Abstract
Pointillistic approach based super-resolution techniques, such as photoactivated localization microscopy (PALM), involve multiple cycles of sequential activation, imaging and precise localization of single fluorescent molecules. A super-resolution image, having nanoscopic structural information, is then constructed by compiling all the image sequences. Because the final image resolution is determined by the localization precision of detected single molecules and their density, accurate image reconstruction requires imaging of biological structures labeled with fluorescent molecules at high density. In such image datasets, stochastic variations in photon emission and intervening dark states lead to uncertainties in identification of single molecules. This, in turn, prevents the proper utilization of the wealth of information on molecular distribution and quantity. A recent strategy for overcoming this problem is pair-correlation analysis applied to PALM. Using rigorous statistical algorithms to estimate the number of detected proteins, this approach allows the spatial organization of molecules to be quantitatively described.
Introduction
Fluorescence microscopy has become an invaluable tool for cell biologists due to its ability to noninvasively image the distribution and dynamics of fluorescently tagged proteins in cells[1–3]. The phenomenon of diffraction, however, imposes a fundamental limit to the resolution of fluorescent images[4,5]. When light from a fluorescent point emitter passes through the lens of a microscope, diffraction spreads the light rays so the source of light appears as a blurry spot instead of a sharp point. Consequently, the maximum resolution of a conventional light microscope is limited to optical wavelength (λ) divided by twice the numerical aperture (N.A.) of the objective (Resolution~λ/2N.A.). Since most modern, highly corrected microscopes objectives have a N.A. of ~1.5, the best spatial resolution for distinguishing two objects when imaged by visible light is ~200 nm.
Single molecule super-resolution imaging techniques based on sequential photoactivation (or photoswitching) of genetically encoded proteins or organic dyes have revolutionized optical imaging by allowing the imaging of biological structures and proteins at a resolution several times less than the wavelength of visible light. A large number of techniques, including photoactivated localization microscopy (PALM)[6–8], fluorescence PALM (fAPLM)[9], PALM with independently running acquisition (PALMIRA)[10], stochastic optical reconstruction microscopy (STORM)[11], ground state depletion microscopy (GDSIM)[12], spectral precision distance microscopy (SPDM)[13], and direct STORM (dSTORM)[14] have been developed. They rely on the same fundamental principle of spatial and temporal separation of emission of molecules, but use different photoactivable (or photoswitchable) probes and imaging algorithms. Studies using these super-resolution techniques to image cellular structures labeled with a high density of probes with sub-diffraction resolution have delineated new structural features at high resolution [15–21]. Despite these successes, only a handful of studies have quantitatively analyzed PALM, STORM and other super-resolution datasets [22–28]. Such datasets contain localization information about hundreds of thousands (sometimes even millions) of molecules, offering the potential for unprecedented insights into spatial organization. The difficulty in precisely estimating the localization of single molecules helps explain why single molecule super-resolution images have so infrequently been quantitatively analyzed. A promising approach discussed here for quantifying such images involves pair-correlation analysis. This approach uses robust statistical algorithm to analyze quantitatively the complex point-patterns resulting from single molecule super-resolution imaging experiments.
Diffraction limits the resolution of images containing high density of fluorescent molecules
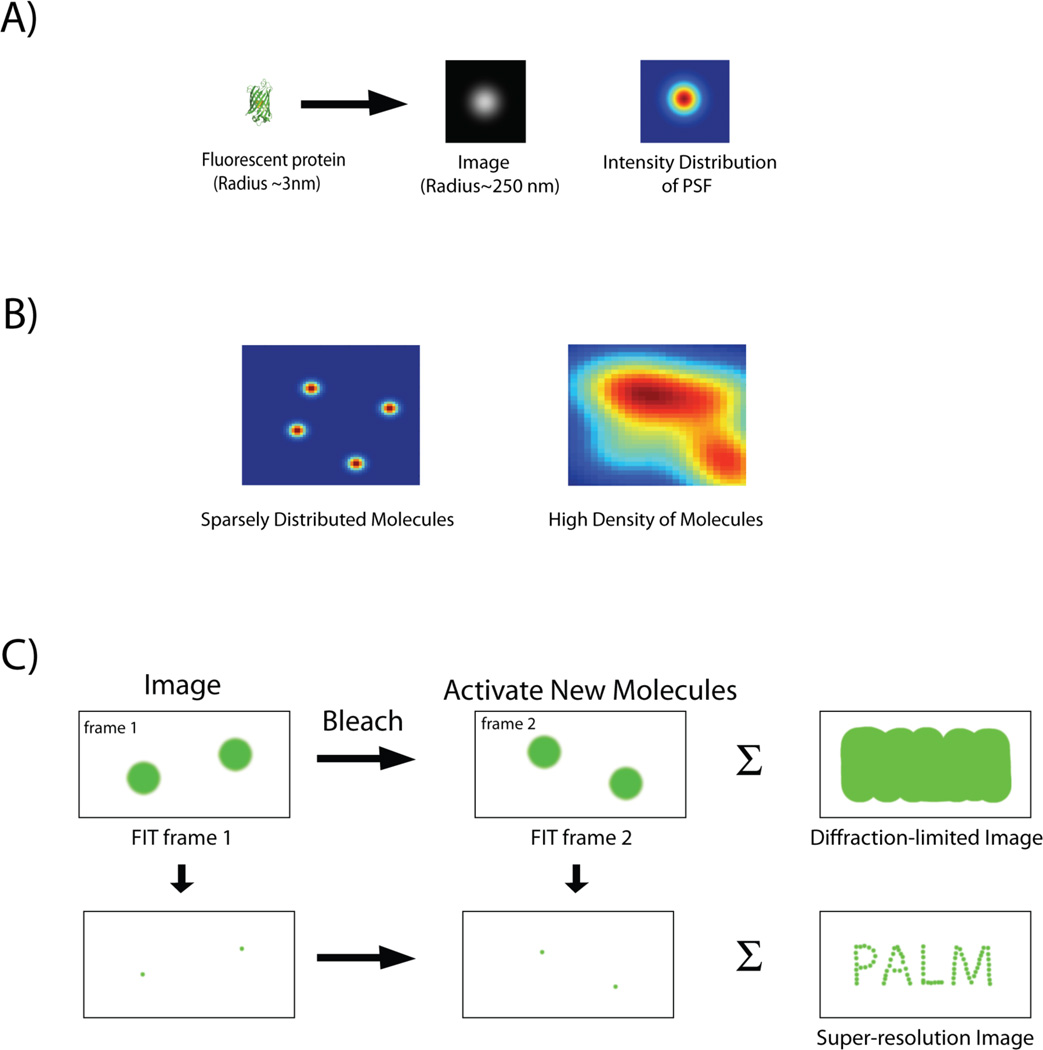
Due to diffraction, a point source of light appears as an extended spot when it is imaged with an optical microscope (Fig. 1A). Point spread function (PSF) describes this response of the imaging system to a point source of radiation. A single, fluorescent protein has dimensions approaching a point source (i.e., a few nm), so its image spatially approximates the PSF of the microscope. The PSF of an optical microscope can be reasonably approximated with a model function, like a two dimensional (2-D) Gaussian. By measuring the PSF from the image of a single molecule and fitting this to an appropriate mathematical function, therefore the position of the center of the PSF can be determined to a much greater precision than the diffraction limit[29–33]. Thus, although the details within the PSF and the size of the molecule cannot be resolved, the location of the emitting molecule can be ascertained to a high degree of precision (i.e., tens of nm) from the position of the center of the PSF.
Figure 1.
A: Image of a single fluorescent protein is convoluted by the point spread function (PSF) of the imaging system and appears as an extended blob. B: Intensity distribution of fluorescent proteins expressed at different densities. Images of single fluorescent proteins that are spatially separated by distances greater then the width of the PSF can be individually fit and localized with high precision. In contrast, PSFs of closely spaced molecules merge (high density of molecules), and individual molecules can no longer be distinguished. C: Imaging protocol for PALM. A sparse subset of photoactivable proteins is activated, imaged and their localization is determined with subdiffraction precision by a 2-D Gaussian fit. Following photobleaching of the activated proteins, a new set of molecules is activated, and the cycle is repeated. Finally the individual localizations are combined together to construct a super-resolution image, which depicts structural details unobservable by conventional, diffraction-limited imaging. The summation of the diffraction-limited spots, in contrast, gives back an image, which is equivalent to one captured by conventional optical microscope.
Such mathematical fitting of the PSF has been used in many studies to obtain positional information about an object far below the diffraction limit[34–39]. The localization precision of a single molecule whose image has been fit to a 2-D Gaussian function can be estimated by the following equation[29]:
| Eqn. 1 |
where σx,y2 is the localization precision, s is the sigma of the fit 2-D Gaussian, a is the size of the pixel, N is the number of photons detected, and b is the background signal. For a single molecule, s is approximately equal to the sigma of the PSF of the imaging system. Usually, for an experiment, the pixel size (a) is constant and the background signal (b) has small variations. Thus the localization precision, σx,y2, primarily depends on the total number of detected photons, N, and it scales inversely with N. Consequently, the uncertainty in position determination decreases with greater numbers of photons collected from the same emitter.
To obtain useful information about a structure, it is necessary to image structures labeled with a sufficiently high density of emitters so that the entire structure can be faithfully reconstructed from the image. However, at high density, the PSFs of neighboring molecules tend to overlap in space (Fig. 1B). This is a situation commonly encountered during conventional fluorescence imaging, when multiple fluorescent proteins within a diffraction-limited spot appear as a single bright spot. In such situations, it becomes impossible to distinguish individual molecules and estimate their locations.
PALM achieves super-resolution by combining multiple images of spatially separated single fluorescent molecules
The generation of photoactivable fluorescent proteins (PA-FPs)[40] and the subsequent development of PALM [6,9] provided an elegant method to circumvent the diffraction barrier. Using PALM, biological structures labeled with a high density of probe molecules are imaged at considerably higher resolution than the wavelength of visible light [6]. In PALM, fluorescence emission is temporally regulated by labeling proteins of interest with photoactivatable (or photoswitchable) flurophores. Upon illumination with specific wavelength of light, photoactivable probes undergo transition from a dark (or “off”) state to a fluorescent (or “on”) state. Photoswitcable proteins undergo interconversion between states that fluoresce at different wavelengths. Subdiffraction resolution is achieved by distinguishing fluorescent molecules from neighboring molecules either,
by activation state, (i.e. a photoactivated molecule in the midst of unactivated molecules), or,
by distinct emission spectra, (i.e. a photoswitched molecule fluorescing in a different spectral window with respect to the neighboring molecules).
A random, sparse subset of fluorescent proteins is photoactivated (or photoswitched) at any given time. Thus, the image being viewed consists of a collection of sparse, activated (or photoswitched) molecules, which are separated by distances significantly greater than the PSF of the microscope (Fig. 1C). Under this condition, it becomes possible to separately fit the images of each molecule to a 2-D Gaussian function (or some other appropriate function), and determine the positions of each individual molecule to a much higher degree of precision than the wavelength of visible light (Fig. 1C). Once the sparse subset of molecules is photobleached, a new subset is turned on, and the entire process is repeated. Iteration of these steps of activation, imaging and deactivation by photobleaching allows the mapping of the locations of many fluorophores. Finally, the localized positions of the molecules from all the frames are compiled to build up a super-resolution image (Fig. 1C). Using this strategy, it has been possible to localize single molecules with a precision of ~20–30 nm[6].
Single fluorescent molecules appear multiple times during the course of an experiment
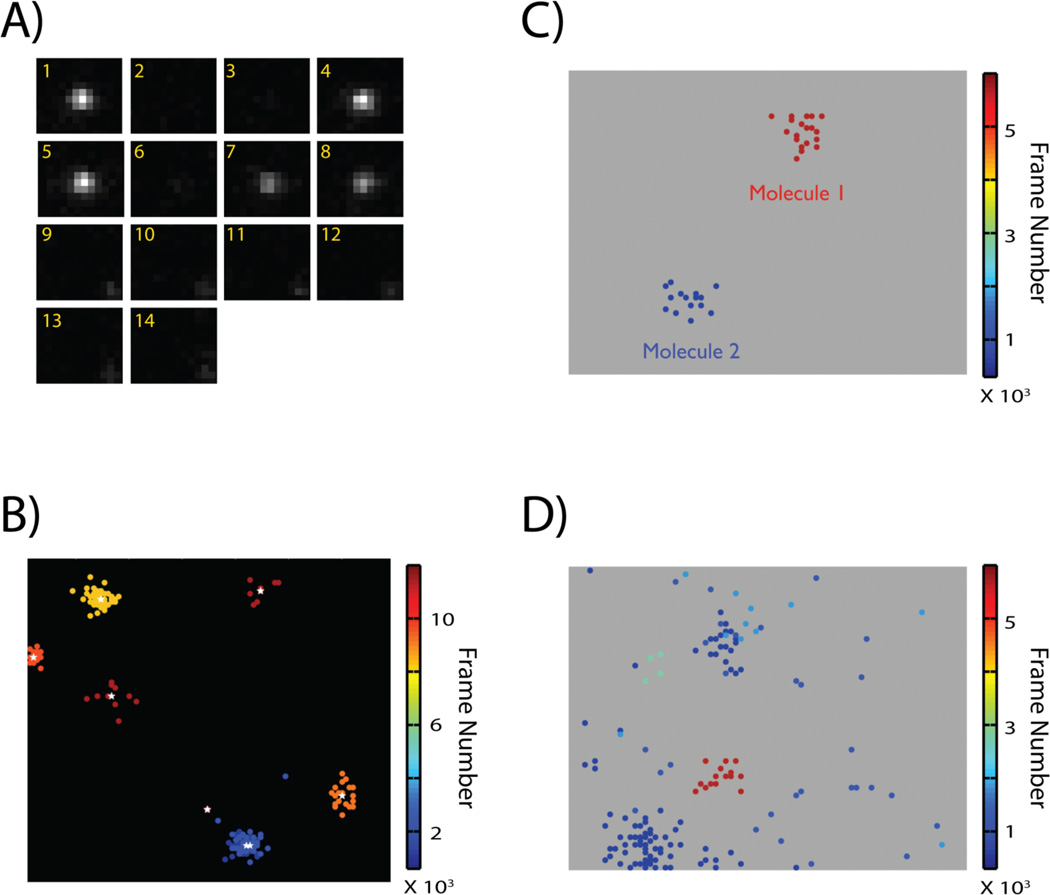
Single fluorescent molecules, including PA-FPs, have specific photophysical properties that are not directly observed in ensemble measurements. Some of these properties complicate the precise identification and quantitative analysis of single molecules. For example, the fluorescence signal from a single fluorescent protein often lasts longer than the usual frame rate of acquisition. A single PA-FP molecule thus appears multiple times across successive frames (Fig. 2A). Each individual image of the single PA-FP in a particular frame is usually designated as a peak. Each of these peaks is individually fit to obtain its location and positional uncertainty. However, the number of photons emitted by a single activated PA-FP varies stochastically over time. As a consequence, the number of photons detected for each peak image, which determines the precision of localization, also varies considerably. Due to this stochastic variation in photon counts, the positions determined for the peaks derived from the same molecule do not exactly coincide. This leads to a single molecule being represented by a cluster of closely spaced peaks[24,41] (Fig. 2B).
Figure 2.
A: A single molecule appears multiple times punctuated by variable dark intervals before it is terminally photobleached. The image frame is indicated in yellow at the top right corner of each sub-image. B: Image of single molecule peaks localized in a PALM experiment. Peaks are color-coded based on their time of appearance during the course of the experiment. Stochastic variation in photon count during individual appearances results in the spatial dispersal of the estimated positions for the multiple appearances of a single molecule. Consequently, in a compiled super-resolution image, a single molecule appears as a cluster of peaks. Peaks arising from the same molecule appear within a time-window, and can be assigned to a single molecule if the molecules are sparsely distributed and the individual clusters are well separated. The actual position and localization uncertainty of the molecule can be calculated as a weighted average of the positions and localization uncertainty of the individual peaks in the cluster (white asterisk represents estimated position of single molecule from the cluster of peaks). C: Cluster of peaks arising from two molecules can be easily distinguished if they are spatially and temporally well separated. D: Localized peaks from a sample with densely labeled fluorescent molecules. Grouping of cluster of peaks and assigning them to single molecule is complicated by the smaller spatial and temporal separation of peaks belonging to different molecules. Images adapted from Reference [23].
Blinking of single fluorescent proteins complicates quantitative analysis of PALM data
An analytical method for quantifying PALM images would have to perform the following tasks:
precisely identify single molecules, and,
evaluate the spatial distribution of single molecules to provide an assessment of the characteristic size, occupancy and density of heterogeneities.
Using the average value of positional uncertainty of the peaks in a PALM image, it is possible to identify peaks appearing in successive frames and within a certain radius as arising from the same molecule. Next, the location and positional uncertainty of the molecule can be estimated as a weighted average of the values corresponding to the individual peaks (Fig. 2B). However, single fluorescent protein molecules exhibit a photophysical process called blinking[42–44], which makes such estimation of molecular location complicated (Fig. 2A). Once activated, a PA-FP reversibly switches between a fluorescent and a non-fluorescent state on timescales ranging between microseconds to seconds. Furthermore, a single PA-FP often undergoes multiple cycles of blinking before converting to an irreversible dark state [23,41].
Blinking has two major consequences:
When it occurs on timescales shorter than the integration time of acquisition (or frame rate), its effect is averaged and not directly observed; but such blinking causes the overall photon-count to fluctuate and adds to uncertainty in position determination.
There can be significant blinking occurring on timescales similar to or longer than the frame rate. This, in turn, temporally isolates the peaks belonging to the same molecule. Thus, dark frames, in which the molecule is invisible, often punctuate the multiple appearances of a single molecule. (Fig. 2A).
Consequently, each molecule is represented by a cluster of peaks that are, (i) spatially separated due to the stochastic uncertainty in position determination in successive frames, and, (ii) temporally separated due to the blinking of the single PA-FPs (Fig. 2B). The situation is further complicated by the variability in number of intervening dark frames due to variations in blinking time. Any kind of quantitative analysis of the spatial organization of the molecules has to include a method to accurately group these peaks and assign them to appropriate molecules. Without rigorous and objective evaluation, there could be detection of spurious organizational patterns.
Grouping and assigning of peaks to single molecules is challenging for high labeling density
If it is possible to estimate a maximum time frame over which blinking occurs, then this time window can be used to identify peaks belonging to the same molecule. Let us define this time-window as the group gap. We can designate group radius as the average distance within which peaks from a single molecule are likely to be distributed (this radius is informed by the average localization uncertainty of peaks). In such a scenario, an algorithm can be devised in which peaks can be grouped together as belonging to the same molecule if they appear
within a distance less than group radius of each other, and,
in a time window less than equal to the group gap,
This cluster of peaks can then be replaced by a single peak (representing the actual molecule) whose positional coordinates and uncertainty in localization are calculated as a weighted average of the position and localization uncertainties of the all the peaks in that group (Fig. 2B).
This grouping approach, however, can be used only for biological samples with sparsely labeled proteins (Fig. 2C). The blinking photophysics of photoactivable proteins is not well understood. Careful experiments done in our lab, as well as other research groups clearly indicate that most of the commonly used photoactivable proteins can blink over timescales as long as few seconds. Thus, very often, a long, group gap (on the order of tens of seconds) has to be used in order to collect and correctly assign all the peaks belonging to a single molecule. Using such a long group gap increases the probability for activation of a second molecule within a distance less than the group radius of the original molecule. This, in turn, can lead to misassignment of the peaks (arising from the second molecule) to the first molecule. This problem becomes especially severe when dealing with biological samples where structures containing high density of proteins need to be imaged (Fig. 2D).
The localization precision of 20–30 nm for single molecule is not directly translated into actual structural resolution, and the actual resolution is a function of both the positional uncertainty and sampling density. According to Nyquist-Shannon sampling theorem[45], in order to accurately describe a biological structure, at least two molecules must be detected for each resolution unit. Thus, in order to achieve sub-diffraction resolution with PALM, it is essential to image biological structures with high sampling density of PA-FPs. In such a scenario, it is difficult to get a good estimate of blinking time that will group the peaks properly. This often leads to either identification of artifactual clustering, or under estimation of clustering.
Pairwise-correlation analysis can identify peaks arising from a single molecule using their unique spatial signature
Due to variability of blinking time and its dependence on experimental conditions like the laser power, an algorithm that can perform spatial analysis of peaks independent of the temporal variability of intervening dark frames needs to be devised. Luckily, the cluster of peaks belonging to a particular protein has a well-defined spatial signature: the peaks are distributed over a 2-D Gaussian surface centered at the actual position of the protein. This 2-D Gaussian surface represents the effective PSF of uncertainty in position determination for the PALM experiment. This unique spatial signature of the cluster of peaks arising from a single molecule can, in principle, be used to estimate the contribution of multiple appearances of single molecule to the overall spatial distribution.
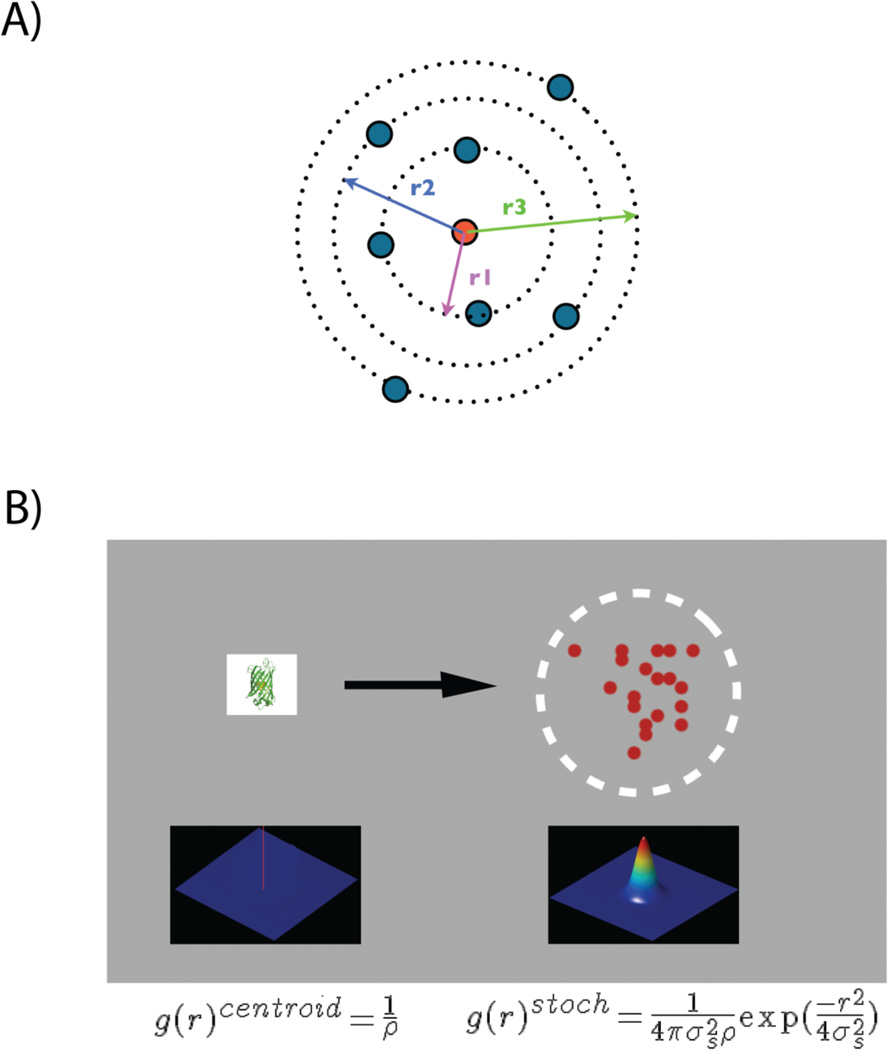
Pair-correlation (PC) analysis can be used to obtain an objective description of the spatial scales of density fluctuation. This information can be used to construct a functional model for the spatial organization of proteins. Autocorrelation function measures the probability of finding a protein at a given distance from another protein (Fig. 3A). Since the average density of proteins is used to normalize the autocorrelation function, it quantifies the increased probability with respect to that expected from random distribution of proteins. The defined spatial signature of peaks arising from multiple appearances of a single protein will have a characteristic autocorrelation function. Specifically, the autocorrelation function of the 2-D Gaussian function describing the distribution of peaks is another 2-D Gaussian function with increased width ( times).
Figure 3.
A: Pair-correlation analysis quantifies the probability of finding a second protein at a certain distance r (represented by r1, r2 and r3) away from a given protein (central red circle) compared to random distribution. B: A single protein is represented by a cluster of peaks distributed over a 2D-surface representing the PSF of localization uncertainty of the PALM experiment. Autocorrelation function of the protein at r=0, g(r)centroid, is a delta function represented by red spike at the centroid of the protein. g(r)centroid is convolved with the autocorrelation function of the PSF. The resulting Gaussian function, g(r)stoch, quantifies the spatial distribution of multiple peaks arising from a single molecule.
Thus, once, the autocorrelation function for all detected peaks in a PALM dataset is computed, it is possible to identify the contribution of correlation function corresponding to the multiple appearances of peaks to total correlation function. The derivation of the models for fitting computed correlation function is described in detail in a recently published paper[23].
Model for fitting computed pair-wise autocorrelation function
Briefly, the radial autocorrelation function of all the peaks (g(r)) essentially quantifies the correlation function of all the protein molecules at different spatial scales (i.e. radii). Autocorrelation of protein at r = 0 measures the correlation of the protein centroids with themselves, and it is a delta function that scales inversely with the density of proteins (Fig. 3B). Because of multiple appearances and the uncertainty in position determination, each protein centroid is represented by a cluster of peaks distributed over a 2D-gaussian, the effective PSF of the PALM measurements. Thus, the delta function at r = 0 is blurred by this effective PSF of the system (Fig. 3B). The PSF-convoluted autocorrelation function corresponding to the centroid of the protein is designated as g(r)stoch, and it takes the following form:
| Eqn. 2 |
where σs is the average uncertainty in position determination, and ρ is the average density of proteins.
The second part of the total calculated correlated function (i.e. g(r)) is the correlation function at r > 0. This represents the correlation arising from the relative spatial organization of the proteins, and is designated as g(r)protein. g(r)protein essentially provides a measurement of the clustering of the protein. Thus the total correlation function, g(r), can be written as:
| Eqn. 3 |
Note that the protein correlation function, g(r)protein, is also convolved (*) with the correlation function of the PSF (g(r)PSF) of the PALM experiment.
Under conditions where the proteins are randomly distributed, g(r)protein is equal to 1, and the total correlation function is given by:
| Eqn. 4 |
Estimation of parameters required for fitting of measured correlation function
To fit the computed correlation function to the models described above, estimates of (i) the mean localization precision (σs), and, (ii) the protein density (ρ), are required. The mean localization precision can be determined from the distribution of the localization precision of the peaks in the PALM dataset. The total protein density can be calculated by dividing the total number of detected peaks by the average appearances of a single protein[23].
Careful characterization of the photoactivable (or photoswitcable) proteins needs to be carried out to obtain an estimate of the average number of appearances. This value depends on experimental conditions like laser power density, ambient surrounding of the probe molecules, and the photoactivable protein being studied. By imaging a sample with sparsely labeled samples and performing careful grouping operations, it is possible to obtain an estimate of average number of appearances under the particular experimental conditions. It is also advisable to perform in vitro characterization of the photoactivable proteins under conditions where the density of the proteins can be precisely controlled.
Recently, it has been reported that photoswitchable cerulean fluorescent protein 2[46] (PS-CFP2) undergoes irreversible photobleaching without blinking[25], and this assumption has been used to quantify spatial distribution of PS-CFP2 labeled proteins in T-cells. However, in experiments performed in our lab, we observed that PS-CFP2 undergoes reversible cycles of blinking before undergoing terminal photobleaching, though the number of reappearances of PS-CFP2 appears to be less than other photoactivable (or photoswitchable) proteins. A photoactivable probe which undergoes irreversible conversion to dark state without blinking will be an immensely useful probe for quantitative PALM experiments, and will make analysis of point-pattern much more simpler. However, it is important to do careful experiments to ensure that the photoactivable protein being used does not blink under the given set of experimental conditions. Careful optimization of experimental conditions might minimize or eliminate blinking of PS-CFP2. In such a scenario, PS-CFP2 can become a valuable probe for interrogating the nanoscale spatial organization of proteins.
Estimation of number of photoactivable (or photoswitchable) proteins in a sample
A recent paper discusses an elegant algorithm for calculating the total number of proteins present in PALM images [24]. The authors demonstrate that it is possible to get an estimate of the number of molecules present in a PALM dataset by fitting the plot of number of molecules versus various maximum dark time intervals (i.e. group gap windows) in the low dark-time threshold region. The actual number of molecular reappearances depend on the photophysical parameters like rate of activation and photobleaching, which in turn is determined largely by the experimental conditions including the laser power, photoactivable proteins used and the immediate microenvironment of the PA-FPs. Thus, there can be large variations in average number of reappearances from experiment to experiment due to variations in these parameters. If the molecular count for the specific experiment can be estimated accurately by analyzing the PALM dataset using this algorithm, it will allow for more accurate determination of the density of molecules. Thus the combination of this algorithm, proposed by Radenovic and colleagues, with pair-correlation analysis can provide more robust description of spatial distribution of single molecules.
Model for pair-wise autocorrelation function for non-randomly organized protein molecules
Once an estimate of the parameters for the correlation function is obtained, the total correlation function can be fit to either Eqn.3 or Eqn.4. This would provide a quantitative description of the spatial organization. Eqn.4 should adequately describe the correlation function of randomly distributed molecules, e.g. proteins immobilized on a glass cover slip. In contrast, to obtain a description of the spatial scales of organization when proteins are distributed non-randomly, the correlation function has to be fit to Eqn. 3 (Fig. 4A). In order to perform such a fitting, a model function for the protein correlation function has to be used. Existing knowledge about the organization of the protein should inform the choice of this model function. The correlation function of proteins organized into randomly distributed domains with no specific shape can be modeled with a simple exponential function,
| Eqn.5 |
where exp designates exponential function, A is the amplitude of the protein correlations extrapolated to r = 0, and, ξ is the correlation length. ξ gives a measure of the domain size.
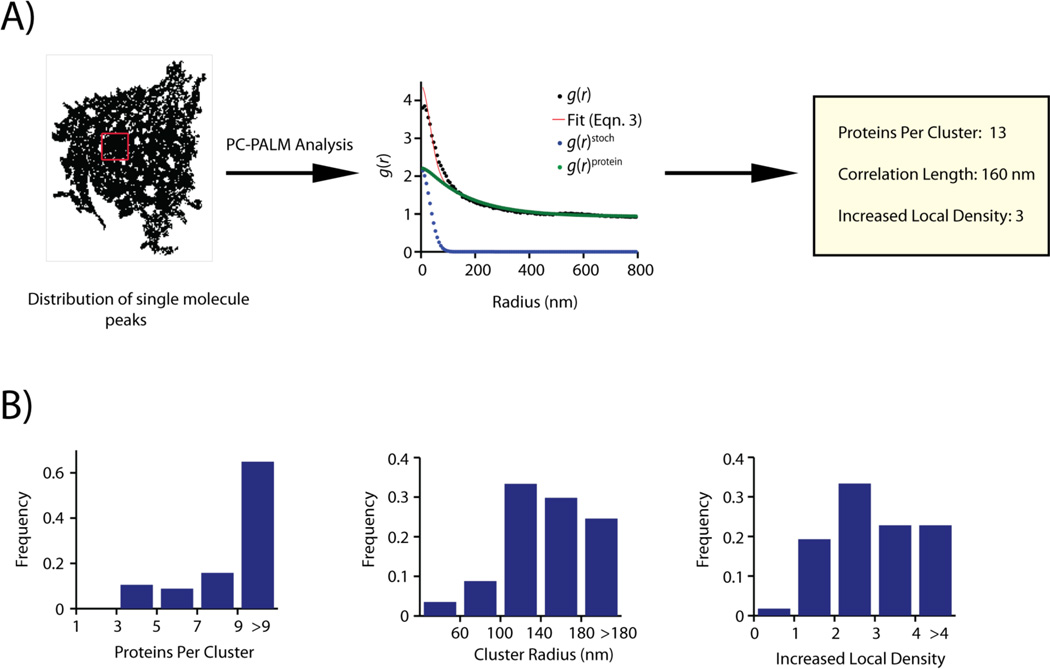
Figure 4.
A: Distribution of single molecule peaks of Lyn-PAGFP across the plasma membrane of a COS-7 cell. Peaks collected in individual frames are combined together. A sub-section of the cell, shown in the red box, is used for pair-correlation analysis. Correlation function of the distribution of peaks (black circles) could be fit to a clustered model using Eqn. 3 (red line). Correlation describing multiple appearances of a single fluorescent protein (g(r)stoch, blue circle) was evaluated from the fit. The protein correlation function (g(r)protein, green circles) is calculated by subtracting g(r)stoch from the measured correlation function. Using g(r)protein, physical parameters describing the nanoscale organization of Lyn-PAGFP could be determined. B: Distribution of cluster parameters evaluated by pair-correlation analysis of spatial distribution of Lyn-PAGFP across plasma membrane of COS-7 cells. Lyn-PAGFP is organized into clusters of 3 or more proteins, and the clusters have a wide range of sizes (60nm – >180nm). Most clusters of Lyn have increased local density ranging from 2 to > 4. Images adapted from Reference [23].
Without any information about the organization of the proteins, an exponential function is a good starting model. We used such a function to model the distribution of plasma membrane proteins, and could obtain satisfactory fits of the correlation functions. The fitting enabled us to describe the details of the spatial organization of plasma membrane proteins by obtaining parameters like protein cluster size (ξ), number of detected proteins in cluster (Ncluster), and density of proteins in cluster (ψcluster)[23] (Fig. 4B)
Pairwise cross-correlation analysis can provide insights about relative spatial organization of two proteins
Various biological processes including receptor signaling, cell polarization, transport intermediate biogenesis and viral budding, involve critical interactions between two or more proteins[47,48]. In order to obtain mechanistic insight into these processes, it is crucial to characterize the spatial and temporal scales of such intermolecular interactions. Pair-correlation analysis can be performed to interrogate the relative spatial distribution of two different proteins, which are labeled with distinct photoactivable probes. Cross-correlation function, c(r), quantifies the probability of finding a second kind of probe molecule in the vicinity of a probe. Mathematical fitting of cross-correlation data is simpler than autocorrelation data since the multiple appearances of a single protein do not contribute to the cross-correlation function. Thus, the calculated cross-correlation function can be described as the protein cross-correlation convoluted with the PSF of the PALM measurements, and is represented as:
| Eqn.6 |
By choosing an appropriate model for the protein cross-correlation function, it is possible to assess the spatial scales of interaction between the proteins of interest.
Choice of photoactivable proteins for two-color PALM experiments
Careful considerations of the photophysical properties must inform the choice of PA-FPs as probes used in cross-correlation PALM experiments. Imaging conditions need to be calibrated such that there is minimal crosstalk in the emission channels of the two probes. Separation of spectral profiles is critical for multi-color single molecule experiments since there is always some spontaneous activation and excitation of the photoactivable proteins. Such excitation can lead to significant misassignment of probes unless the emission spectra of the probes are well separated. Additionally, a two-color PALM experimental protocol should ensure that there is minimal loss of signal due to cross-activation. Ideally, the two PA-FPs should have different sensitivities to the activation laser. This will ensure that the probes can be differentially activated, and imaged separately.
Using photoactivable green fluorescent protein[40] (PAGFP) and photoactivable mCherry1[49] (PAmCh1), it is possible to satisfy the above criteria. The emission spectra of these two PA-FPs are well separated (PAGFP emission maximum is at 515 nm, whereas emission maximum of PAmCh1 is at 595 nm). PAGFP can be both simultaneously activated and imaged by a 488 nm laser[23,50]. PAmCh1 on the other hand requires a pulse of 405 nm for activation, and has to be subsequently imaged using a green laser (~560 nm). Thus, it is possible to devise the following imaging protocol for these two probes, which should minimize the loss of signal due to cross-activation of PA-FPs:
Image PAGFP exhaustively by activating and exciting with 488 nm laser.
Next, activate PAmCh1 with 405 nm laser and image with 561 nm laser line.
We used this protocol to examine the spatial relationship between PAGFP-labeled GPI-anchored protein and PAmCh1-labeled actin in COS-7 cells under different conditions[23]. PAmCh1 is not efficiently activated by low power 405 nm laser line. So, a short, low power 405 nm laser pulse can be used in between the two acquisition steps to collect the residual PAGFP fluorescence. Following this, PAmCh1 can be activated with higher power density of 405 nm laser line. The laser powers to be used in such a protocol should to be decided by doing careful standardization of the experimental conditions.
An important consideration for selection of PA-FPs is the contrast between post-activation and pre-activation images (or that following photoswitching). This parameter, commonly called contrast ratio, is a measure of the brightness (i.e. number of photons emitted) by the activated form relative to the initial one. A higher contrast ratio translates into a higher signal-to-noise ratio, and thus improves the localization precision of the single molecules. PAGFP does not have a good contrast ratio (~100) and has low photon count compared to some of the newly developed PA-FPs[7]. Additionally, since both PAGFP and PAmCh1 are dark before activation, it becomes difficult to choose cells and regions of interest during imaging with these proteins.
New and improved probes for PALM experiments
Among the recently developed probes, PS-CFP2[46] and monomeric Eos2[51,52] (mEos2) are promising pair for two color PALM experiments. Both PSCFP2 and mEos2 are photoswitchable fluorescent proteins. PSCFP2 has cyan fluorescence before activation, and its emission switches to green upon illumination with violet (~405 nm) light. Emission of mEos2, in contrast, switches from green to red upon activation with 405 nm laser line. Both PSCFP2 and mEos2 have a large contrast ratio (the contrast ratio of PSCFP2 is ~2000) and photon count. Thus, the PSCFP2-mEos2 pair should allow imaging of single molecules with higher precision. The increased number of photons detected from these proteins and the higher contrast ratio should also ensure detection of larger number of single molecules. This, in turn, should increase the effective resolution by enabling imaging of structures at higher sampling density. mEos2 also undergoes spontaneous activation with 561 nm laser line (unpublished result), and can be exhaustively photoactivated with low power 405 nm laser. PSCFP2 requires higher power 405nm laser for activation. Thus it should be possible to devise an optimal imaging protocol in the following way:
mEos2 is first activated and imaged with 561 nm laser.
mEos2 is imaged further with a combination of low power 405 nm and 561 nm lasers. This step should allow exhaustive imaging of mEos2 labeled protein with minimal activation/excitation of PSCFP2 molecules.
Finally, PSCFP2 can be activated with high power 405 nm and imaged with 488 nm laser.
Laser intensities to be used in each step of this protocol would have to be carefully calibrated to ensure minimal cross-activation and optimal detection of single molecules.
Photoswitchable monomeric orange [53](PSmOrange), with its orange to far-red switching of emission profile, can also be used with PSCFP2 for two color PALM experiments. PSmOrange undergoes photoswitching with blue light. In a two-color experiment involving these two proteins, an imaging protocol can be set up such that PSmOrange is initially activated and imaged using a combination of blue (~488 nm) and far-red laser (~640 nm) lines. Following this, PSCFP2 can be photoswitched and imaged using violet (~405 nm) and blue lasers, respectively. The laser powers in the first segment of the experiment have to be meticulously optimized to minimize the activation of PSCFP2 molecules. The wide separation of the excitation and emission spectra of the photoconverted form of these two proteins should ensure minimal cross talk between these proteins (excitation and emission maximum of photoconverted PSCFP2 are 468 nm and 511 nm, respectively; excitation and emission maximum of photoswitched form of PSmOrange are 636 nm and 662 nm, respectively).
Conclusion
In recent years, the development of various single molecule super-resolution imaging techniques has helped to circumvent the diffraction limit of visible light. Consequently, it has become possible to improve the resolution of images of biological structures by more than an order of magnitude. Using imaging techniques like PALM, it is now possible to interrogate a wide range of length scales (~30–250nm). This, in turn, is furnishing valuable information about fundamental biological structures and processes at molecular level, hitherto unattainable by conventional light microscopy. The super-resolution images contain vast amount of information organized into complex patterns with complicated spatial and temporal relationships. These datasets have necessitated the development of appropriate statistical algorithms, which can comprehensively and accurately analyze the spatial information contained in them. Such analysis would provide invaluable mechanistic insights about various biological processes, and help us to realize the full potential of PALM and related super-resolution imaging techniques.
Multiple appearances of a single protein with associated stochastic uncertainty in position, and reversible blinking of individual fluorophores complicate quantitative analysis of image sequences of single molecules. Pair-correlation analysis, described in this review, provides an objective analysis method to quantify complex patterns of single molecule images. This method can characterize the spatial features of nanoscale protein distribution across different spatial scales (~30–250 nm), and provides estimates including size, number of molecules and molecular density of clusters. The analysis can describe the organization of both small protein oligomers as well as larger domains of proteins, thus making it a versatile tool for studying organizational remodeling during various physiological processes. Furthermore, the general concepts of the algorithm discussed here can be adapted for analysis of high-resolution data from other single molecule techniques where multiple appearances of single probe molecules confound accurate description of spatial organization[54].
Acknowledgements
We thank and D. T. Burnette and S. B. van Engelenburg (National Institute of Child Health and Development) for help with the illustrations.
References
- 1.Lippincott-Schwartz J, Patterson GH. Development and use of fluorescent protein markers in living cells. Science. 2003;300:87–91. doi: 10.1126/science.1082520. [DOI] [PubMed] [Google Scholar]
- 2.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 3.Lippincott-Schwartz J. Emerging in vivo analyses of cell function using fluorescence imaging. Annu Rev Biochem. 2011;80:327–332. doi: 10.1146/annurev-biochem-121010-125553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abbe E. Beitrage zur Theorie des Mikroskops und der mikroskopischen Wahrnehmung. Archiv fur mikroskopische Anatomie. 1873;9:413–418. [Google Scholar]
- 5.Rayleigh L. On the theory of optical images, with special reference to microscope. Philos. Mag. 1896;42:167–195. [Google Scholar]
- 6.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 7.Lippincott-Schwartz J, Patterson GH. Photoactivatable fluorescent proteins for diffraction-limited and super-resolution imaging. Trends Cell Biol. 2009;19:555–565. doi: 10.1016/j.tcb.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patterson G, Davidson M, Manley S, Lippincott-Schwartz J. Superresolution imaging using single-molecule localization. Annu Rev Phys Chem. 2010;61:345–367. doi: 10.1146/annurev.physchem.012809.103444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hess ST, Girirajan TPK, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egner A, Geisler C, von Middendorff C, Bock H, et al. Fluorescence nanoscopy in whole cells by asynchronous localization of photoswitching emitters. Biophys J. 2007;93:3285–3290. doi: 10.1529/biophysj.107.112201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rust MJ, Bates M, Zhuang XW. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Folling J, Bossi M, Bock H, Medda R, et al. Fluorescence nanoscopy by ground-state depletion and single-molecule return. Nat Methods. 2008;5:943–945. doi: 10.1038/nmeth.1257. [DOI] [PubMed] [Google Scholar]
- 13.Lemmer P, Gunkel M, Weiland Y, Muller P, et al. Using conventional fluorescent markers for far-field fluorescence localization nanoscopy allows resolution in the 10-nm range. J Microsc. 2009;235:163–171. doi: 10.1111/j.1365-2818.2009.03196.x. [DOI] [PubMed] [Google Scholar]
- 14.Wombacher R, Heidbreder M, van de Linde S, Sheetz MP, et al. Live-cell super-resolution imaging with trimethoprim conjugates. Nat Methods. 2010;7:717–719. doi: 10.1038/nmeth.1489. [DOI] [PubMed] [Google Scholar]
- 15.Shroff H, Galbraith CG, Galbraith JA, White H, et al. Dual-color superresolution imaging of genetically expressed probes within individual adhesion complexes. Proc Natl Acad Sci U S A. 2007;104:20308–20313. doi: 10.1073/pnas.0710517105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shtengel G, Galbraith JA, Galbraith CG, Lippincott-Schwartz J, et al. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc Natl Acad Sci U S A. 2009;106:3125–3130. doi: 10.1073/pnas.0813131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bates M, Huang B, Dempsey GT, Zhuang X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science. 2007;317:1749–1753. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenfield D, McEvoy AL, Shroff H, Crooks GE, et al. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 2009;7:e1000137. doi: 10.1371/journal.pbio.1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.York AG, Ghitani A, Vaziri A, Davidson MW, et al. Confined activation and subdiffractive localization enables whole-cell PALM with genetically expressed probes. Nat Methods. 2011;8:327–333. doi: 10.1038/nmeth.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pavani SR, Thompson MA, Biteen JS, Lord SJ, et al. Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proc Natl Acad Sci U S A. 2009;106:2995–2999. doi: 10.1073/pnas.0900245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manley S, Gillette JM, Patterson GH, Shroff H, et al. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 22.Owen DM, Rentero C, Rossy J, Magenau A, et al. PALM imaging and cluster analysis of protein heterogeneity at the cell surface. J Biophotonics. 2010;3:446–454. doi: 10.1002/jbio.200900089. [DOI] [PubMed] [Google Scholar]
- 23.Sengupta P, Jovanovic-Talisman T, Skoko D, Renz M, et al. Probing protein heterogeneity in the plasma membrane using PALM and pair correlation analysis. Nat Methods. 2011;8:969–975. doi: 10.1038/nmeth.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Annibale P, Vanni S, Scarselli M, Rothlisberger U, et al. Quantitative photo activated localization microscopy: unraveling the effects of photoblinking. PLoS One. 2011;6:e22678. doi: 10.1371/journal.pone.0022678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lillemeier BF, Mortelmaier MA, Forstner MB, Huppa JB, et al. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat Immunol. 2009;11:90–96. doi: 10.1038/ni.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherman E, Barr V, Manley S, Patterson G, et al. Functional nanoscale organization of signaling molecules downstream of the T cell antigen receptor. Immunity. 2011;35:705–720. doi: 10.1016/j.immuni.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hess ST, Gould TJ, Gudheti MV, Maas SA, et al. Dynamic clustered distribution of hemagglutinin resolved at 40 nm in living cell membranes discriminates between raft theories. Proc Natl Acad Sci U S A. 2007;104:17370–17375. doi: 10.1073/pnas.0708066104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu CJ, Baumgart T. Spatial association of signaling proteins and F-actin effects on cluster assembly analyzed via photoactivation localization microscopy in T cells. PLoS One. 2011;6:e23586. doi: 10.1371/journal.pone.0023586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kubitscheck U, Kuckmann O, Kues T, Peters R. Imaging and tracking of single GFP molecules in solution. Biophys J. 2000;78:2170–2179. doi: 10.1016/S0006-3495(00)76764-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gelles J, Schnapp BJ, Sheetz MP. Tracking Kinesin-Driven Movements with Nanometre-Scale Precision. Nature. 1988;331:450–453. doi: 10.1038/331450a0. [DOI] [PubMed] [Google Scholar]
- 32.Cheezum MK, Walker WF, Guilford WH. Quantitative comparison of algorithms for tracking single fluorescent particles. Biophys J. 2001;81:2378–2388. doi: 10.1016/S0006-3495(01)75884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin walks hand-over-hand. Science. 2004;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 34.Lacoste TD, Michalet X, Pinaud F, Chemla DS, et al. Ultrahigh-resolution multicolor colocalization of single fluorescent probes. Proc Natl Acad Sci U S A. 2000;97:9461–9466. doi: 10.1073/pnas.170286097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lacoste TD, Michalet X, Pinaud FF, Chemla DS, et al. Super resolution molecular ruler using single quantum dots. Biophysical Journal. 2000;78:402A-A. [Google Scholar]
- 36.Heilemann M, Herten DP, Heintzmann R, Cremer C, et al. Highresolution colocalization of single dye molecules by fluorescence lifetime imaging microscopy. Anal Chem. 2002;74:3511–3517. doi: 10.1021/ac025576g. [DOI] [PubMed] [Google Scholar]
- 37.Gordon MP, Ha T, Selvin PR. Single-molecule high-resolution imaging with photobleaching. Proc Natl Acad Sci U S A. 2004;101:6462–6465. doi: 10.1073/pnas.0401638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burnette DT, Sengupta P, Dai YH, Lippincott-Schwartz J, et al. Bleaching/blinking assisted localization microscopy for superresolution imaging using standard fluorescent molecules. Proc Natl Acad Sci U S A. 2011;108:21081–21086. doi: 10.1073/pnas.1117430109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qu XH, Wu D, Mets L, Scherer NF. Nanometer-localized multiple single-molecule fluorescence microscopy. Proc Natl Acad Sci U S A. 2004;101:11298–11303. doi: 10.1073/pnas.0402155101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 41.Annibale P, Vanni S, Scarselli M, Rothlisberger U, et al. Identification of clustering artifacts in photoactivated localization microscopy. Nat Methods. 2011;8:527–528. doi: 10.1038/nmeth.1627. [DOI] [PubMed] [Google Scholar]
- 42.Dickson RM, Cubitt AB, Tsien RY, Moerner WE. On/off blinking and switching behaviour of single molecules of green fluorescent protein. Nature. 1997;388:355–358. doi: 10.1038/41048. [DOI] [PubMed] [Google Scholar]
- 43.Schwille P, Kummer S, Moerner WE, Webb WW. Fluorescence correlation spectroscopy (PCS) of different GFP mutants reveals fast light-driven intramolecular dynamics. Biophys J. 1999;76:A260-A. [Google Scholar]
- 44.Schwille P, Kummer S, Heikal AA, Moerner WE, et al. Fluorescence correlation spectroscopy reveals fast optical excitation-driven intramolecular dynamics of yellow fluorescent proteins. Proc Natl Acad Sci U S A. 2000;97:151–156. doi: 10.1073/pnas.97.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shannon CE. Communication in the Presence of Noise. Proceedings of the Institute of Radio Engineers. 1949;37:10–21. [Google Scholar]
- 46.Chudakov DM, Verkhusha VV, Staroverov DB, Souslova EA, et al. Photoswitchable cyan fluorescent protein for protein tracking. Nat Biotechnol. 2004;22:1435–1439. doi: 10.1038/nbt1025. [DOI] [PubMed] [Google Scholar]
- 47.Lagerholm BC, Weinreb GE, Jacobson K, Thompson NL. Detecting microdomains in intact cell membranes. Annual Review Of Physical Chemistry. 2005;56:309–336. doi: 10.1146/annurev.physchem.56.092503.141211. [DOI] [PubMed] [Google Scholar]
- 48.Batista FD, Treanor B, Harwood NE. Visualizing a role for the actin cytoskeleton in the regulation of B-cell activation. Immunological Reviews. 2010;237:191–204. doi: 10.1111/j.1600-065X.2010.00943.x. [DOI] [PubMed] [Google Scholar]
- 49.Subach FV, Patterson GH, Manley S, Gillette JM, et al. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat Methods. 2009;6:153–159. doi: 10.1038/nmeth.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Testa I, Mazza D, Barozzi S, Faretta M, et al. Blue-light (488 nm)-irradiation-induced photoactivation of the photoactivatable green fluorescent protein. Appl Phys Lett. 2007;91:133902–133904. [Google Scholar]
- 51.McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, et al. A bright and photostable photoconvertible fluorescent protein. Nat Methods. 2009;6:131–133. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiedenmann J, Ivanchenko S, Oswald F, Schmitt F, et al. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc Natl Acad Sci U S A. 2004;101:15905–15910. doi: 10.1073/pnas.0403668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subach OM, Patterson GH, Ting LM, Wang Y, et al. A photoswitchable orange-to-far-red fluorescent protein, PSmOrange. Nat Methods. 2011;8:771–777. doi: 10.1038/nmeth.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Veatch SL, Machta B, Shelby S, Chiang E, Holowka D, Baird B. Correlation functions quantify super-resolution images and estimate apparent clustering due to over-counting. arXiv:1106.6068. 2011 doi: 10.1371/journal.pone.0031457. [DOI] [PMC free article] [PubMed] [Google Scholar]