant grown under short-day conditions; (b) young wild-type flower; (c) mature wild-type flower; (d) opened developing wild-type silique; (e) rosette of a 4-week-old kd-usp plant grown under short-day conditions; (f) young kd-usp flower; (g) mature kd-usp flower; (h) opened developing kd-usp silique; (i) pollen from wild-type plants; (j) pollen from kd-usp plants.
The pollen from wild-type and kd-usp mutants differ in size (Figure 5). The average size of wild-type pollen is 26.2 μm in length and 21.2 μm in width, whereas the pollen from kd-usp plants are smaller, with a length of 22 μm and a width of 19.1 μm. The flat surface area of the pollen is 437 μm2 in the wild type and 330 μm2 in kd-usp.
Figure 5.
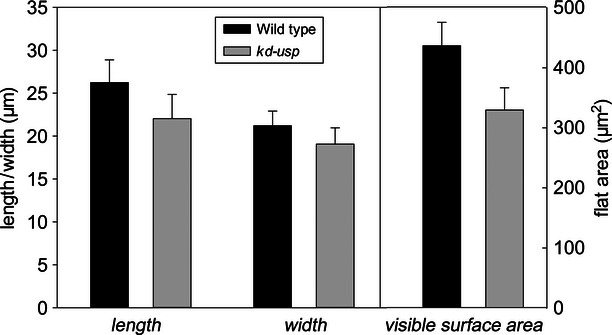
Pollen diameters. Pollen from wild-type plants and kd-usp mutants were spread on a microscopic slide and photographed; the length and width was determined using ImageJ.
The kd-usp phenotype during vegetative growth already suggests an important function of USP for normal plant development beyond the requirement in pollen growth. We also compared gene expression data for USP and other enzymes related to nucleotide sugar recycling pathways from microarray studies in an organ-specific manner (Figure S1). The USP gene is expressed in all plant organs, with a preference for pollen. The sugar-1-kinases, acting in concert with USP, are also expressed during the life cycle of the plant, pointing to nucleotide sugar recycling not only in pollen but in the whole plant. As USP accepts many sugar-1-phosphates as substrates, we were interested to find out which of them are of major importance for normal metabolism. We anticipated that sugars and sugar-1-phosphates should accumulate in the cell if the salvage pathway to UDP sugars is blocked in kd-usp plants. Indeed, we found the accumulation of two sugars, arabinose and xylose, in metabolite extracts of kd-usp. Arabinose-1-phosphate and xylose-1-phosphate could not be measured directly: the electrochemical detection has a low sensitivity and reference compounds are not commercially available. All other sugars remain at the same level in wild-type and mutant plants (Figure 6).
Figure 6.
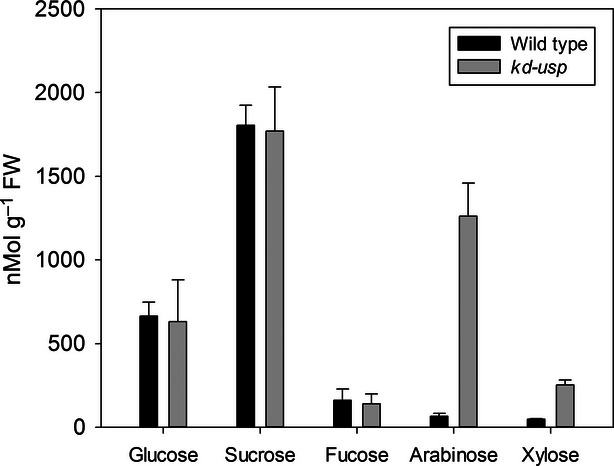
Sugar metabolites. Extracts from 4-week-old leaves were separated on HPLC and soluble sugars were determined by pulsed amperometric detection. Data are shown for the wild type and kd-usp mutants (n = 4).
Next we compared the cell wall composition and found only a small, but significant, reduction in arabinose in kd-usp, suggesting that in wild-type plants part of the arabinose from polymers is liberated during development and recycled to UDP-arabinose via USP (Figure 7). For xylose we did not see statistically significant differences in the cell wall composition of wild type and kd-usp plants, although xylose clearly accumulates in kd-usp mutants. Interestingly, GlcA as an intermediate of the MIOX pathway to UDP-GlcA does not accumulate in kd-usp mutants.
Figure 7.
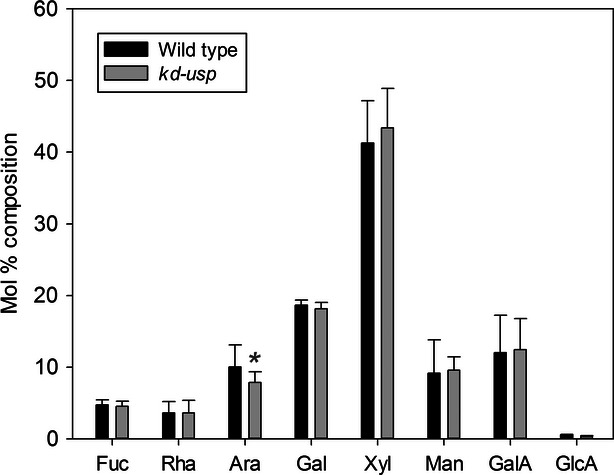
Cell wall sugar composition. Cell walls were hydrolyzed in trifluoroacetic acid (TFA), and liberated sugars were separated on HPLC and detected by pulsed amperometric detection. Data are shown for wild-type plants and kd-usp mutants (n = 8); *P < 0.1.
The mur4 mutant has a lower arabinose content, which results in shorter sugar chains of arabinogalactanproteins (AGPs; Burget and Reiter, 1999). We therefore isolated total AGPs from the wild type, an ara1-1 mutant with a defect in arabinokinase (Dolezal and Cobbett, 1991) and kd-usp plants, and size-separated the glycoproteins on an agarose gel (Figure S2). The average size of AGPs is, however, similar in wild type, ara1-1 and kd-usp mutants.
Discussion
The recycling of sugars into nucleotide sugars usually requires two steps: a phosphorylation at the C1 position, which is catalyzed by sugar-specific kinases, and USP, which takes at least five different sugar-1-phosphates as substrates, and converts them into the corresponding UDP sugars. The most important substrates for USP in vivo are therefore unknown. USP is encoded by a single-copy gene in Arabidopsis, and a knock-out in the USP gene is lethal, as pollen carrying a usp allele do not develop normally (Schnurr et al., 2006), which we confirmed in our initial analysis.
Here, we show that a strong knock-down mutant with <5% residual activity is compromised in vegetative growth. This suggests that the recycling of nucleotide sugars occurs during the vegetative growth phase, and that blocking this recycling inhibits the normal growth of, for example, rosette leaves. Thus, the function of USP is not limited to metabolism in pollen development, but is presumably necessary throughout the life cycle of the plant. This is also backed up by gene expression data for enzymes involved in nucleotide sugar recycling, which are usually expressed in all plant organs, although often with a pollen preference (Figure S1).
The observation that USP can convert many different sugar-1-phosphates into their UDP sugars does not necessarily imply an equal importance of the recycling of all sugars. Pollen tubes are fast-growing cells with a high concentration of pectins in their cell wall. The direct pectin precursor is UDP-galacturonic acid, which is derived from UDP-GlcA by epimerization (Gu and Bar-Peled, 2004). It has been shown previously by Kanter et al. (2005) and Kroh and Loewus (1968) that the myo-inositol pathway, which requires the USP as the final enzyme to produce UDP-GlcA, is highly active in pollen. Microarray data reveal a high expression of the genes for myo-inositol oxygenase, glucuronokinase and USP (Figure S1). Thus, the knock-out of USP would interrupt the MIOX pathway to UDP-GlcA, and could severely reduce pectin precursor availability; however, a strong knock-down quadruple mutant in all four MIOX genes shows no visible phenotype changes, but an upregulation of the genes for UDP-glucose dehydrogenase, which probably compensates for a reduction in the metabolite UDP-GlcA caused by MIOX downregulation (Endres and Tenhaken, 2011). Our approach to overexpress UDP-glucose dehydrogenase in kd-usp mutants to increase the level of UDP-GlcA failed to complement the usp mutation, suggesting that the cause of pollen lethality in kd-usp mutants is not the non-functional MIOX pathway to UDP-GlcA, but a defect in the recycling of other sugars.
Another function of nucleotide sugar recycling pathways in pollen could be the usage of pistil exudates, as suggested by Labarca and Loewus (1972). This role of USP may well be important for the better growth of pollen tubes, but cannot explain the lethal pollen phenotype that we observed in kd-usp mutants. The pollen in usp do not develop properly (Figure 2), and must therefore have a problem in early maturation. Schnurr et al. (2006) localized the defect in usp to a missing intine layer. The possible nutrition function of USP would come into play far later, when germinated pollen grows through the style.
In Oryza sativa (rice), pollen development requires an active UDP-Glc pyrophosphorylase, as co-suppressed plants develop their pollen normally until meiosis, but fail to deposit callose shortly afterwards (Chen et al., 2007), leading to male-sterile rice plants. We tried a complementation with the Arabidopsis UDP-Glc pyrophosphorylase, but failed to obtain homozygous usp plants, suggesting that this function is not the critical step in usp mutants.
Unexpectedly we did not find an accumulation of GlcA in the cell, indicating that alternative pathways for GlcA usage exist. Most likely GlcA is converted to ascorbic acid along an animal-like pathway (Linster and Van Schaftingen, 2007), which at least is functional in feeding studies (Davey et al., 1999) in Arabidopsis cell cultures, and acts independently from the regular pathway via GDP-mannose.
We found an accumulation of arabinose and xylose in kd-usp lines. For technical reasons we could not measure arabinose-1-phosphate and xylose-1-phosphate, which will most likely also accumulate in the cells. Nevertheless, this allows several conclusions about the function of USP in plants. First of all, arabinose and xylose are substrates for the salvage pathway via USP in vivo, and are most likely derived from the turnover of cell wall polymers. As we are working with knock-down lines, the accumulation of arabinose and xylose would be far higher in a true knock-out line. The cell wall composition of kd-usp plants show a small but significant reduction in arabinose, suggesting that the recycling of arabinose contributes to the cellular UDP-arabinose pool.
Is the reduction of arabinose causing the kd-usp phenotype?
Work from the Reiter laboratory (Burget et al., 2003) has identified UDP-xylose-4-epimerase in plants, the enzyme that contributes most to the pool of UDP-arabinose in the novel biosynthesis pathway. The mur4-1 mutant with a defect in UDP-xylose-4-epimerase has strongly reduced levels of arabinose in the cell wall, but shows almost no visible phenotype compared with wild-type plants. Therefore the reduction in UDP-arabinose alone is unlikely to be responsible for the phenotype of kd-usp mutants. One important difference between mur4-1 and kd-usp, however, is the cellular compartment, in which UDP-arabinose is generated by the respective pathway. UDP-xylose-4-epimerase is located in the Golgi apparatus, and thus mainly contributes to the Golgi pool of UDP-arabinose used to synthesize arabinans, petic side chains and arabinogalactan proteins. The enzymes for the salvage pathway, arabinokinase (Dolezal and Cobbett, 1991) and USP are localized in the cytoplasm, and thus contribute to a cytosolic pool of UDP-arabinose. Putative targets in the cytoplasm requiring UDP-arabinose are glycosyltransferases for secondary compounds (Yonekura-Sakakibara et al., 2008), or unknown enzymes synthesizing soluble heteroglycans, with an arabinose, galactose and xylose core, involved in starch metabolism (Fettke et al., 2005).
The mutant ara1-1, which has a defect in the cytoplasmic enzyme arabinokinase, dies during seedling development when low mM concentrations of arabinose are applied to mutant plants (Dolezal and Cobbett, 1991). This might suggest a problem for plants when arabinose accumulates in the cytoplasm. Whether the accumulation of arabinose in kd-usp mutants is responsible for the phenotype remains to be shown.
Free galactose was shown to inhibit growth in plants (Ordin and Bonner, 1957), suggesting that the removal of free sugars is critical for plant growth. The role of USP is then to convert the intermediate sugar-1-phosphates into nucleotide sugars as precursors for novel polysaccharides. Whether galactose or galactose-1-phosphate is toxic remains to be shown, but a recent paper by Egert et al. (2012) suggests that it is not the free galactose, but galactose-1-phosphate or a misbalance in the sugar-1-phosphate and nucleotide-sugar network, that causes the growth defects.
Many mutations in Arabidopsis cause a stop in pollen development. Recently, Levitin et al. (2008) identified two arabinogalactan proteins (AGP6 and AGP11) that are essential for pollen growth. We analyzed AGPs to compare the average size of total arabinogalactan chains and found no difference to wild-type plants; however, mur4-1 mutants clearly show shorter sugar chains of AGPs on average (Burget and Reiter, 1999). Therefore, it is rather unlikely that a modification of AGPs is causing the phenotype of kd-usp mutants.
Finally, the accumulation of xylose in kd-usp is surprising, as it suggests a so far unknown recycling pathway for xylose via xylose-1-phosphate. Based on feeding radioactive precursor to Lilium pollen, it was assumed until now that xylose recycling occurs via xylulose and the pentose phosphate pathway into fructose/glucose, which would not require USP activity (Rosenfield and Loewus, 1978; Rosenfield et al., 1978). Nucleotide sugar pathways vary between pollen and leaves. A conversion of xylose into glucose via the pentose phosphate pathway in pollen does not allow a generalization for the whole plant. We are not aware of any publication in which the existence of a xylose kinase was excluded. In a recent review, xylose to xylose-1-phosphate conversion was described as an unresolved pathway (Bar-Peled and O'Neill, 2011). From the accumulation of xylose in kd-usp mutants we conclude that a xylosekinase exists in plants, but remains to be identified in future work. Consistent with this, Kotake et al. (2004) and Litterer et al. (2006) have already shown that xylose-1-phosphate is a substrate of USP.
Experimental procedures
Plant material
Wild-type Arabidopsis thaliana plants (Col), and two SALK T-DNA mutant lines (SALK_092647; SALK_030169)(Alonso et al., 2003) were obtained from the Arabidopsis stock center (NASC, http://arabidopsis.info). Plants were grown in standard soil (Einheitserde ED73) in a growth chamber at 22°C with ∼130 μm photons m–2 s–1 and a 10-h (16-h) light period for short (long) day plants.
PCR
DNA from Arabidopsis leaves was extracted by the standard cetyl trimethylammonium bromide (CTAB)-buffer method. Genotyping of T-DNA mutants was performed by PCR with primers USP-WT-Salk_030169F and USP-WT-Salk_030169R, or with USP-WT-Salk_092647F and USP-WT-Salk_092647R for wild-type alleles. The T-DNA was detected with primers Salk_030169-T-DNA and Salk-LB or Salk_092647-T-DNA and SalkLB using the following conditions: 94°C for 3 min, followed by 32 cycles of 95°C for 10 s, 59°C for 20 s and 72°C for 1 min. Primer sequences are listed in Table S1.
Constructs for complementation
All PCR primers used to generate constructs are listed in Table S1. The gateway-compatible plant expression vector pMDC84 (Curtis and Grossniklaus, 2003) was used to construct complementation vectors for the usp mutant. The CaMV35S promotor was removed by restriction with HinDIII and SpeI, and replaced by either the pMiox4-(1480 bp), pLAT52- (1540 bp) or pUbiquitin10-(1934 bp) promotor flanked by HinDIII and SpeI restriction sites. The promoters were amplified from genomic Arabidopsis DNA by PCR using the primers pMiox4-F and pMiox4-R, pLAT52-F and pLAT52-R, pUbq10-F and pUbq10-R, and Phusion (Thermo Scientific, http://www.thermoscientific.com) proof-reading polymerase under conditions recommended by the manufacturer.
The USP gene was amplified from a first-strand cDNA from Arabidopsis RNA with Phusion DNA-polymerase using primers attB1-AtUSP and attB2-AtUSP-with-stop or attB2-AtUSP-without-stop to allow Gateway recombination later. The gene for UDP-glucose dehydrogenase from Glycine max (soybean; Tenhaken and Thulke, 1996) was amplified with primers attB1-UDP-GlcDH and attB2-UDP-GlcDH-with-stop or attB2-UDP-GlcDH-without-stop. The gene for Arabidopsis UDP-Glc pyrophosphorylase (At5 g17310) was amplified with primers attB1-UGPase and attB2-UGPase-with-stop or attB2-UGPase-without-stop.
Clones without terminal stop codons were used for translational fusions with GFP in pMDC84. The attB1 and attB2 sites were extended in a second PCR reaction with primers attB1-adaptor and attB2-adaptor to give full-length attB1 and attB2 sites.
Constructs were verified by restriction mapping and sequencing, and then transformed into Agrobacterium (GV3101 pMP90). Colonies from Agrobacterium were tested for the plasmid by colony-PCR, and used for transformation of heterozygous SALK_030169 and SALK_092647 plants using the floral-dip standard method. Seeds were collected and selected for transformants on half-strength MS-phytagel plates with 5 g l–1 sucrose and 30 μg ml–1 hygromycin. Primary transformants were transferred to soil and genotyped for the presence of the complementation construct, the SALK_030169 (092647) T-DNA and the absence of the USP wild-type allele. Typically more than 40 siblings were analyzed for each construct.
Plants with the construct containing the dexamethasone-inducible gene for UDP-glucose dehydrogenase from soybean were sprayed with 20 μm dexamethasone every day during flowering.
USP enzyme assay
Leaves from 4-week-old rosettes, grown under short-day conditions, were extracted in the cold with buffer [50 mm Tris/Cl, pH 7.8, 2 mm MgCl2, 10% glycerol, 0.5 mm phenylmethylsulfonyl fluoride (PMSF)]. After centrifugation (13 000 g for 5 min at 4°C), the supernatant was desalted in the same buffer using a NAP5 size-exclusion column (GE Healthcare, http://www.gehealthcare.com). USP activity was measured in the forward reaction using glucuronic acid-1-phosphate as substrate. The substrate was synthesized during the assay by the addition of recombinant glucuronokinase from Arabidopsis. The USP assay (200 μl in total) consisted of 50 mm Tris/Cl, pH 8, 2 mm MgCl2, 50 μl of desalted leaf extract, 1 mm ATP, 1 mm UTP, 1 mm glucuronic acid and 0.01 units of recombinant glucuronkinase (Pieslinger et al., 2010). The assay was performed at 30°C for 20 min and stopped by heat inactivation (95°C for 3 min). A 1:3 dilution of the assay was applied to a Partisil SAX10 HPLC-column. Products were separated by a phosphate buffer gradient: buffer A, 10 mM Na-phosphate, pH 2.8; buffer B, 750 mm Na-phosphate, pH 3.7. The HPLC-program used was t0 2% B, t13 9% B, t22 9% B, t27 36% B and t50 60% B. The flow rate was 0.5 ml min–1. The product UDP-GlcA eluted around 30 min and was quantified at 262 nm using authentic standards (Sigma-Aldrich, http://www.sigmaaldrich.com) as a reference.
Cell wall and metabolite analysis
Cell walls were prepared from 4-week-old plant leaves. Material was homogenized in liquid N2, extracted with different solvents, acid hydrolyzed and finally separated on HPLC, as described by Reboul et al. (2011). Metabolites from leaves (40–60 mg fresh weight) were extracted from liquid N2 homogenized material in 250 μl methanol/chloroform (7:3) for 1 h at 4°C. Sugars were recovered by the addition of 300 μl H2O and a 10-min extraction with shaking. The upper phase was dried in an Eppendorf vacuum concentrator to dryness and redissolved in 200 μl H2O for HPLC analysis. The quantities of metabolites were compared with authentic sugar standards.
AGP analysis
Total AGP proteins were extracted from homogenized leaves according to the method described by Burget and Reiter (1999). Samples were separated by electrophoresis on a 1% agarose gel (5 V cm–1 for 2 h) and stained for several hours in 15 μm β-Glc-Yariv. Samples were slightly destained in 1% NaCl solution and photographed.
Acknowledgments
This project was funded by the Austrian Science foundation (FWF; P20297). We would like to thank Doris Wittman for excellent technical help and Wolf-Dietrich Krautgartner for his support with the scanning electron microscope.
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article.
Figure S1. Heat map of expression data in different plant organs for genes involved in nucleotide sugar biosynthesis.
Figure S2. Yariv staining of gel-separated arabinogalactan proteins from wild type, ara1-1 and kd-usp mutant plants.
References
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide Insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Bar-Peled M, O'Neill MA. Plant nucleotide sugar formation, interconversion, and salvage by sugar recycling. Annu. Rev. Plant Biol. 2011;62:127–155. doi: 10.1146/annurev-arplant-042110-103918. [DOI] [PubMed] [Google Scholar]
- Burget EG, Reiter WD. The mur4 mutant of arabidopsis is partially defective in the de novo synthesis of uridine diphospho L-arabinose. Plant Physiol. 1999;121:383–389. doi: 10.1104/pp.121.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burget E-G, Verma R, Molhoj M, Reiter W-D. The biosynthesis of L-Arabinose in plants: molecular cloning and characterization of a Golgi-localized UDP-D-Xylose 4-epimerase encoded by the MUR4 gene of Arabidopsis. Plant Cell. 2003;15:523–531. doi: 10.1105/tpc.008425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Zhao X, Shao Z, Wei Z, Wang Y, Zhu L, Zhao J, Sun M, He R, He G. Rice UDP-glucose pyrophosphorylase1 is essential for pollen callose deposition and its cosuppression results in a new type of thermosensitive genic male sterility. Plant Cell. 2007;19:847–861. doi: 10.1105/tpc.106.044123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey MW, Gilot C, Persiau G, Ostergaard J, Han Y, Bauw GC, Van Montagu MC. Ascorbate biosynthesis in Arabidopsis cell suspension culture. Plant Physiol. 1999;121:535–543. doi: 10.1104/pp.121.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezal O, Cobbett CS. Arabinose Kinase-Deficient Mutant of Arabidopsis thaliana. Plant Physiol. 1991;96:1255–1260. doi: 10.1104/pp.96.4.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egert A, Peters S, Guyot C, Stieger B, Keller F. An Arabidopsis T-DNA Insertion Mutant for Galactokinase (AtGALK, At3 g06580) Hyperaccumulates Free Galactose and is Insensitive to Exogenous Galactose. Plant Cell Physiol. 2012;53:921–929. doi: 10.1093/pcp/pcs036. [DOI] [PubMed] [Google Scholar]
- Endres S, Tenhaken R. Down-regulation of the myo-inositol oxygenase gene family has no effect on cell wall composition in Arabidopsis. Planta. 2011;234:157–169. doi: 10.1007/s00425-011-1394-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschrich W. Der Calloseabbau bei den Pollentetraden von Cucurbita ficifolia. Z. Pflanzenphysiol. 1966;54:463–471. [Google Scholar]
- Feingold DS, Avigad G. 1980. Sugar Nucleotide Transformations In Plants.
- Fettke J, Eckermann N, Tiessen A, Geigenberger P, Steup M. Identification, subcellular localization and biochemical characterization of water-soluble heteroglycans (SHG) in leaves of Arabidopsis thaliana L.: distinct SHG reside in the cytosol and in the apoplast. Plant J. 2005;43:568–585. doi: 10.1111/j.1365-313X.2005.02475.x. [DOI] [PubMed] [Google Scholar]
- Gu XG, Bar-Peled M. The biosynthesis of UDP-galacturonic acid in plants. Functional cloning and characterization of Arabidopsis UDP-D-glucuronic acid 4-epimerase. Plant Physiol. 2004;136:4256–4264. doi: 10.1104/pp.104.052365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanter U, Usadel B, Guerineau F, Li Y, Pauly M, Tenhaken R. The inositol oxygenase gene family of Arabidopsis is involved in the biosynthesis of nucleotide sugar precursors for cell-wall matrix polysaccharides. Planta. 2005;221:243–254. doi: 10.1007/s00425-004-1441-0. [DOI] [PubMed] [Google Scholar]
- Kleczkowski LA, Geisler M, Ciereszko I, Johansson H. UDP-Glucose pyrophosphorylase. An old protein with new tricks. Plant Physiol. 2004;134:912–918. doi: 10.1104/pp.103.036053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake T, Yamaguchi D, Ohzono H, Hojo S, Kaneko S, Ishida HK, Tsumuraya Y. UDP-sugar pyrophosphorylase with broad substrate specificity toward various monosaccharide 1-phosphates from pea sprouts. J. Biol. Chem. 2004;279:45728–45736. doi: 10.1074/jbc.M408716200. [DOI] [PubMed] [Google Scholar]
- Kotake T, Hojo S, Yamaguchi D, Aohara T, Konishi T, Tsumuraya Y. Properties and physiological functions of UDP-sugar pyrophosphorylase in Arabidopsis. Biosci. Biotechnol. Biochem. 2007;71:761–771. doi: 10.1271/bbb.60605. [DOI] [PubMed] [Google Scholar]
- Kroh M, Loewus F. Biosynthesis of pectic substance in germinating pollen: labeling with myoinositol-2-14C. Science. 1968;160:1352–1354. doi: 10.1126/science.160.3834.1352. [DOI] [PubMed] [Google Scholar]
- Labarca C, Loewus F. Nutritional Role of Pistil Exudate in Pollen-Tube Wall Formation in Lilium-Longiflorum .1. Utilization of Injected Stigmatic Exudate. Plant Physiol. 1972;50:7–14. doi: 10.1104/pp.50.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitin B, Richter D, Markovich I, Zik M. Arabinogalactan proteins 6 and 11 are required for stamen and pollen function in Arabidopsis. Plant J. 2008;56:351–363. doi: 10.1111/j.1365-313X.2008.03607.x. [DOI] [PubMed] [Google Scholar]
- Linster CL, Van Schaftingen E. Vitamin C - Biosynthesis, recycling and degradation in mammals. FEBS J. 2007;274:1–22. doi: 10.1111/j.1742-4658.2006.05607.x. [DOI] [PubMed] [Google Scholar]
- Litterer LA, Schnurr JA, Plaisance KL, Storey KK, Gronwald JW, Somers DA. Characterization and expression of Arabidopsis UDP-sugar pyrophosphorylase. Plant Physiol. Biochem. 2006;44:171–180. doi: 10.1016/j.plaphy.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Ordin L, Bonner J. Effect of Galactose on Growth and Metabolism of Avena Coleoptile Sections. Plant Physiol. 1957;32:212–215. doi: 10.1104/pp.32.3.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieslinger AM, Hoepflinger MC, Tenhaken R. Cloning of Glucuronokinase from Arabidopsis thaliana, the last missing enzyme of the myo-inositol oxygenase pathway to nucleotide sugars. J. Biol. Chem. 2010;285:2902–2910. doi: 10.1074/jbc.M109.069369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboul R, Geserick C, Pabst M, Frey B, Wittmann D, Lutz-Meindl U, Leonard R, Tenhaken R. Down-regulation of UDP-glucuronic acid biosynthesis leads to swollen plant cell walls and severe developmental defects associated with changes in pectic polysaccharides. J. Biol. Chem. 2011;286:39982–39992. doi: 10.1074/jbc.M111.255695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter WD. Biochemical genetics of nucleotide sugar interconversion reactions. Curr. Opin. Plant Biol. 2008;11:236–243. doi: 10.1016/j.pbi.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Rosenfield CL, Loewus FA. Metabolic studies on intermediates in myoinositol oxidation pathway in lilium-longiflorum pollen .2. Evidence for participation of uridine diphosphoxylose and free xylose as intermediates. Plant Physiol. 1978;61:96–100. doi: 10.1104/pp.61.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfield CL, Fann C, Loewus FA. Metabolic Studies on Intermediates in Myoinositol Oxidation Pathway in Lilium-Longiflorum Pollen .1. Conversion to Hexoses. Plant Physiol. 1978;61:89–95. doi: 10.1104/pp.61.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnurr JA, Storey KK, Jung HJ, Somers DA, Gronwald JW. UDP-sugar pyrophosphorylase is essential for pollen development in Arabidopsis. Planta. 2006;224:520–532. doi: 10.1007/s00425-006-0240-1. [DOI] [PubMed] [Google Scholar]
- Seifert GJ. Nucleotide sugar interconversions and cell wall biosynthesis: how to bring the inside to the outside. Curr. Opin. Plant Biol. 2004;7:277–284. doi: 10.1016/j.pbi.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Sharples SC, Fry SC. Radioisotope ratios discriminate between competing pathways of cell wall polysaccharide and RNA biosynthesis in living plant cells. Plant J. 2007;52:252–262. doi: 10.1111/j.1365-313X.2007.03225.x. [DOI] [PubMed] [Google Scholar]
- Tenhaken R, Thulke O. Cloning of an enzyme that synthesizes a key nucleotide-sugar precursor of hemicellulose biosynthesis from soybean: UDP-glucose dehydrogenase. Plant Physiol. 1996;112:1127–1134. doi: 10.1104/pp.112.3.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Bar-Peled M. Identification of a novel UDP-sugar pyrophosphorylase with a broad substrate specificity in Trypanosoma cruzi. Biochem J. 2010;429:533–543. doi: 10.1042/BJ20100238. [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tohge T, Matsuda F, Nakabayashi R, Takayama H, Niida R, Watanabe-Takahashi A, Inoue E, Saito K. Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell. 2008;20:2160–2176. doi: 10.1105/tpc.108.058040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


