The pluripotent cell-purging assay validated herein demonstrates that pluripotent cells are selectively hypersensitive to DNA damage-induced apoptosis as a function of the specific apoptotic inducer protein Puma. Risk of dysregulated growth is decreased and the safety profile of transplant-ready, bioengineered progenitor cells is augmented.
Keywords: Apoptosis, Stem cells, Pluripotent stem cells, Stem cell transplantation, Tumor cell purging
Abstract
Pluripotent stem cells have been the focus of bioengineering efforts designed to generate regenerative products, yet harnessing therapeutic capacity while minimizing risk of dysregulated growth remains a challenge. The risk of residual undifferentiated stem cells within a differentiated progenitor population requires a targeted approach to eliminate contaminating cells prior to delivery. In this study we aimed to validate a toxicity strategy that could selectively purge pluripotent stem cells in response to DNA damage and avoid risk of uncontrolled cell growth upon transplantation. Compared with somatic cell types, embryonic stem cells and induced pluripotent stem cells displayed hypersensitivity to apoptotic induction by genotoxic agents. Notably, hypersensitivity in pluripotent stem cells was stage-specific and consistently lost upon in vitro differentiation, with the mean half-maximal inhibitory concentration increasing nearly 2 orders of magnitude with tissue specification. Quantitative polymerase chain reaction and Western blotting demonstrated that the innate response was mediated through upregulation of the BH3-only protein Puma in both natural and induced pluripotent stem cells. Pretreatment with genotoxic etoposide purged hypersensitive pluripotent stem cells to yield a progenitor population refractory to teratoma formation upon transplantation. Collectively, this study exploits a hypersensitive apoptotic response to DNA damage within pluripotent stem cells to decrease risk of dysregulated growth and augment the safety profile of transplant-ready, bioengineered progenitor cells.
Introduction
Pluripotent stem cells hold great promise for therapeutic intervention of a wide variety of conditions, ranging from heart disease [1, 2] to neurodegenerative disorders [3]. Although differentiation plasticity accounts for unique therapeutic utility, the pluripotent capacity also illuminates the risk of the formation of teratomas, dysregulated tumors arising from cells containing all three germ layers. Although most teratomas are benign, the genomic instability of pluripotent stem cells could contribute to genomic aberrations that further lead to malignant teratocarcinomas [4, 5]. As stem cells undergo sequential differentiation and lose pluripotency, daughter cells acquire a gene expression profile that decreases the risk for disruptive teratoma formation [6]. Therefore, to decrease teratoma formation, stem cell-based therapeutic strategies have focused on using partially differentiated or tissue-specific progenitor cells. However, heterogeneous early-stage progenitor populations may still harbor residual undifferentiated pluripotent cells, challenging the safety of therapeutic applications.
Strategies to improve the safety of cell therapy have focused on separation or depletion of remaining pluripotent stem cells [7]. A recent study has demonstrated the utility of this approach with stem cell-specific surface antigens and fluorescence-activated cell sorting to reduce the burden of pluripotency within a heterogeneously differentiated cell population [7]. Although this protocol was able to diminish large teratoma formation and significantly reduce the size of formed teratomas, the technical burdens and specialized reagents/equipment required for these protocols remains a challenge for widespread clinical applications. Additionally, proposed mechanisms to purge teratoma-forming cells out of heterogeneous populations include the use of “stop” mechanisms via suicide genes [4, 8] or apoptotic induction in stem cells [9, 10]. A pluripotent stage-specific characteristic that would allow selective pharmacologic-induced elimination without adverse effects to differentiated progeny would advance pluripotent stem cell-derived technologies toward feasible therapeutic interventions. Furthermore, a phenotype universal to pluripotent stem cell populations for both natural and bioengineered sources would enable a broadly applicable strategy agnostic to the starting material or genetic background. Herein we hypothesize that DNA-damage repair mechanisms and inducible apoptotic response to genomic insults represent a feasible target for purging pluripotent stem cells.
Maintenance of genomic stability is dependent on high-fidelity DNA repair [11], as cells continuously encounter low levels of spontaneous genomic insults as a result of processes such as replication, energy metabolism, and environmental stresses. Correspondingly, the potential for devastatingly deleterious effects upon transmission of genetic errors to progeny cannot be tolerated by embryonic stem cells underscored in their high capacity to repair genomic aberrations through hyperactive DNA damage response and repair [11–15]. This defining characteristic of dependence on high-fidelity repair in the pluripotent ground state is further highlighted by studies showing that lack of the Fanconi anemia DNA repair proteins [16] prevents the ability of somatic cells to be successfully reprogrammed to a pluripotent state [17–19]. Furthermore, embryonic stem cells also react differently than somatic cells when faced with large genomic insults. Instead of inducing a prominent cell cycle arrest and undergoing a process to repair damaged DNA, embryonic stem cells rapidly hyperinduce both p53-dependent and -independent apoptosis to prevent accumulated mutational load [20–26]. The DNA damage response/repair and apoptosis induction differences between embryonic stem cells and differentiated cells have also recently been recapitulated in induced pluripotent stem (iPS) cells [27, 28]. Thus, exploiting the natural mechanisms of DNA damage response may provide a specific strategy to segregate pluripotent stem cells from differentiated progeny.
The present study defines the DNA damage-induced apoptotic sensitivity of a panel of embryonic stem cell lines, three-factor and four-factor derived iPS cell lines. The apoptotic-sensitive phenotype is attributed to an apoptotic “priming” of cells through the upregulation of Puma protein. Moreover, this hypersensitive phenotype is selective and disappears upon in vitro differentiation. Quantitative reverse transcription-polymerase chain reaction (PCR) and in vivo teratoma studies demonstrate that pharmacologic pretreatment of partially differentiated heterogeneous cell populations with a DNA-damaging agent purges the population of residual teratoma-forming progenitors. This treatment effectively eliminates teratoma formation within the progenitor population of cells upon in vivo transplantation without causing genomic instability in the surviving progeny. Collectively, this study demonstrates that the Puma-induced phenotype of a hypersensitive apoptotic response to DNA damage within pluripotent stem cells can be exploited to decrease risk of aberrant tumor formation. Therefore, a targeted pharmacologic strategy based on stage-specific apoptotic induction advances the therapeutic potential and safety profile of pluripotent stem cell-based technologies toward feasible regenerative medicine applications.
Materials and Methods
Materials
Allophycocyanin (APC)-annexin V was from BD Pharmingen (San Diego, CA, http://www.bdbiosciences.com), etoposide and hydroxyurea from Sigma-Aldrich (St. Louis, MO, http://www.sigmaaldrich.com), and the broad-spectrum caspase inhibitor Q-VD-OPhe from SM Chemicals (Anaheim, CA, http://www.smbiochemicals.com). Troponin T was obtained from Thermo Scientific (Billerica, MA, http://www.thermoscientific.com), α-actinin from Sigma-Aldrich, Alexa Fluor 568 goat anti-mouse from Invitrogen (Carlsbad, CA, http://www.invitrogen.com), and all other antibodies from Cell Signaling Technology (Beverly, MA, http://www.cellsignal.com). Quantitative PCR reagents and primer/probes were from Applied Biosystems (Foster City, CA, http://www.appliedbiosystems.com) and Integrated DNA Technologies (IDT, Skokie, IL, http://www.idtdna.com), respectively.
Cell Culture
iPS1–5 and four-factor (4F)-iPS1–3 were created by introduction of lentiviral vectors expressing Oct4, Sox2, and Klf4 (and c-Myc in 4F) into mouse embryonic fibroblasts (viPS Vector Kit; Open Biosystems) followed by isolation and expansion of individual clones. All pluripotent lines were maintained in EmbryoMax Dulbecco's modified Eagle's medium (Millipore, Billerica, MA, http://www.millipore.com) containing penicillin G, streptomycin, l-glutamine (Invitrogen), sodium pyruvate (Lonza, Basel, Switzerland, http://www.lonza.com), nonessential amino acids (Mediatech, Herndon, VA, http://www.cellgro.com), 2-mercaptoethanol (Sigma-Aldrich), 15% embryonic stem cell qualified fetal calf serum (FCS) (Invitrogen) and leukemia inhibitor factor (LIF) (Millipore). During and after differentiation cells were cultured in Dulbecco's modified Eagle's medium containing penicillin G, streptomycin, l-glutamine, sodium pyruvate, nonessential amino acids, and 20% FCS.
Annexin V Staining and Analysis
Cells were seeded in gelatin-coated plates 1 day before treatment. Twenty-four hours later cells were treated with the indicated drug concentrations for an additional 24 hours. Cells were then analyzed for apoptosis using APC-coupled annexin V. Cells were resuspended in binding buffer (1 mM HEPES, 0.14 M NaCl, 2.5 mM CaCl2) with 5% (vol/vol) APC annexin V and incubated in the dark for 30 minutes. After collection on a FACSCalibur flow cytometer (Becton, Dickinson and Company, Franklin Lakes, NJ, http://www.bd.com), data were analyzed by assessing percentage of APC-annexin V-positive using CellQuest software (BD Biosciences, San Jose, CA, http://www.bdbiosciences.com). Data are plotted as normalized percentage of annexin V-negative calculated as 100 − ([(observed − baseline)/(100 − baseline)] × 100). Half-maximal inhibitory concentration (IC50) values were calculated using online ReaderFit software (http://www.readerfit.com) and a four-parameter logistic model equation F(x) = ([A − D]/[1 + (x/C)B]) + D, where C is equivalent to the half-maximal inhibitory concentration.
Detection of Apoptotic Cells
Cells were sedimented at 1,000 rpm for 10 minutes, washed with ice-cold phosphate-buffered saline (PBS), fixed in 3:1 (vol/vol) methanol:acetic acid for 1 hour at room temperature, deposited onto microscope slides, stained with 1 μg/ml Hoechst 33258, examined by fluorescence microscopy, and analyzed for apoptotic morphological changes.
Protein and Message Levels
Cells were treated with 5 μM etoposide or vehicle in the presence of 5 μM of the broad-spectrum caspase inhibitor Q-VD-OPhe for 24 hours. Cells were then collected and washed with PBS; either they were lysed in radioimmunoprecipitation protein lysis buffer (50 mM Tris, 150 mM NaCl, 0.1% SDS, 1% Triton, 0.5% sodium deoxycholate) with protease (Roche Applied Science, Indianapolis, IN, https://www.roche-applied-science.com) and phosphatase (Thermo Scientific) inhibitors or total RNA was extracted using an RNeasy kit (Qiagen, Valencia, CA, http://www.qiagen.com). Total protein concentration was determined using a bicinchoninic acid assay (Pierce, Rockford, IL, http://www.piercenet.com), and 50 μg of total protein was subjected to SDS-polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride, and probed with antibodies as indicated. For quantification, band intensities were determined using ImageJ software, and then values were normalized to loading controls then control lanes. cDNA was synthesized from extracted RNA using a SuperScript III First-Strand Synthesis System (Invitrogen). Quantitative PCR (qPCR) was performed in triplicate using 50 ng of RNA and TaqMan Universal PCR Master Mix (Applied Biosystems). PCR was performed on an ECO Real-Time PCR system (Illumina) using a program that consisted of 50°C for 2 minutes and 95°C for 10 minutes, then 40 cycles of 95°C for 10 seconds and 60°C for 30 seconds using Puma (mm.PT.42.7446951), Bim (mm.PT.49a.15907147), Nanog (mm.PT.42.10788230), Oct4 (mm.PT.42.7439100.g), Sox2 (mm.PT.42.12958650.g), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (mm.PT.39.1) probe sets. Data analysis was performed using the following equations: ΔCt = Ct (sample) − Ct (GAPDH), ΔΔCt = ΔCt (sample) − ΔCt (control/standard sample), and are expressed as relative fold change = 2−ΔΔCt.
Short Hairpin RNA Knockdown
Puma short hairpin RNA (shRNA) experiments were performed using Mission TRC1 predesigned shRNAs directed against mouse Puma (TRCN0000009711) or a nontargeting control (Sigma-Aldrich). 293T cells were transfected with 3 μg of the indicated shRNA plasmid plus 2.25 and 0.75 μg, respectively, of psPAX2 and pMDG packaging vector plasmids using Lipofectamine 2000 (Invitrogen). Twenty-four hours after transfection, the medium was replaced with fresh medium, and 48 hours later the supernatant was harvested, filtered, and applied to R1 cells plated in gelatin-coated plates. Twenty-four hours after infection, virus-containing media were replaced, and 24 hours later cells were treated with vehicle or the indicated concentrations of etoposide for an additional 24 hours. Cells were then assayed for annexin V binding by flow cytometry as described above or harvested for Western blotting. Four different shRNAs directed against Puma were screened and produced varying levels of knockdown; repeated analysis was done with the single shRNA that produced the best knockdown. Control experiments using fluorescent-tagged shRNA molecules indicated an approximately 40% transfection efficiency into R1 cells under these conditions.
In Vitro Differentiation
Cells were differentiated into three-layer embryoid bodies (EBs) using the hanging-drop method. Indicated cells were harvested and resuspended in differentiation medium (20% FCS without LIF) to a concentration of 8 × 104 cells per milliliter. Twenty-five-microliter drops were deposited on the lids of plates and incubated for 48 hours. Cells were then transferred to floating suspension for another 48 hours to allow for spontaneous differentiation. Cells were then transferred to gelatin-coated plates and allowed to adhere and further differentiate for the indicated number of days.
For DNA damage response experiments, EBs at day 5 were dissociated using trypsin, replated into gelatin-coated wells, and allowed to adhere for 24 hours; these cells were treated with the indicated drug and concentration on day 6 for an additional 24 hours and then assayed as described above. For survival experiments, cells were differentiated as above, plated onto gelatin-coated plates on day 5, treated with the indicated concentration of etoposide on day 6 for 24 hours, and then washed once with drug-free media and further incubated in drug-free media for up to 10 additional days.
For mouse spectral karyotyping, the indicated samples were incubated with colcemid, and metaphase spreads were prepared 3 hours later. Slides from each indicated specimen were pretreated, hybridized with mouse SKYPaint probe (Applied Spectral Imaging, Vista, CA, http://www.spectral-imaging.com), incubated for 72 hours, and then washed and detected. For immunofluorescence, spontaneously contracting day 15 EBs were observed via light microscopy. Then cells were stained for troponin T and α-actinin using Alexa Fluor 568 as a secondary antibody and 4′,6-diamidino-2-phenylindole (DAPI) as a nuclear stain. For qPCR, samples were assayed as above with vascular endothelial growth factor receptor 2 (KDR) (mm.PT.45.5869721), troponin I (Tnni3) (mm.PT.45.16007114.g), C-X-C chemokine receptor 4 (CXCR4) (mm.PT.45.17237585), α-feto protein (AFP) (mm.PT.49.10942304), myocardin (MyoCD) (mm.PT.45.10793490), Nkx2.5 (mm.PT.45.15814021), myocyte-specific enhancer factor 2c (Mef2c) (mm.PT.45.16471049), Tbx5 (mm.PT.45.9001572), Gata4 (mm.PT.45.12430274), myosin heavy chain β (Myh7) (mm.PT.45.8913378), myosin 6 (Myh6) (mm.PT.45.7820254), and α-actin (Actc1) (mm.PT.45.11428881) primer/probes (Integrated DNA Technologies).
For flow cytometry, day 15 EBs were fixed in 2% paraformaldehyde, permeabilized with ice-cold 90% methanol, and stained with a cardiac troponin T antibody in phosphate-buffered saline with 0.5% bovine serum albumin. Cells were analyzed on a FACSCalibur flow cytometer (Becton Dickinson), and data were analyzed by assessing percentage troponin T-positive using CellQuest software. Data are represented as normalized to cells stained with an IgG control.
Teratoma Formation
Cells at different stages of differentiation were harvested through trypsinization and washed with PBS, and the number of total viable cells was determined using trypan blue and a Countess Automated Cell Counter (Invitrogen). Cells were resuspended in medium containing 10% FCS at a concentration of 2 × 107 live cells per milliliter, and 50 μl was injected subcutaneously into nude mice. Tumor size was monitored every other day through measurement with vernier calipers, and mice were sacrificed when tumor volume reached 10% of the initial animal body weight. All mouse experiments were conducted according the procedures approved by the Institutional Animal Care and Use Committee at the Mayo Clinic (Rochester, MN).
Results
Pluripotent Cells Are Hypersensitive to Apoptosis Induction in Response to DNA Damage
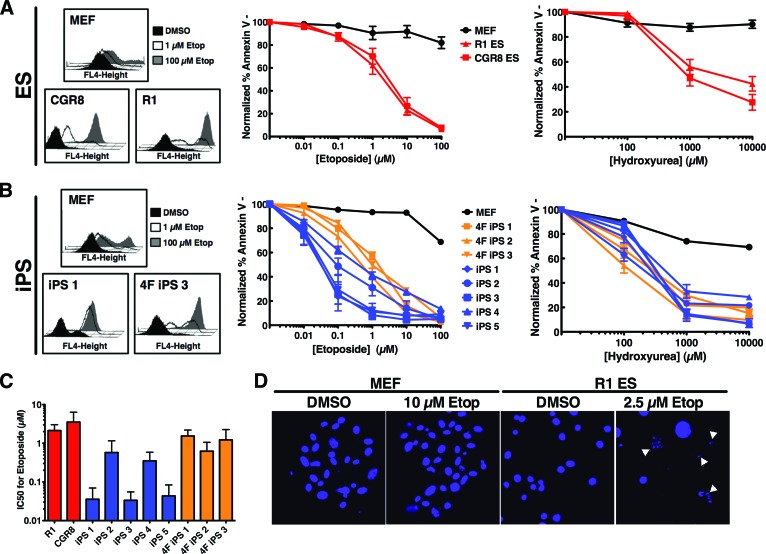
Since maintaining the fidelity of the genome within cells capable of creating a complete organism is crucial, specific mechanisms are in place to ensure proper transmission of error-free DNA to embryonic progeny. One such mechanism in pluripotent stem cells is the rapid activation of the apoptotic machinery in response to DNA double-strand breaks [21]. We investigated the sensitivity of various mouse embryonic stem (ES) cell lines compared with their somatic counterparts (mouse embryonic fibroblasts [MEFs]) to increasing dosages of DNA-damaging agents. In response to the DNA-damaging agents etoposide or hydroxyurea, ES cell lines exhibited dose-dependent apoptosis, assessed through annexin V binding and staining, with mean IC50 values for etoposide of 2.1 and 3.5 μM (R1 and CGR8, respectively) compared with an IC50 of >100 μM in MEFs (Fig. 1A). A similar hypersensitive response of ES cells compared with MEFs to hydroxyurea suggested that the response was not solely a drug-specific phenotype but rather an innate response to extensive DNA double-strand breaks. Additionally, apoptosis was assessed through Hoechst nuclei staining to demonstrate that this phenotype was not limited to the annexin V binding assay. Although MEFs exhibit mainly intact nuclei after high-dose etoposide, R1 ES cell nuclei exhibit ∼50% apoptotic morphology (marked by white arrowheads) upon treatment with a lower dose of etoposide (Fig. 1D), consistent with the annexin V binding assay data. Furthermore, assessment of the apoptotic response to etoposide or hydroxyurea of five independent three-factor (Oct4, Klf4, and Sox2) reprogrammed iPS cell clones (Fig. 1B, 1C) demonstrated mean IC50 values for etoposide ranging from 0.03 to 0.5 μM within this class of stem cells. These data indicate that the phenotype of apoptotic hypersensitivity to DNA damage is shared among pluripotent stem cells. However, the response of iPS cells to DNA damage is not a unique phenotype restricted to cells reprogrammed without the use of potentially oncogenic c-Myc, as the apoptotic responses of three independent four-factor (Oct4, Klf4, Sox2, and c-Myc) reprogrammed iPS cell lines were also more sensitive to apoptosis induction when compared with MEFs (Fig. 1B, 1C). These four-factor reprogrammed iPS cell lines had mean IC50 ranges, 0.6–1.5 μM, similar to those of the ES cell lines. Collectively these results extend previous studies [24, 27] and demonstrate the ability of various pluripotent cells to undergo hyperactive apoptosis in response to drug-induced DNA damage.
Figure 1.
Pluripotent cells are hypersensitive to apoptosis induction in response to DNA damage. (A): MEFs and two embryonic stem cell lines, CGR8 and R1, were treated with 0, 1, or 100 μM etoposide for 24 hours; stained with annexin V-allophycocyanin (APC); and analyzed by fluorescence-activated cell sorting (FACS). Histograms from a representative experiment are shown at left. The average apoptotic response of the indicated cells to increasing concentrations of etoposide (middle) or hydroxyurea (right) is also shown. Curves are the mean of three independent experiments; error bars represent ±SEM. (B): Embryonic fibroblasts from green fluorescent protein/luciferase mice (MEF), three-factor reprogrammed induced pluripotent stem cells (iPS1), or four-factor reprogrammed induced pluripotent stem cells (4F iPS3) were treated with 0, 1, or 100 μM etoposide for 24 hours; stained with annexin V-APC; and analyzed by FACS. Histograms from a representative experiment are shown at left. Average apoptotic response of the indicated cells (five 3-factor iPS clones or three 4-factor iPS clones) to increasing concentrations of etoposide (middle) or hydroxyurea (right) is shown at right. Curves are the mean of three independent experiments; error bars represent ±SEM. (C): IC50 for each of the indicated cell lines to etoposide was calculated using online ReaderFit software and four-parameter logistic model equation. Data are IC50 ± SD. (D): Twenty-four hours after treatment of MEFs or R1 ES cells with DMSO or 10 or 2.5 μM etoposide, respectively, cells were stained with Hoechst 33258 and examined by fluorescence microscopy. Examples of cells exhibiting apoptotic morphology are marked with white arrowheads. Abbreviations: 4F, four-factor; DMSO, dimethyl sulfoxide; ES, embryonic stem; Etop, etoposide; IC50, half-maximal inhibitory concentration; iPS, induced pluripotent stem; MEF, mouse embryonic fibroblast.
Pluripotent Cells Have Elevated Levels of the Proapoptotic BH3-Only Protein Puma, Which Is Further Induced in Response to DNA Damage
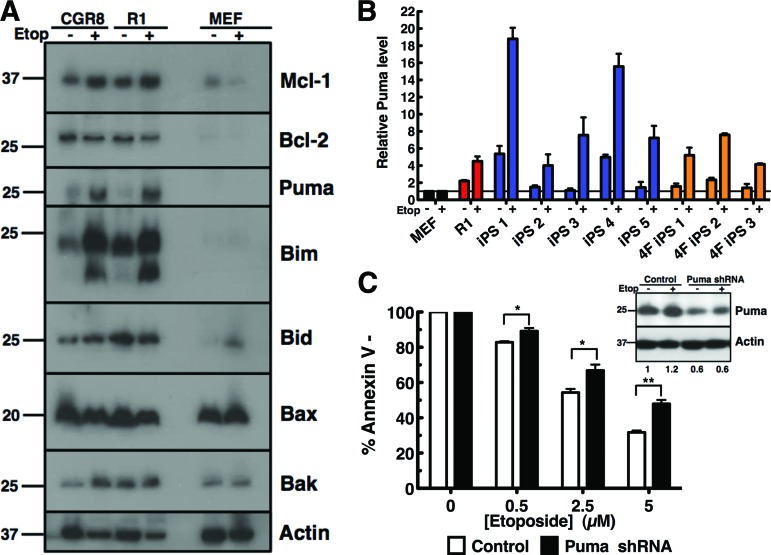
Although hyperactivation of apoptosis induction is consistent for pluripotent cells, we aimed to further dissect the downstream mechanism of this stage-specific activation. To determine which apoptotic proteins are responsible for the induction of DNA damage-induced cell death, levels of intrinsic apoptotic proteins before and after etoposide treatment were determined by Western blotting somatic (MEF) and ES cell (CGR8 and R1) lysates (Fig. 2A). The proapoptotic multidomain proteins Bak and Bax and the antiapoptotic proteins Bcl-2 and Mcl-1 did not correlate with or account for differential sensitivity of pluripotent cells over somatic cells to DNA damage. However, the BH3-only proteins Bim and Puma were expressed higher in the ES cell lines and were further upregulated in response to etoposide treatment (Fig. 2A). This basal and induced upregulation of Puma protein was also observed in the induced pluripotent stem cell lines (supplemental online Fig. 1). qPCR confirmed higher relative Puma message levels in all pluripotent cell lines, 1.5- to 5-fold, and a 4.5- to 15-fold increase upon etoposide treatment (Fig. 2B); Bim, however, showed no consistent correlation in mRNA level between cell lines before or after drug treatment (supplemental online Fig. 2A).
Figure 2.
The BH3-only protein Puma is responsible for the hypersensitivity of pluripotent cells to apoptosis induction following DNA damage. (A): After CGR8 or R1 embryonic stem cells or MEFs were treated for 24 hours with dimethyl sulfoxide (DMSO) (−) or 5 μM etoposide (+) for 24 hours in the presence of 5 μM Q-VD-OPhe, whole cell lysates were subjected to SDS-polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride, and probed with the indicated antibodies against various apoptotic proteins. (B): Total RNA was extracted from MEF, R1, iPS1–5, and 4F-iPS1–3 cell lines treated for 24 hours with DMSO or 5 μM etoposide for 24 hours in the presence of 5 μM Q-VD-OPhe. Real-time polymerase chain reaction was performed on cDNA made from each sample using a primer/probe for mouse Puma. Results are expressed as fold change compared with corresponding MEF sample (see Materials and Methods). Error bars represent ±SD of technical triplicates; results are representative of three independent experiments. (C): Forty-eight hours after infection with the indicated shRNA, R1 cells were treated with indicated concentration of etoposide for 24 hours and then stained with allophycocyanin-annexin V and analyzed by flow cytometry. Results are the average of three independent experiments. Error bars indicate ±SEM. *, p < .05; **, p < .005 in an unpaired t test. Western blot inset in (C) shows immunoblot of shRNA transfected cells treated with DMSO (−) or 5 μM etoposide (+). Numbers under lanes represent approximate Puma level as determined through quantification and normalization to loading control using ImageJ software. Abbreviations: 4F, four-factor; Etop, etoposide; iPS, induced pluripotent stem; MEF, mouse embryonic fibroblast; shRNA, short hairpin RNA.
To determine whether Puma is necessary to induce apoptosis in pluripotent stem cell lines after DNA damage, the hypersensitive phenotype was examined upon shRNA-mediated Puma downregulation. As Puma was decreased approximately 40% (determined via Western blotting; Fig. 2C, inset), annexin V staining following 5 μM etoposide treatment demonstrated that R1 ES cells had acquired resistance (p = .0025) to DNA damage with a decrease from 68.3% apoptosis in control cells to 52% apoptosis in Puma shRNA cells (Fig. 2C). However, loss of Bim through shRNA-mediated downregulation did not affect the apoptotic response upon etoposide treatment in ES cells (supplemental online Fig. 2B), suggesting that it is not a necessary mediator of apoptosis induction in this setting. These data indicate that Puma is a critical downstream mediator that regulates induction of apoptosis in response to DNA damage in pluripotent stem cells.
Differentiated Progeny Decrease Puma Expression and Acquire Resistance to Apoptosis Induction in Response to DNA Damage
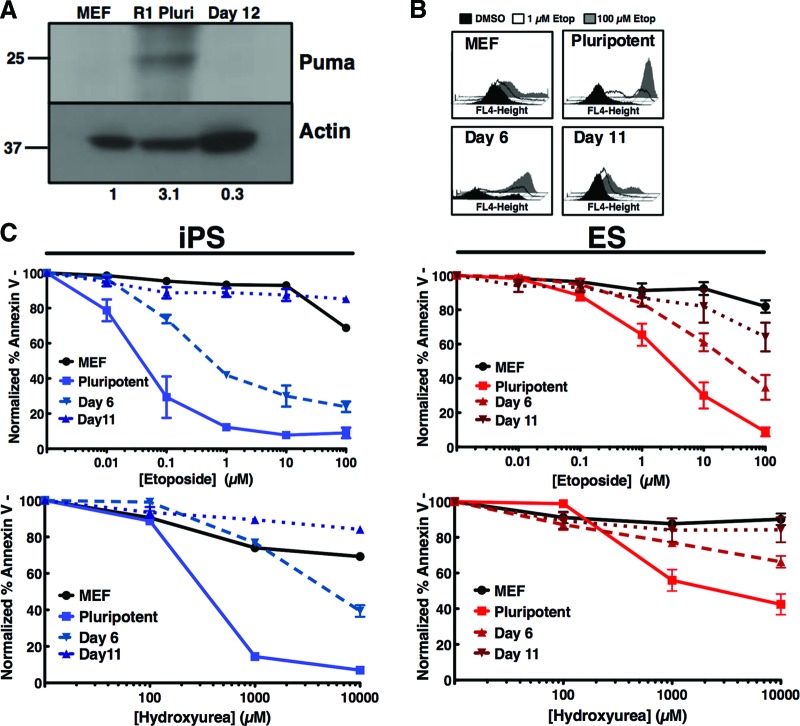
As elevated levels of the BH3-only protein Puma are a major mediator of apoptotic hypersensitivity of pluripotent cells, we hypothesized that spontaneous differentiation of pluripotent cells would reduce expression of this protein. Indeed, Puma protein levels seen in pluripotent R1 cells decreased to levels seen in somatic cells in progeny subjected to spontaneous differentiation in embryoid bodies for 12 days (Fig. 3A). Upon treatment of R1 ES cells (right, red graphs) with increasing doses of etoposide (top graph) a decrease in apoptosis occurred, shifting the IC50 12-fold from 2.1 to 25.8 μM, in day 6 post-embryoid body formation (Fig. 3B). This change was further exacerbated at day 11 post-embryoid body formation, with the IC50 shifting to >100 μM (Fig. 3B). In the iPS5 clone (left, blue graphs), the IC50 for etoposide increased fivefold from 0.044 to 0.223 μM at day 6 and >100 μM at day 11 post-embryoid body formation (Fig. 3C). These results were recapitulated with hydroxyurea treatment (bottom graphs). Collectively, these data indicate that the hypersensitivity of pluripotent stem cells to DNA damage, mediated through the BH3-only protein Puma, is lost upon tissue-specific differentiation and is associated with natural expression levels of Puma.
Figure 3.
Differentiation initiates resistance to apoptosis induction in response to DNA damage. (A): MEFs, pluripotent R1 cells, and R1 cells at day 12 post-embryoid body (EB) differentiation were isolated, and whole cell lysates were subjected to SDS-polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride, and probed with the indicated antibodies. Numbers under lanes represent approximate Puma level as determined through quantification and normalization to loading control using ImageJ software. (B): Progeny isolated on day 6 or day 11 post-EB formation were treated with 0, 1, or 100 μM etoposide for 24 hours, stained with annexin V-allophycocyanin, and analyzed by fluorescence-activated cell sorting. Histograms from a representative experiment are shown at the top right (histograms and curves from MEF and pluripotent R1 from Fig. 1A). The average apoptotic response of the indicated cells to increasing concentrations of etoposide (top) or hydroxyurea (bottom) is shown at right. Curves are mean of three independent experiments; error bars represent ±SEM. (C): Three-factor reprogrammed induced pluripotent stem cell clone 5 (iPS5) was differentiated to day 6 or day 10 and treated and assayed as in (A) (curves from green fluorescent protein/luciferase MEF and iPS5 from Fig. 1B). Shown is the average apoptotic response of the indicated cells to increasing concentrations of etoposide (top) or hydroxyurea (bottom) for 24 hours. Curves are mean of three independent experiments ±SEM. Abbreviations: DMSO, dimethyl sulfoxide; Etop, etoposide; iPS, induced pluripotent stem; MEF, mouse embryonic fibroblast; Pluri, pluripotent.
Pharmacologic Purging of Pluripotent Cells Without Functional Disruption of Daughter Cell Genomic Stability or Cardiogenic Potential
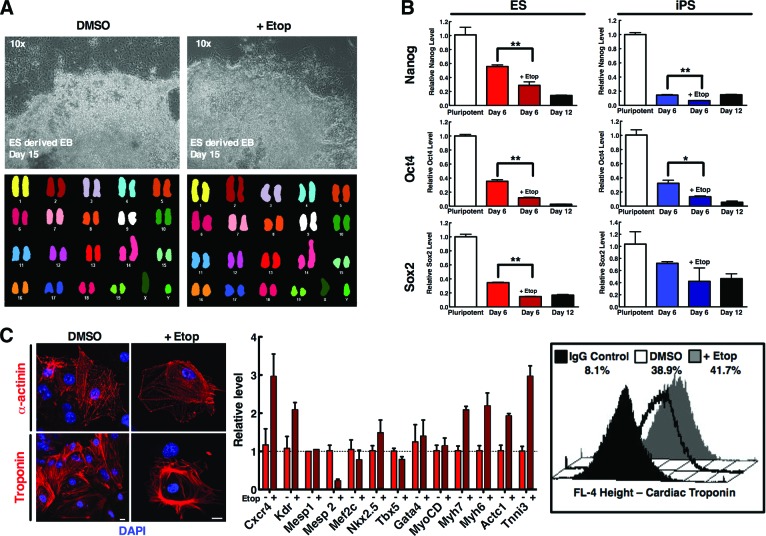
A concern limiting the use of DNA-damaging strategies is the risk of a pharmacologic agent eliciting genotoxic effects and causing permanent damage to the genome of the tissue-specific progeny. The consequential effect of pluripotent cell purging could produce accumulation of excessive DNA damage either reducing the progenitor cell differentiation capacity or unintentionally intensifying their tumorigenic potential. Progenitor cells after 5 days of spontaneous differentiation were treated with etoposide for 24 hours and then incubated in drug-free medium for an additional 10 days. Progeny formed colonies of surviving daughter cells similar to those of untreated stem cells (Fig. 4A, upper panels). Spectral karyotyping revealed no clonal structural or chromosomal gains or losses (defined as being present in at least two or three metaphase spreads, respectively) in the cells subjected to treatment (Fig. 4A), demonstrating a normal karyotype in surviving cells.
Figure 4.
Pharmacologic treatment of a progenitor population of semidifferentiated cells purges out pluripotent cells while preserving differentiation capability. (A): R1 ES cells were subjected to differentiation through the hanging drop method. Cells on day 6 post-EB formation were treated with DMSO or 2.5 μM etoposide for 24 hours. Twenty-four hours after treatment, cells were washed and further incubated in drug-free media for another 9 days. Surviving cells were harvested at day 9 post-etoposide treatment and analyzed via spectral karyotyping of mouse chromosomes. (B): R1 (left, red) and iPS5 (right, blue) cells were subjected to differentiation. Cells on day 6 post-EB formation were treated with DMSO or 2.5 or 0.05 μM etoposide (R1 and iPS5, respectively) for 24 hours. RNA was harvested from pluripotent starting cells, day 6 post-EB with and without treatment, and day 12 post-EB cells (untreated). cDNA was synthesized and subjected to quantitative polymerase chain reaction (qPCR) for the pluripotency markers Nanog, Oct4, and Sox2. Data are average fold change of the indicated message of cells from triplicates ± SEM and are representative of three independent experiments. *, p < .05; **, p < .005 in an unpaired t test. (C): R1 cells were spontaneously differentiated and treated as in (A) but subjected to 10 μM etoposide treatment, to provide a more stringent level of characterization; cells were then harvested 10 days after treatment for RNA extraction and subsequent qPCR with the indicated primer/probe or harvested and subjected to flow staining for cardiac troponin and analyzed via fluorescence-activated cell sorting. For microscopy, cells were dissociated and replated onto coverslips on day 13 post-EB for troponin or α-actinin fluorescence staining 48 hours later on day 15. qPCR data are average fold change of the indicated message of cells from triplicates ± SEM. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; DMSO, dimethyl sulfoxide; EB, embryoid body; ES, embryonic stem; Etop, etoposide; iPS, induced pluripotent stem.
Because this strategy preserved the genomic stability of the selected cells, the cardiogenic efficacy of purged progenitor cells within a spontaneous differentiation population was assessed. R1 ES and iPS5 cells were subjected to spontaneous differentiation for 6 days and then treated with or without an IC50 of etoposide. Live cells were harvested 24 hours later, and qPCR analysis of several pluripotency markers (Oct4, Sox2, and Nanog) revealed significantly lower levels of pluripotency markers than their untreated counterparts after the pulse treatment of etoposide (Fig. 4B). Comparatively, the pluripotency levels in treated cells at day 6 (Fig. 4B, darker red or blue bars) approached those of cells at day 12 of differentiation (Fig. 4B, black bars), demonstrating the reduction in pluripotent capacity induced by the selective purging treatment.
Furthermore, an extensive analysis of the potential of untreated and purged surviving cells to differentiate into a cardiac lineage demonstrated that both populations were able to equally yield cardiomyocytes, as visualized by ability of EBs to spontaneously beat (data not shown), as well as cardiac troponin and α-actinin expression (Fig. 4C, left). qPCR analysis demonstrated the retention of cells after etoposide treatment that expressed the cardiac markers CXCR4, KDR, Mesp1, Mesp2, Mef2C, Nkx2.5, TBX5, Gata4, MyoCD, Myh7, Myh6, Actc1, and Tnni3, similar to that of untreated cells (Fig. 4C, middle), whereas flow analysis demonstrated similar percentages of these populations that were cardiac troponin-positive (Fig. 4C, right). Together these data indicate that treatment of a progenitor population of cells with etoposide limits pluripotent cells in vitro without causing genomic instability or loss of daughter cell differentiation capability.
Pharmacologic Purging of a Progenitor Population Decreases Teratoma Formation
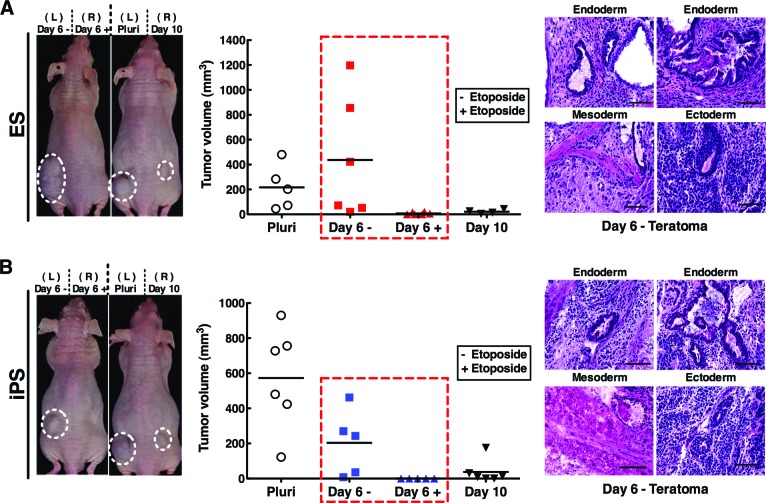
To functionally validate the utility of the DNA damage-induced purging assay, propensity for teratoma formation upon transplantation was assessed. Pluripotent cells compared with day 6 spontaneously differentiated progeny with or without etoposide treatment were injected through subcutaneous transplantation into immunodeficient nude mice. After 4 weeks of in vivo growth of engrafted cytotypes, the size of teratomas formed from R1 ES cells injected mice was reduced from a median tumor volume of 557 mm3 in the untreated group to 25 mm3 in the treated group (Fig. 5A). Examining the iPS cell line, there was also a notable reduction from 203 mm3 in tumor volume without pretreatment of etoposide to no detectable tumors with treatment (Fig. 5B). These results, recapitulated in both embryonic and induced pluripotent stem cell lines, indicate the ability to pharmacologically purge pluripotent cells and eradicate dysregulated cell growth and subsequent teratoma formation.
Figure 5.
Pharmacologic purging of a therapeutic progenitor population decreases teratoma formation. R1 (A) and iPS5 (B) cells were subjected to differentiation through the hanging drop method and treated with dimethyl sulfoxide or 2.5 or 0.05 μM etoposide (R1 and iPS5, respectively) on day 6 post-embryoid body (EB) formation. Twenty-four hours later these cells, as well as pluripotent cells and cells at day 10 post-EB formation, were harvested through trypsinization and washed before 1 × 106 cells were injected subcutaneously into nude mice. Tumor size was monitored every other day through measurement with vernier calipers. Images at left show representative mice, graphs in the middle plot tumor size of animals in different treatment groups at day of sacrifice, and images at right show hematoxylin- and eosin-stained histological sections from a representative day 6 without treatment teratoma. Scale bars = 50 μm. Abbreviations: ES, embryonic stem; iPS, induced pluripotent stem; L, left; Pluri, pluripotent; R, right.
Discussion
Pluripotent stem cells offer unprecedented potential for regenerative medicine based on an unlimited potential to differentiate into tissue-specific lineages. However, harnessing this unique therapeutic potential requires a fail-safe strategy to minimize the risk of dysregulated growth of residual pluripotent cells in the preparation of therapeutic progeny. In this study, we aimed to use a genotoxic strategy that could selectively purge pluripotent stem cells from tissue-specific progenitors in vitro and avoid the risk of uncontrolled cell growth upon in vivo transplantation. Herein we demonstrated that pluripotent stem cells are hypersensitive to apoptosis induction in response to DNA damage when compared with their somatic counterparts. We have attributed this hypersensitivity to a “priming” of the pluripotent cells for apoptosis induction through upregulation of the BH3-only protein Puma. This selective apoptotic phenotype and the causative agent Puma are both rapidly lost upon early stages of spontaneous differentiation. Notably, we then demonstrated that teratoma-forming cells could be purged out of a heterogeneous population of differentiating progeny through treatment with a DNA-damaging agent in order to reduce the undesirable effect of dysregulated growth upon transplantation. As the pharmacologic treatment is genotoxic, we confirmed that the dose and duration of etoposide in vitro did not affect survival or differentiation capacity of remaining tissue-specific or differentiating progeny. Collectively, these observations not only demonstrate the utility of exploiting the DNA damage hypersensitivity of pluripotent cells to enhance the safety of transplantable progenitor cells as therapeutics, but also identify a critical protein and inducible mechanism of apoptosis responsible for this stage-specific phenotype.
Accumulating genomic mutations in embryonic stem cells is unacceptable because of the devastating consequences of transmitting secondary genomic mutations to rapidly expanding progenitor cells within the embryo. Therefore ES cells possess the phenotype of hyperactive DNA repair to reduce mutational load by quickly removing any acquired aberrations through error-free repair [12, 24]. Additionally, upon overwhelming amounts of DNA damage, the balance within stem cells shifts from survival through repair to death by apoptosis of damaged cells as an organism-wide protective mechanism [29]. This massive, rapid apoptosis induction occurred consistently in all pluripotent cell types tested herein and was greatly diminished in somatic and differentiated cells of the same genotype. The mechanism of selective hypersensitive apoptotic response to DNA damage of pluripotent stem cells, especially to the topoisomerase poison etoposide, has previously been attributed to the tumor suppressor protein p53 [29]. However, the actual downstream apoptotic mediators have not been completely elucidated. Puma is a BH3-only protein that when activated, through either transcriptional upregulation or other mechanisms, neutralizes antiapoptotic proteins and activates the proapoptotic multidomain proteins Bax and Bak [30]. Western blotting and qPCR data indicated that unstimulated pluripotent cells have higher basal levels of Puma compared with their somatic counterparts [31] and that these levels are further induced in response to drug treatment. This differential expression could explain why pluripotent cells are significantly more sensitive to DNA damage than even embryonic fibroblasts. In support of this hypothesis, recent studies have reported the involvement of Puma in DNA damaged induced apoptosis during reprogramming [32]. These data suggest that the BH3-only protein Puma is a critical downstream mediator regulating induction of apoptosis in response to DNA damage in pluripotent cells.
Concordant with the idea that the BH3-only protein Puma is the main downstream mediator of hypersensitive, pluripotent-specific DNA damage-induced apoptosis, this protein was demonstrated to be highly downregulated upon spontaneous differentiation. This decrease should then shift the balance in somatic or in vitro differentiated cell types back to repair rather than immediate apoptosis in response to DNA damage. Accordingly, decreased apoptotic hypersensitivity to DNA damage was observed when pluripotent cells were subjected to spontaneous differentiation. Presumably this loss of apoptosis in these differentiated cells correlates with a concordant increase in repair activity and cell cycle arrest in these cells after induction of DNA double-strand breaks and thus warrants further investigation. The data herein support the idea that intolerance to DNA-damaging agents is an acquired, selective characteristic of pluripotent cells that is readily lost upon tissue-specific differentiation.
The genomic instability of pluripotent cells has been well documented [33–35] and could possibly be further exacerbated upon introduction of a potentially mutagenic pharmacologic agent when assessed through spectral karyotyping [36]. However, when a heterogeneous population of progenitor cells was subjected to a pulse of etoposide, spectral karyotyping demonstrated that surviving cells have karyotypes similar to those of cells not subjected to treatment. These results demonstrate not only that surviving cells do not accumulate genomic aberrations but also that treatment does not impose a selective pressure and/or advantage on the remaining cells. Interestingly, the IC50 values for etoposide of the pluripotent cell lines are near the circulating levels attainable in the plasma of children treated for acute lymphoblastic leukemia (1–2 μM) [37]. Because this hypersensitive apoptotic induction in response to DNA damage is an inherent pluripotent-specific phenotype and results in robust elimination of pluripotent cells, these data indicate that the selective phenotype could be exploited as a titratable assay to target stage-specific elimination of unwanted cell populations.
In this study, we demonstrated that eradication of “contaminating” pluripotent cells through etoposide treatment does not cause karyotypic changes in surviving differentiated cells and does not disrupt the capacity for tissue-specific differentiation. The stage-specific progenitors obtained by 6 days after in vitro differentiation yielded consistent cardiogenic progenitors with structural and functional characteristics expected from bioengineered progenitors despite the low dose of genotoxic purging. Furthermore, this purging assay significantly reduced the potential for dysregulated cell growth, as mice injected with cells at day 6 post-embryoid body formation and etoposide treatment demonstrated a significant reduction in teratoma formation. However, several mice in the untreated group did form large teratomas, demonstrating the presence of contaminating pluripotent cells within day 6 progenitors, highlighting the unacceptable risk of untreated pluripotent stem cell-based strategies. Ultimately, these data demonstrate the efficiency of using DNA damage-induced selective purging of residual pluripotent cells out of a heterogeneous therapeutic cell population to increase the safety of tissue-specific, transplant-ready progenitors by eliminating the risk of teratoma formation inherent to pluripotent stem cell sources.
Conclusion
Although the potential for therapeutic use of pluripotent stem cells for debilitating diseases is appropriately highlighted, the risk of dysregulated cell growth will continue to prevent therapeutic translation until safe and effective strategies are established as a fail-safe for clinical-grade application. Collectively, our pluripotent cell-purging assay validated herein demonstrates that pluripotent cells are selectively hypersensitive to DNA damage-induced apoptosis as a function of the specific apoptotic inducer protein Puma. This strategy, primed for clinical translation, establishes the feasibility and utility of a teratoma-purging assay based on natural mechanisms required for maintenance of genomic stability and ensuring healthy embryogenesis.
Acknowledgments
We thank the Mayo Clinic Cancer Center for the use of the Cytogenetics Core, which provided mouse spectral karyotyping services. We also thank Dr. Xing Li for assistance with statistics, Lois Rowe for assistance with histological analysis, and Elizabeth Bruinsma for technical assistance. The Mayo Clinic Cancer Center is supported in part by National Cancer Institute Cancer Center Support Grant 5P30 CA15083-36. This work was supported by the Todd and Karen Wanek Family Program for Hypoplastic Left Heart Syndrome, NIH New Innovator Award OD007015-01 (to T.J.N.), and a Mayo Clinic Center for Regenerative Medicine accelerated research grant (to T.J.N. and A.J.S.).
Author Contributions
A.J.S.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; N.G.N. and S.O.: collection and/or assembly of data; K.A.H.: provision of study material, collection and/or assembly of data; C.D.F.: provision of study material; A.T.: data analysis and interpretation; T.J.N.: conception and design, financial support, data analysis and interpretation, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Segers VFM, Lee RT. Stem-cell therapy for cardiac disease. Nature. 2008;451:937–942. doi: 10.1038/nature06800. [DOI] [PubMed] [Google Scholar]
- 2.Nelson TJ, Martinez-Fernandez A, Yamada S, et al. Repair of acute myocardial infarction by human stemness factors induced pluripotent stem cells. Circulation. 2009;120:408–416. doi: 10.1161/CIRCULATIONAHA.109.865154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freed CR, Greene PE, Breeze RE, et al. Transplantation of embryonic dopamine neurons for severe Parkinson's disease. N Engl J Med. 2001;344:710–719. doi: 10.1056/NEJM200103083441002. [DOI] [PubMed] [Google Scholar]
- 4.Goldring CEP, Duffy PA, Benvenisty N, et al. Assessing the safety of stem cell therapeutics. Cell Stem Cell. 2011;8:618–628. doi: 10.1016/j.stem.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 5.Ben-David U, Benvenisty N. The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat Rev Cancer. 2011;11:268–277. doi: 10.1038/nrc3034. [DOI] [PubMed] [Google Scholar]
- 6.Bjorklund LM, Sanchez-Pernaute R, Chung S, et al. Embryonic stem cells develop into functional dopaminergic neurons after transplantation in a Parkinson rat model. Proc Natl Acad Sci USA. 2002;99:2344–2349. doi: 10.1073/pnas.022438099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang C, Lee AS, Volkmer J-P, et al. An antibody against SSEA-5 glycan on human pluripotent stem cells enables removal of teratoma-forming cells. Nat Biotechnol. 2011;29:829–834. doi: 10.1038/nbt.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng F, Ke Q, Chen F, et al. Protecting against wayward human induced pluripotent stem cells with a suicide gene. Biomaterials. 2012;33:3195–3204. doi: 10.1016/j.biomaterials.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 9.Knoepfler PS. Deconstructing stem cell tumorigenicity: A roadmap to safe regenerative medicine. Stem Cells. 2009;27:1050–1056. doi: 10.1002/stem.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bieberich E, Silva J, Wang G, et al. Selective apoptosis of pluripotent mouse and human stem cells by novel ceramide analogues prevents teratoma formation and enriches for neural precursors in ES cell-derived neural transplants. J Cell Biol. 2004;167:723–734. doi: 10.1083/jcb.200405144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francis R, Richardson C. Multipotent hematopoietic cells susceptible to alternative double-strand break repair pathways that promote genome rearrangements. Genes Dev. 2007;21:1064–1074. doi: 10.1101/gad.1522807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cervantes RB, Stringer JR, Shao C, et al. Embryonic stem cells and somatic cells differ in mutation frequency and type. Proc Natl Acad Sci USA. 2002;99:3586–3590. doi: 10.1073/pnas.062527199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donoho G, Jasin M, Berg P. Analysis of gene targeting and intrachromosomal homologous recombination stimulated by genomic double-strand breaks in mouse embryonic stem cells. Mol Cell Biol. 1998;18:4070–4078. doi: 10.1128/mcb.18.7.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saretzki G, Armstrong L, Leake A, et al. Stress defense in murine embryonic stem cells is Superior to that of various differentiated murine cells. Stem Cells. 2004;22:962–971. doi: 10.1634/stemcells.22-6-962. [DOI] [PubMed] [Google Scholar]
- 15.de Waard H, Sonneveld E, de Wit J, et al. Cell-type-specific consequences of nucleotide excision repair deficiencies: Embryonic stem cells versus fibroblasts. DNA Repair. 2008;7:1659–1669. doi: 10.1016/j.dnarep.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 16.Kee Y, D'Andrea AD. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev. 2010;24:1680–1694. doi: 10.1101/gad.1955310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raya A, Rodriguez-Piza I, Guenechea G, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460:53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raya A, Rodriguez-Piza I, Navarro S, et al. A protocol describing the genetic correction of somatic human cells and subsequent generation of iPS cells. Nat Protoc. 2010;5:647–660. doi: 10.1038/nprot.2010.9. [DOI] [PubMed] [Google Scholar]
- 19.Müller LUW, Milsom MD, Harris CE, et al. Overcoming reprogramming resistance of Fanconi anemia cells. Blood. 2012;119:5449–5457. doi: 10.1182/blood-2012-02-408674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aladjem MI, Spike BT, Rodewald LW, et al. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr Biol. 1998;8:145–155. doi: 10.1016/s0960-9822(98)70061-2. [DOI] [PubMed] [Google Scholar]
- 21.Corbet SW, Clarke AR, Gledhill S, et al. p53-dependent and -independent links between DNA-damage, apoptosis and mutation frequency in ES cells. Oncogene. 1999;18:1537–1544. doi: 10.1038/sj.onc.1202436. [DOI] [PubMed] [Google Scholar]
- 22.Sabapathy K, Klemm M, Jaenisch R, et al. Regulation of ES cell differentiation by functional and conformational modulation of p53. EMBO J. 1997;16:6217–6229. doi: 10.1093/emboj/16.20.6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heyer BS, MacAuley A, Behrendtsen O, et al. Hypersensitivity to DNA damage leads to increased apoptosis during early mouse development. Genes Dev. 2000;14:2072–2084. [PMC free article] [PubMed] [Google Scholar]
- 24.Roos WP, Christmann M, Fraser ST, et al. Mouse embryonic stem cells are hypersensitive to apoptosis triggered by the DNA damage O6-methylguanine due to high E2F1 regulated mismatch repair. Cell Death Differ. 2007;14:1422–1432. doi: 10.1038/sj.cdd.4402136. [DOI] [PubMed] [Google Scholar]
- 25.Ardehali R, Inlay MA, Ali SR, et al. Overexpression of BCL2 enhances survival of human embryonic stem cells during stress and obviates the requirement for serum factors. Proc Natl Acad Sci USA. 2011;108:3282–3287. doi: 10.1073/pnas.1019047108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong Y, Stambrook PJ. Restoration of an absent G1 arrest and protection from apoptosis in embryonic stem cells after ionizing radiation. Proc Natl Acad Sci USA. 2004;101:14443–14448. doi: 10.1073/pnas.0401346101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Momcilovic O, Knobloch L, Fornsaglio J, et al. DNA damage responses in human induced pluripotent stem cells and embryonic stem cells. PLoS One. 2010;5:e13410. doi: 10.1371/journal.pone.0013410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marión RM. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grandela C, Pera MF, Wolvetang EJ. p53 is required for etoposide-induced apoptosis of human embryonic stem cells. Stem Cell Res. 2007;1:116–128. doi: 10.1016/j.scr.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Yu J, Zhang L. PUMA, a potent killer with or without p53. Oncogene. 2008;27:S71–S83. doi: 10.1038/onc.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madden DT, Davila-Kruger D, Melov S, et al. Human embryonic stem cells express elevated levels of multiple pro-apoptotic BCL-2 family members. PLoS One. 2011;6:e28530. doi: 10.1371/journal.pone.0028530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lake BB, Fink J, Klemetsaune L, et al. Context-dependent enhancement of induced pluripotent stem cell reprogramming by silencing Puma. Stem Cells. 2012;30:888–897. doi: 10.1002/stem.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Draper JS, Smith K, Gokhale P, et al. Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat Biotechnol. 2004;22:53–54. doi: 10.1038/nbt922. [DOI] [PubMed] [Google Scholar]
- 34.Baker DEC, Harrison NJ, Maltby E, et al. Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol. 2007;25:207–215. doi: 10.1038/nbt1285. [DOI] [PubMed] [Google Scholar]
- 35.Lefort N, Feyeux M, Bas C, et al. Human embryonic stem cells reveal recurrent genomic instability at 20q11.21. Nat Biotechnol. 2008;26:1364–1366. doi: 10.1038/nbt.1509. [DOI] [PubMed] [Google Scholar]
- 36.Bueno C, Catalina P, Melen GJ, et al. Etoposide induces MLL rearrangements and other chromosomal abnormalities in human embryonic stem cells. Carcinogenesis. 2009;30:1628–1637. doi: 10.1093/carcin/bgp169. [DOI] [PubMed] [Google Scholar]
- 37.Edick MJ, Gajjar A, Mahmoud HH, et al. Pharmacokinetics and pharmacodynamics of oral etoposide in children with relapsed or refractory acute lymphoblastic leukemia. J Clin Oncol. 2003;21:1340–1346. doi: 10.1200/JCO.2003.06.083. [DOI] [PubMed] [Google Scholar]